Abstract
DsrA RNA regulates the translation of two global regulatory proteins in Escherichia coli. DsrA activates the translation of RpoS while repressing the translation of H-NS. The RNA-binding protein Hfq is necessary for DsrA to function in vivo. Although Hfq binds to DsrA in vitro, the role of Hfq in DsrA-mediated regulation is not known. One hypothesis was that Hfq acts as an RNA chaperone by unfolding DsrA, thereby facilitating interactions with target RNAs. To test this hypothesis, we have examined the structure of DsrA bound to Hfq in vitro. Comparison of free DsrA to DsrA bound to Hfq by RNase footprinting, circular dichroism, and thermal melt profiles shows that Hfq does not alter DsrA secondary structures, but might affect its tertiary conformation. We identify the site on DsrA where Hfq binds, which is a structural element in the middle of DsrA. In addition, we show that although long poly(U) RNAs compete with DsrA for binding to Hfq, a short poly(U) stretch present in DsrA is not necessary for Hfq binding. Finally, unlike other RNAs, DsrA binding to Hfq is not competed with by poly(A) RNA. In fact, DsrA:poly(A):Hfq may form a stable ternary complex, raising the possibility that Hfq has multiple RNA-binding sites.
Keywords: Small regulatory RNA, translational regulation, RNA-binding protein, Sm protein, Sm-like protein, siRNA
INTRODUCTION
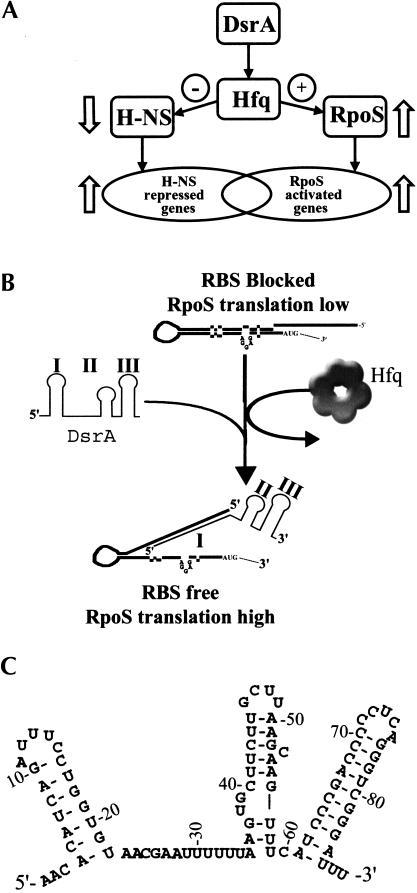
DsrA is an 85-nt RNA that regulates the translation of two global regulatory proteins, RpoS and H-NS (Fig. 1A ▶; Sledjeski and Gottesman 1995; Sledjeski et al. 1996). At low temperature, DsrA increases the translation of RpoS by binding to a complementary sequence in the 5′-untranslated region of the rpoS mRNA (Fig. 1B ▶; Lease et al. 1998; Majdalani 1998). This binding leads to the formation of an alternative secondary structure in the rpoS mRNA that is translationally active (Lease et al. 1998; Majdalani 1998). Two other RNAs, RprA and OxyS, also have roles in the translational regulation of rpoS (Zhang et al. 1998; Majdalani et al. 2001). DsrA has a mild repressing effect on H-NS translation by binding to a complementary region in the H-NS mRNA and presumably blocking ribosome binding (Lease and Belfort 2000). These two different regulatory activities map to separate domains of DsrA.
FIGURE 1.
Overview and model of DsrA/Hfq/RpoS-mediated regulation. (A) Overview of the role of DsrA in H-NS and RpoS-mediated translational regulation. DsrA increases the translation of RpoS mRNA, leading to increased transcription of RpoS-regulated genes. DsrA decreases the translation of H-NS, leading to increased transcription of H-NS-repressed genes (Sledjeski et al. 1996). (B) Model of DsrA/Hfq/RpoS interaction. In this model, Hfq unfolds the RpoS 5′-leader region, allowing DsrA Domain I to base-pair with the RpoS mRNA (Majdalani et al. 1998). This stabilizes an alternative conformer of RpoS mRNA that leads to increased translation by exposing the ribosome-binding site. (C) Sequence and proposed secondary structure of DsrA based on Lease and Belfort (2000).
In vivo, DsrA-mediated regulation is dependent on the RNA chaperone Hfq (Sledjeski et al. 2001). In the cell, 11.2-kD Hfq monomers form heat-stable hexamers that are very abundant (up to 55,000 copies;Azam et al. 1999). Hfq was originally identified as an Escherichia coli host factor essential for the in vivo and in vitro replication of Qβ RNA bacteriophage (Fernandez et al. 1972; Schuppli et al. 1997). Potential homologs of Hfq have been identified in more than 30 species of bacteria (Sun et al. 2002). Hfq targets several mRNAs for degradation by increasing polyadenylation (Hajnsdorf and Regnier 2000) or by interfering with ribosome binding (Vytvytska et al. 2000). In addition, Hfq binds tightly to Qβ RNA (de Haseth and Uhlenbeck 1980b), poly(A) RNA (de Haseth and Uhlenbeck 1980a), OxyS (Zhang et al. 1998), and DsrA RNAs (Sledjeski et al. 2001), as well as a number of recently identified RNAs (Wassarman et al. 2001). The abundance of Hfq in the cell and its ability to bind a large array of RNAs indicates that it may play an important role in cell regulation. Although a large number of Hfq targets and in vivo effects have been identified, its mechanism of action is not known.
In this paper, we identify the Hfq-binding site on DsrA RNA and show that, when bound, Hfq does not alter the secondary structure of DsrA. In addition we show that although long (200–400 nt in length) polydisperse poly(U) competes with DsrA for binding to Hfq, a short poly(U) stretch present in DsrA is not necessary for Hfq binding. Finally, unlike other RNAs, poly(A) RNA does not compete for binding to Hfq. In fact DsrA:poly(A):Hfq may form a ternary complex. The potential for a ternary complex raises the possibility that Hfq has multiple RNA-binding sites.
RESULTS
DsrA Domain 2 competes with full-length DsrA for binding to Hfq
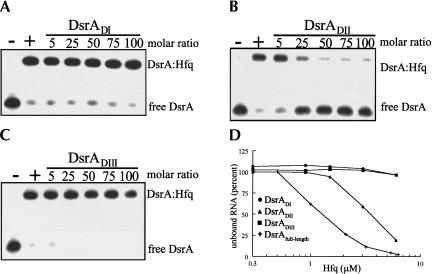
Gel shift assays demonstrated that Hfq and DsrA interact in vitro (Sledjeski et al. 2001). Previous work showed that DsrA contains three folding domains, each corresponding to a distinct function (Majdalani et al. 1998; Lease and Belfort 2000). To probe whether Hfq specifically bound to one of these domains, we tested the individual structural elements of DsrA for their ability to compete with full-length DsrA for binding to purified Hfq. DsrADI (nucleotides 1–25) and DsrADIII (nucleotides 60–85) did not compete with full-length DsrA for binding to Hfq even at 100-fold molar excess over the labeled DsrA (Fig. 2A,C ▶). On the other hand, a 25-fold molar excess of DsrADII (nucleotides 23–60) was sufficient to partially displace full-length DsrA from Hfq, and at a 50-fold molar excess of DsrADII, DsrA was completely displaced (Fig. 2B ▶). The individual domains were also tested for their ability to bind directly to Hfq. Similar to the competition assays, end-labeled DsrADI and DsrADIII were not shifted by Hfq, even at Hfq concentrations well above that necessary to bind DsrA (Fig. 2D ▶). As expected, a DsrADII:Hfq complex was evident on the native gels, but its binding affinity was ∼4 μM, threefold weaker than that of full-length DsrA, which was 1.2 μM (Fig. 2D ▶). Although we did not detect binding or competition of Domains I and III to Hfq, it is still possible that they contribute to the binding of DsrA to Hfq in the context of the whole DsrA molecule.
FIGURE 2.
DsrADII competes with full-length DsrA for binding to Hfq. 5′-end-labeled DsrA was incubated with increasing concentrations of DsrADI, DsrADII, and DsrADIII at 25°C for 5 min, followed by the addition of Hfq and an incubation at 25°C for 5 min. Samples were separated on an 8% native polyacrylamide gel in 1× TBE. (A) DsrADI, (B) DsrADII, and (C) DsrADIII were incubated with full-length 5′-end-labeled DsrA. Samples were incubated without (−) or with (+) 3 μM Hfq. (D) 5′-end-labeled full-length DsrA (+), DsrADI (•), DsrADII (▴), or DsrADIII (▪) were incubated at 25°C for 10 min with the indicated amounts of purified Hfq. The percent of unbound RNA was determined using a phosphorimager (see Materials and Methods) and plotted against Hfq concentration. Only full-length DsrA and DsrADII bound to Hfq.
Poly(U) but not poly(A) competes with DsrA for binding to Hfq
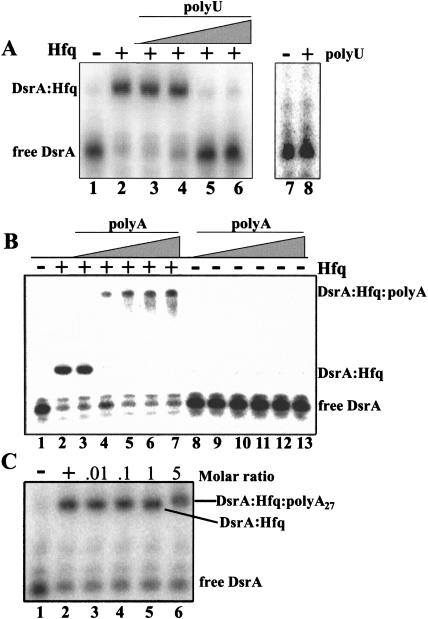
Hfq can bind to both poly(U)- and poly(A)-rich RNA (Senear and Steitz 1976; Zhang et al. 2002); therefore, we tested whether poly(U) and poly(A) could compete with DsrA for binding to Hfq. Long, polydisperse poly(U) (200–400 nt in length) competed with DsrA for binding to Hfq at a concentration of 50 ng/μL (Fig. 3A ▶). In contrast, polydisperse poly(A) caused a supershift of the DsrA–Hfq complex, indicating that Hfq was bound to 5′-end-labeled DsrA and poly(A) simultaneously (Fig. 3B ▶). Because a discrete supershifted band was not resolved, it was possible that the poly(A):Hfq complex was forming aggregates that nonspecifically trapped the labeled DsrA. To distinguish between these two possibilities, a poly(A) 27-mer, poly(A)27, was synthesized and tested for its ability to compete with DsrA for binding to Hfq. As was seen with the polydisperse poly(A), poly(A)27 did not compete for binding, but formed a supershifted complex with Hfq and DsrA (Fig. 3C ▶). We did not observe any binding between poly(A) and DsrA in the absence of Hfq even at very high ratios of poly(A) to DsrA (data not shown). It is also interesting to note that a poly(U) 15-mer did not compete with DsrA for binding to Hfq or cause a supershift of the DsrA–Hfq complex (data not shown). This result indicates that a minimum size is necessary for poly(U) to bind to Hfq.
FIGURE 3.
DsrA binding to Hfq is competed with by poly(U), but not poly(A). 5′-end-labeled DsrA was incubated with increasing concentrations of (A) long polydisperse (200–400 nt) poly(U) (0.5–500 ng, lanes 3–6); (B) long polydisperse poly(A) (0.5–500 ng, lanes 3–7, 8–13); and (C) poly(A)27 (lanes 3–6) at 25°C for 5 min, followed by the addition of 3 μM Hfq (+) or binding buffer (−) and a second incubation at 25°C for 5 min. To control for RNA, RNA interaction DsrA was incubated (A) with (+, lane 8) or without (−, lane 7) 3.6 μg of polydisperse poly(U); or (B) with (0.5–500 ng, lanes 8–13) or without (lane 1) polydisperse poly(A). The top complexes in B are trapped in the well.
The poly(U) region of DsrA Domain II is not required for binding to Hfq
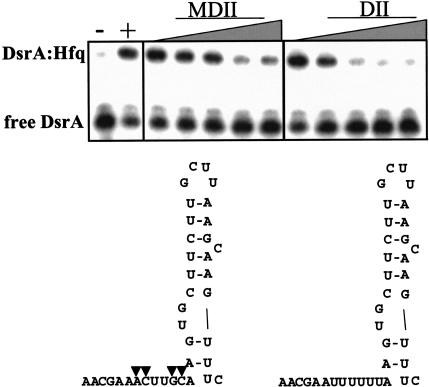
Domain II of DsrA contains a short poly(U) sequence (nucleotides 29–34). Because Domain II of DsrA and poly(U) both compete with DsrA for binding to Hfq, we hypothesized that this stretch of uridine residues might be the Hfq-binding site. One prediction of this model was that changing this U-rich sequence would decrease the binding affinity. We therefore prepared a modified DsrA Domain II RNA (DsrAMDII) in which the sequence AUUUUUUA was changed to AACUUGCA. DsrAMDII was then assayed for its ability to compete with labeled DsrA for Hfq binding. Surprisingly, the DsrAMDII RNA competed with DsrA (Kc = 180 nM) nearly as well as DsrADII (Kc = 100 nM; Fig. 4 ▶), indicating that Hfq was not simply binding to the poly(U) region of Domain II, but was recognizing some more complex structural element.
FIGURE 4.
Modified Domain II (DsrAMDII) competed with full-length DsrA for binding to Hfq. 5′-end-labeled DsrA was incubated with increasing concentrations of DsrAMDII or DsrADII at 25°C for 5 min, followed by the addition of Hfq and an incubation at 25°C for 5 min. The four modified nucleotides are marked by triangles on the structure of DsrAMDII. Samples were incubated without (−) or with (+) 3 μM Hfq. The gels were quantitated with a phosphorimager, and the Kc was calculated for MDII (180 nM) and DII (100 nM).
The poly(A/U) region of DsrA is not sufficient for binding to Hfq
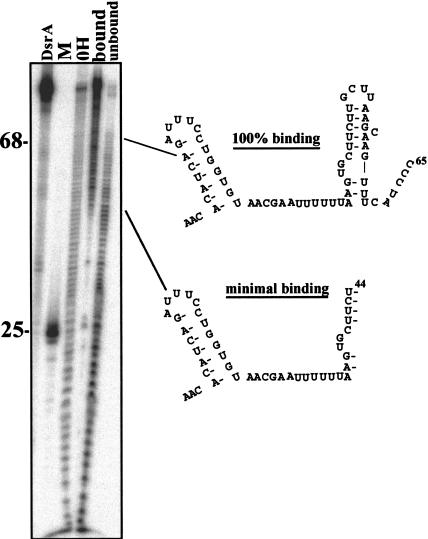
The data described above indicate that higher-order RNA structure is at least as important as simple linear sequence for Hfq binding to DsrA. This result is in agreement with other studies of Hfq RNA binding (Zhang et al. 2002). We used a minimal binding site assay to identify regions of DsrA that are necessary for binding to Hfq. In this assay, 5′-end-labeled DsrA was cleaved by alkaline hydrolysis, resulting in cleavage of ∼10% of the total RNA (Fig. 5 ▶). This partially cleaved RNA was incubated with Hfq, and the bound and unbound fractions were separated on a nondenaturing gel. RNA was eluted from gel slices and subsequently separated on a denaturing polyacrylamide gel. The smallest RNA that bound to Hfq (nucleotides 1–44) encompassed all of Domain I, the A/U-rich region of Domain II, and the ascending portion of the stem of Domain II (Fig. 5 ▶). Fragments between 44 and 65 nt in length were present primarily in the bound fraction; however, these nucleotides were also present in the unbound fraction. These results are consistent with nucleotides 44–65 of DsrA being necessary for binding. Fragments >65 nt in length were present only in the bound fraction. This indicates that tight binding of DsrA to Hfq requires nucleotides past C65. If the A/U-rich region alone were sufficient for binding, then we would have expected fragments as small as 34 nt to be present in the bound fraction.
FIGURE 5.
Minimal binding assay of the DsrA–Hfq complex. 5′-end-labeled DsrA transcript (0.17 pmole) was subjected to limited alkaline hydrolysis to give ∼10% cleavage. The fragments of DsrA were incubated with purified Hfq protein, and bound and unbound fragments were subsequently separated on a native polyacrylamide gel. Bound and unbound DsrA fragments were excised, eluted, and fractionated on a denaturing polyacrylamide gel. Uncleaved DsrA (DsrA), 5′-end-labeled DsrADI (M), DsrA alkaline hydrolysis ladder (OH), DsrA fragments bound to Hfq (bound), and DsrA fragments that are not bound to Hfq (unbound) are represented in the figure. The bands seen in the bound lane at positions 25 and 35 are nonspecific and were likely due to small amounts of RNA degradation during purification of the DsrA bound fraction. These bands were not seen in other experiments
Nuclease footprinting of the DsrA–Hfq complex
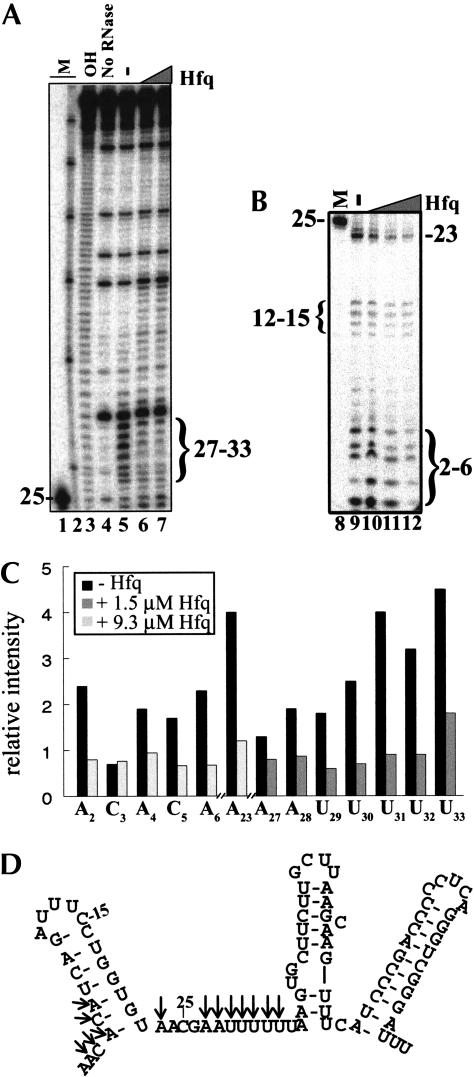
To identify the nucleotides protected by the DsrA–Hfq interaction, we used the single-strand-specific nuclease RNase I to analyze the DsrA–Hfq complex. The RNase I cleavage pattern of DsrA without Hfq was consistent with the previously reported in vitro secondary structure mapping by nuclease (A, T1, S1) cleavage (Lease and Belfort 2000). The most dramatic change in the cleavage pattern with the addition of Hfq occurred in the predicted single-stranded region of Domain II (Fig. 6A,C ▶), which is consistent with Hfq binding to Domain II. Seven nucleotides in this A/U-rich region (ACGAAUUUUUU) were strongly protected from cleavage by RNase I in the presence of Hfq (Fig. 6 ▶). Hfq also protected portions of Domain I from cleavage (Fig. 6B,C ▶). The nucleotides protected were mainly at the bottom of the stem of Domain I (Fig. 6C ▶). We did not observe any strong increase in nuclease sensitivity (single strandedness) in DsrA when bound to Hfq. The modest increase in nuclease sensitivity observed in Domains II and III upon addition of Hfq was probably caused by nonspecific nuclease activity and was not reproducible.
FIGURE 6.
RNase I cleavage of the DsrA–Hfq complex. DsrA in the absence and presence of varying concentrations of Hfq protein was cleaved by RNase I. Samples were analyzed on a denaturing polyacrylamide gel. (A) RNase I cleavage of the DsrA–Hfq complex showing nucleotides 25–85. End-labeled DsrA (16 nM) was mixed with 0 (lane 5), 1.5 μM (lane 6), or 3.1 μM (lane 7) purified Hfq hexamer and incubated at 25°C for 10 min. This was followed by cleavage with 0.05 U of RNase I at 25°C for 2 min. (B) RNase I cleavage of the DsrA–Hfq complex showing nucleotides 2–26. End-labeled DsrA (16 nM) was mixed with 0 (lane 9), 1.5 μM (lane 10), 3.1 μM (lane 11), or 9.3 μM (lane 12) purified Hfq hexamer and treated as above. Markers (M) are 5′-end-labeled DsrADI (lanes 1,8); Decade Marker (Ambion) representing 30, 40, 50, 60, 70, 80, and 90 nt (lane 2); and limited hydrolysis of DsrA (OH, lane 3). (C) Quantitative analysis of protected nucleotides shown in A and B in the absence (lanes 5,9, black bar) or presence (lane 6, dark gray bar) of 1.5 μM Hfq or 9.3 μM Hfq (lane 12, light gray bar). Individual bands were quantitated using a phosphorimager and normalized to the amount of full-length DsrA in the same lane. The data shown are representative of several independent experiments. (D) Structure of DsrA proposed by Lease and Belfort (2000), with arrows showing the nucleotides that are cleaved less by RNase I in the presence of Hfq.
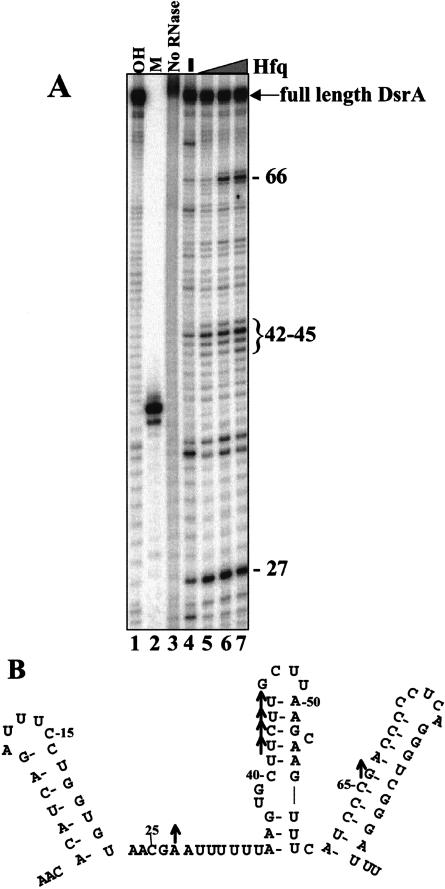
To further investigate whether Hfq affected the double-stranded regions of DsrA, we probed the structure of DsrA with the double-strand-specific nuclease RNase V1 (Lockard and Kumar 1981). RNase V1 recognizes predominantly helical structures at least 4–6 bp in length (Lowman and Draper 1986). These helical structures can be single stranded or double stranded (Lowman and Draper 1986). In the presence of Hfq we noted a strong enhancement in the RNase V1 sensitivity of nucleotide A27 and U42–U45 in Domain II and G66 in Domain III (Fig. 7A,B ▶). All of these cleavages were observed in the absence of Hfq, but they became stronger upon protein binding. There are two possible explanations for these observations. First, Hfq binding might simply stabilize the existing secondary structure. Although this is an attractive hypothesis, the CD spectroscopic data support an argument against it (see below). Alternatively, enhanced cleavage can occur when a portion of DsrA’s double helical elements are sterically protected as part of the RNA–protein complex. This protection, in effect, means that the given amount of nuclease is acting on a smaller pool of substrate. Nuclease cleavage occurred at the same rate in each sample, but because fewer possible sites exist in the DsrA–Hfq complex, more cleavage was observed at a few positions. Three sites (U34, C60, and C73), all at pyrimidine-A steps, displayed decreases in nuclease cleavage. This finding most likely reflects a decrease in adventitious hydrolysis of the Hfq-bound RNA.
FIGURE 7.
RNase V1 cleavage of the DsrA–Hfq complex. DsrA in the presence or absence of Hfq was cleaved by RNase V1. Samples were fractionated on a denaturing polyacrylamide gel. (A) RNase V1 cleavage of the DsrA–Hfq complex. (Lanes 1,2) Markers: the limited alkaline hydrolysis ladder of DsrA (OH) and 38-nt DsrADII (see Materials and Methods). (Lane 3) 5′-end-labeled DsrA (16 nM) that was not treated with RNase V1. DsrA (16 nM) was treated with RNase V1 with (+) or without (−) purified Hfq protein (3.1 μM). (B) Structure of DsrA proposed by Lease and Belfort (2000), with upward arrows showing the nucleotides that have increased cleavage in the presence of Hfq. The data shown are representative of several independent experiments.
Circular dichroism and thermal melt profile of DsrA bound to Hfq
There are two alternative models to explain the RNase footprinting experiments of the DsrA:Hfq complex. Hfq could bind to and alter the structure of DsrA or Hfq could be protecting DsrA via steric interactions. To distinguish these models, we measured the circular dichroism (CD) spectra of DsrA alone or in the presence of saturating amounts of Hfq. CD is a powerful method for examining changes in RNA secondary structure in solution (Sosnick et al. 2000). Because the significant spectral features of the RNA and protein occur at different wavelengths, this technique can be used to examine structural changes in either an RNA or a protein upon complex formation (Fasman 1996).
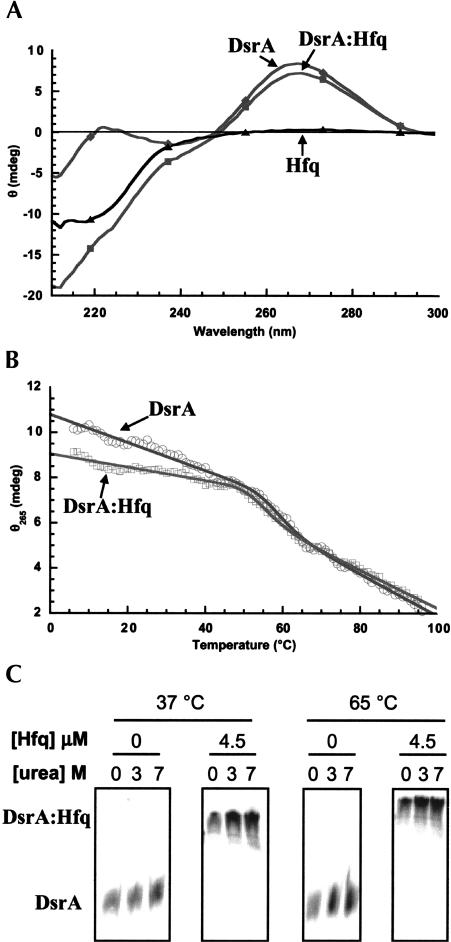
The CD spectrum of DsrA alone shows a positive peak at 265 nm and a negative peak at 209 nm. These features are indicative of significant A-form helix in the folded state (Fig. 8A ▶). Addition of Hfq did not significantly alter the position or magnitude of the 265-nm feature (Fig. 8A ▶). The spectra show a slight decrease in intensity within this region that may be indicative of an alteration in RNA tertiary structure but is inconsistent with significant unfolding of helical elements. Titration experiments (data not shown) show that this behavior occurs with an overall stoichiometry of 1:6 (DsrA to Hfq) and thus clearly represents RNA binding to the hexameric Hfq. The spectral changes for the negative feature at 209–225 nm result from the superposition of the protein signal in that region but are not indicative of large-scale conformational changes in either the RNA or protein components when viewed as difference spectra (data not shown). Together these data indicate that the global structures of both DsrA and Hfq are largely unaltered by the binding event. Based on the gel shift data under the conditions used for spectroscopy, all of the RNA was bound to Hfq (Fig. 8C ▶).
FIGURE 8.
Circular dichroism studies of DsrA and the DsrA:Hfq complexes. (A) Circular dichroism spectra of 0.5 μM DsrA with (▪) and without (♦) 3.0 μM Hfq, in 50 mM cacodylate buffer (pH 6.6), 250 mM NaCl at 37°C. The difference spectrum (▴) corresponds very closely to the spectrum of Hfq alone (data not shown). (B) Trace of circular dichroism intensity at 263 nm as a function of temperature (0°–100°C) for 0.5 μM DsrA and with (□) and without (○) 3.0 μM Hfq in 3 M urea, 50 mM cacodylate buffer (pH 6.6), and 250 mM NaCl. Lines represent fits of the data to a unimolecular two-state folding model. (C) Binding of DsrA to Hfq in the presence of urea and at high temperature. DsrA was incubated with or without 3.0 μM Hfq in 50 mM cacodylate buffer (pH 6.6), 250 mM NaCl, with 0, 3 M, or 7 M urea. Samples were incubated at 37°C or 65°C and run on gels at the same temperature. We observed 100% binding of DsrA to Hfq irrespective of the urea concentration or temperature of incubation.
We further verified that the structure of the RNA was unaltered by looking at the thermal melting profile of DsrA in the presence and absence of Hfq (Fig. 8B ▶). Urea (3 M) was added to these samples to lower the Tm and promote better resolution of the high-temperature baseline (Shelton et al. 1999). Because the gel shift of DsrA by Hfq is stable even in the presence of 7 M urea and at elevated temperatures (Fig. 8C ▶), we were not concerned with loss of stability of the DsrA:Hfq interaction under these conditions. Nevertheless, no difference was apparent in the Tm or enthalpy of the observed RNA unfolding transition between the free and bound forms, the magnitude of which is consistent with a secondary structure unfolding transition. Therefore, initial spectroscopic characterization of DsrA in the presence and absence of Hfq strongly indicated that Hfq binding does not result in a significant modulation of DsrA secondary structure, but does not preclude some effect of binding on DsrA tertiary structure. Based on the total CD signal observed, a maximum of 2–3 bp worth of secondary structure might be disrupted by the interaction with Hfq, and that assumes that all of the spectroscopic changes report on secondary rather than tertiary structure interactions.
DISCUSSION
Hfq and DsrA RNA interact in vitro and in vivo (Sledjeski et al. 2001; Wassarman et al. 2001). This interaction is necessary for DsrA to bind to the RpoS mRNA and activate translation (Majdalani et al. 1998). It has been proposed that Hfq functions as an RNA chaperone to “melt out” RNA secondary structure (Muffler et al. 1997; Schuppli et al. 1997; Tsui et al. 1997). Domain I of DsrA is the region that must interact with the 5′-UTR of RpoS mRNA by forming a stable secondary structure element that inhibits cis pairing of RpoS mRNA. One function of Hfq could be to melt out this domain, freeing it to pair with RpoS mRNA. We used three different techniques, RNase footprinting (single and double stranded), steady-state CD, and CD thermal melt profiles to assay changes in DsrA secondary structure in the presence of Hfq. Domain I of DsrA exhibits no increased sensitivity to the single-strand nuclease RNase I in the presence of Hfq; in fact, a slight decrease in sensitivity was observed. It is possible that this domain is melted out when bound to Hfq, but steric effects prevent the RNase from cleaving the RNA. Such a melted structure seems unlikely, however, because RNase I can still cleave the loop region of Domain I of DsrA. The CD spectra and thermal melt profiles of DsrA in the presence and absence of Hfq further support the conclusion that the secondary structure of DsrA remains unchanged upon binding to Hfq. It is, therefore, doubtful that the role of Hfq in DsrA-mediated regulation is to unfold DsrA, but it is possible that Hfq unfolds the 5′-untranslated region of rpoS mRNA. DsrA would then act to trap the rpoS mRNA in the highly translated conformer. In this model, the function of DsrA is analogous to a doorstop, preventing the rpoS from refolding into the poorly translated closed conformer (Fig. 1B ▶).
Even though we did not detect changes in the secondary structure of Domain I of DsrA in the presence of Hfq, nucleotides at the base of the Domain I stem were protected from RNase I cleavage. Given that chemically synthesized DsrADI alone does not bind to Hfq in gel shift assays (see Fig. 2 ▶), the interaction of Domain I of DsrA with Hfq is likely to be weak and may be dependent on Hfq binding of Domain II. One explanation for the results is that when the base of the Domain II stem binds Hfq, it orients Domain I such that it protrudes from the RNA/protein interface. This orientation may increase the accessibility of Domain I, thereby facilitating its interaction with rpoS mRNA.
Among the RNAs that interact with Hfq, DsrA is unique in that poly(A) did not compete for binding to Hfq. Neither polydisperse poly(A) nor a poly(A)27 affected formation of the DsrA–Hfq complex. Instead, it appears that DsrA and poly(A) can simultaneously bind to Hfq. We do not know whether poly(A) is bound indirectly to Hfq via DsrA or if Hfq has two or more RNA-binding sites. Investigations of Hfq:poly(A) interactions support the possibility of multiple RNA-binding sites in Hfq. Tight binding of Hfq to poly(A) is dependent on the length of the RNA, as Hfq binds much more tightly to a poly(A) 18-mer than to a poly(A) 16-mer (de Haseth and Uhlenbeck 1980a). This length dependence indicates that Hfq could contain multiple weak binding sites or that a poly(A) 18-mer forms a secondary structure that binds to Hfq. Although we were unable to detect a direct interaction between DsrA:poly(A) in the absence of Hfq, it is possible that, as reported for DsrA and other RNAs (R. Buchanan and O. Sledjeski, in prep.; Moller et al. 2002; Zhang et al. 2002), Hfq can facilitate RNA interactions. More work is necessary to reveal the nature of this ternary complex.
In addition to binding to A/U-rich sequences (Senear and Steitz 1976), Hfq binds to a large number of different RNAs with various affinities (Wassarman et al. 2001). Because many of the RNAs that bind to Hfq contain short A/U-rich regions (Senear and Steitz 1976; Zhang et al. 2002), it was reasonable to propose that the A/U-rich sequence between Domains I and II is the site of Hfq binding. RNase footprinting and competition experiments with the individual domains of DsrA localized the main Hfq-binding site to Domain II of DsrA. Given that poly(U) competed with DsrA for binding to Hfq and Domain II of DsrA contained an A/U-rich region that was protected from RNase I cleavage by Hfq, we predicted that altering the sequence of this region would decrease Hfq binding. Surprisingly, DsrAMDII RNA competed nearly as well as wild-type DsrADII for binding to Hfq (see Fig. 4 ▶). DsrAMDII may have assumed an alternate conformation, exposing additional A/U-rich single-stranded regions. However, two additional pieces of data support the dispensability of the poly(U) region. First, the minimal binding assay showed that at least 10 nt beyond the poly(U) region were necessary for weak binding and even more nucleotides were needed for strong binding. Second, although long polydisperse poly(U) (200–400 nt in length) could compete well with DsrA, a poly(U) 15-mer did not (data not shown). Therefore it is possible that a secondary structure formed by poly(U) (Rabczenko and Shugar 1971; Young and Kallenbach 1978) actually competed for binding to Hfq and not a linear sequence. Thus, we conclude that Hfq requires other structural elements (e.g., secondary or tertiary) for tight binding to DsrA. In support of this model, we observed increased RNase V1 sensitivity (cleaves double-stranded nucleotides) of 4 nt in Domain II upon binding to Hfq (Fig. 7 ▶). This increased double-stranded character could be owing to Hfq binding to and stabilizing the bottom portion of the stem or to fewer numbers of nuclease-binding sites. Because these nucleotides are accessible to RNase V1, we propose that Hfq is binding to the lower portion of the stem and the single-stranded region of DsrA (Fig. 9 ▶). The interaction with Hfq could also alter coaxial stacking between Domains II and III and explain the enhancement of an RNase V1 cleavage site at nucleotide G66 in Domain III (Fig. 9 ▶). Cleavage of nucleotide A27 by RNase V1 has been noted before (Lease and Belfort 2000). Because the region is predicted to be unstructured, it is not known with which nucleotide A27 might be interacting. Given A27 and G66 are close to the boundaries of the Hfq-binding site and both show enhanced cleavage, it is possible that these bases somehow interact.
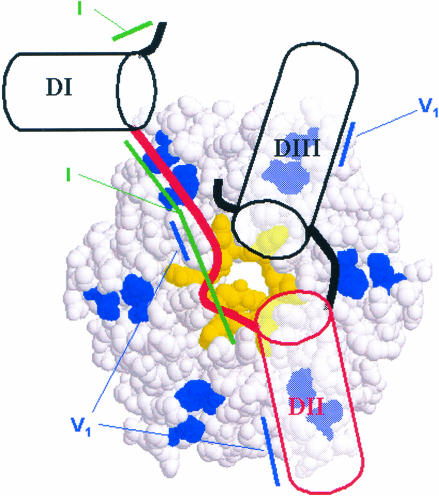
FIGURE 9.
Schematic model of interaction between DsrA and Hfq. In our present model all three DsrA domains are located on one face of the hexameric complex. The A/U-rich single-stranded region between Domains I and II (red line) might wrap around the inner diameter of the torus by analogy to the interaction between Sm-proteins and short poly(U) substrates (Toro et al. 2001). If both Domains I and II lie on the same face of Hfq, only a portion of the central cavity can be used for interaction with the U-rich region to avoid a steric clash between the 5′- and 3′-extensions from this primary binding site. The additional structural requirements we observe for tight binding of larger RNAs probably result from the need of RNA secondary structure elements to lie down against the rest of the hexamer face of Hfq. Two highly conserved residues (R15 and F41, shown in blue) could assist in this process by providing ionic interactions with the RNA. In our model, the formation of a dodecamer might bring together two RNAs, one bound to either face of the torus, thereby increasing the chances of interactions between the RNAs. Tertiary structure showing the coaxial stacking of Domains II and III of DsrA is purely speculative at this point. The regions of increased RNase V1 nuclease sensitivity (blue lines) or decreased RNase I cleavage (green lines) in the presence of Hfq are indicated. Note that the cleavage of nucleotide A27 by RNase V1 could be interpreted as this region forming a single-stranded helix (Lowman and Draper 1986).
It is not clear where DsrA (or any RNA) is located on the surface of Hfq. In our present model all three domains are located on one face of the hexameric complex (Fig. 9 ▶). The crystal structure of the phylogenetically related archeabacterial Sm-like proteins indicates that RNAs bind within a central cationic pore (Collins et al. 2001; Mura et al. 2001; Toro et al. 2001). Recently, a cocrystal structure of the Staphylococcus aureus Hfq protein complexed with a short oligonucleotide (5′-AUUUUUG-3′) was also reported (Schumacher et al. 2002). In this structure, the RNA once again interacts with the central cationic pore, so this binding mode is highly conserved among Sm and Sm-like proteins. Because the natural RNA substrates for Hfq are substantially longer than the one used crystallographically, one must consider the steric constraints of these molecules. One possibility is that the A/U-rich region in Domain II passes through the central pore of the Hfq hexamer and that Domains I and II lie on the opposites side of the Hfq hexamer. Analysis of the electrostatic surface of the Hfq hexamer in which one face is rather hydrophobic supports an argument against this mode of binding unless two hexamers dimerize to form a dodecamer similar to that observed in the crystal structure.
Alternatively, one can imagine facial binding of DsrA, with the U-rich region of Domain II binding to the central cavity. If both Domains I and II lie on the same face of Hfq, only a portion of the central cavity can be used for interaction with the U-rich region to avoid a steric clash between the 5′- and 3′-extensions from this primary binding site. In the crystal structure, the 5′- and 3′-ends are 7.1 Å apart and lie on the same face of the hexamer. The RNA-binding mode observed in the crystals is undoubtedly related to the natural binding site, but it is likely that only a subset of the crystallographically observed base-stacking sites are occupied, allowing the 5′- and 3′-ends to escape the central cavity. The additional structural requirements we observe for tight binding of larger RNAs probably result from the need of RNA secondary structure elements to lie down against the rest of the hexamer face of Hfq. In this case, the cationic bands observed in the electrostatic surface might provide ionic interactions with the phosphodiester backbone of a helical structure. In this model, the formation of a dodecamer might bring together two RNAs, one bound to either face of the torus. Such a model was recently proposed based on a modeling study of Hfq (Arluison et al. 2002). Facilitation of the RNA–RNA contacts between DsrA and an mRNA target such as RpoS mRNA might then require Hfq sequence elements along the outer face of the Hfq ring. Residues R15 and F41 (corresponding to K16 and Y39 in the Streptococcus aureus structure) are likely residues to participate in the surface for binding based on their strong conservation among all bacterial Hfq homologs.
The hypothesis that Hfq recognizes a higher-order structural element of DsrA is also consistent with the lack of sequence homology between the known Hfq sub-strates (Zhang et al. 2002) and Qβ (Senear and Steitz 1976). All of the RNAs known to bind Hfq contain elements of secondary structure, but not all RNA helices bind Hfq with equal affinity. Therefore, it seems likely that higher-order structures may be involved in these interactions, but further work will be required to map out the exact specificity elements regulating the formation of these RNA/protein complexes. Identification of the RNA structural element(s) to which Hfq binds and the role of Hfq in DsrA-mediated regulation will provide valuable insight into the function of this highly conserved protein in cellular physiology.
MATERIALS AND METHODS
In vitro synthesized RNA
DsrA RNA for gel shift assays and footprinting experiments was in vitro synthesized using the T7-MEGAshortscript In Vitro Transcription Kit (Ambion) according to the protocol suggested by the manufacturer. We used the plasmid pDDS164 (Sledjeski and Gottesman 1995) as the PCR template for generation of a DNA template carrying a T7 promoter for DsrA. The following primer set was used: DsrA, 5′-TAATACGACTCACTATAGGACACAT CAGATTTCC-3′ and 5′-ATCCCGACCCTGAGG-3′. Following in vitro transcription, the full-length RNA transcripts were purified on an 8% polyacrylamide–7 M urea gel and eluted into elution buffer (20 mM Tris-HCl at pH 7.5, 0.5 M sodium acetate, 10 mM EDTA, 1% SDS) at 65°C for 1 h.
Large-scale preparations of DsrA RNA for spectroscopic studies were prepared by in vitro run-off transcription with T7 RNA polymerase. dsDNA of the DsrA sequence properly positioned behind a T7 promoter and flanked by EcoRI and BamHI restriction sites was assembled from two long synthetic DNA oligonucleotides (IDT) by using standard procedures for primer extension (Sambrook et al. 1989). The resulting dsDNA was cloned into pUC19 and used to transform XL-10 supercompetent E. coli (Stratagene). The resulting plasmid (pBAU-001) was verified by standard automated sequencing protocols. Large-scale isolation of pBAV-001 was performed using a Giga-prep Kit (Qiagen) and the manufacturer’s protocol. Plasmid DNA was further purified by phenol–chloroform–isoamyl alcohol extraction and ethanol precipitation. The plasmid was prepared for run-off transcription by digestion with SspI and further purified by phenol–chloroform extraction and ethanol precipitation. Transcriptions were performed on the 10 mL scale (40 mM Tris at pH 8.1, 10 mM MgCl2, 0.2% TX-100, 10 mM DTT, 1.5 mM of each NTP, 0.063 μg/mL plasmid template, 30 μg/mL T7 RNAP) and allowed to react for 3–4 h. Upon conclusion of the transcription, precipitate was removed by centrifugation, and the resulting solution was ethanol-precipitated in the presence of 10 mM EDTA and 250 mM NaCl. RNA isolated from the ethanol precipitation was gel-purified by PAGE on 8% denaturing gels. The product was identified by UV shadowing, and the appropriate band was eluted into 500 mM NH4OAc at 4°C overnight. After ethanol precipitation, the DsrA RNA was resuspended in an appropriate volume of water, and the RNA concentration was determined based on its optical absorbance at 260 nm.
RNAs for competition experiments were chemically synthesized (Dharmacon Research, Inc.). The RNA contained the following sequences: (1) DsrADI, AACACAUCAGAUUUCCUGGUGUAAC; (2) DsrADII, AACGAAUUUUUUAAGUGCUUCUUGCUUAAG CAAGUUUC; (3) DsrADIII, CAUCCCGACCCCCUCAGGGUCG GGAUUU; MDII (modified Domain II), AACGAAACUUGCA AGUGCUUCUUGCUUAAGCAAGUUUC; and a poly(A) RNA 27-mer. The Decade marker (Ambion) was also used.
Labeling of RNA
DsrA RNA was radioactively labeled at the 5′ end with [γ-32P]ATP (Amersham Life Science, Inc.) and T4 Kinase (GIBCO Life Technologies) at 37°C for 45 min and 60°C for 10 min. The labeled RNA was then purified using Centri-Spin−10 columns (Princeton Separations) and accompanying protocol. Following labeling, DsrA RNA was heated to 80°C for 2 min and slowly cooled to 25°C. The RNA concentration was determined by measuring the OD260 prior to labeling.
Hfq purification
Hfq was purifed from E. coli essentially as described previously (Zhang et al. 2002). Briefly, cultures containing pET21b-hfq plasmid were grown to OD600 = 0.6 at 37°C. Hfq expression was induced by 5 mM IPTG, and after further growth, cells were harvested by centrifugation and resuspended in lysis buffer (50 mM Tris at pH 7.5, 1 mM EDTA, 50 mM NH4Cl, 5% glycerol). Cells were lysed by ultrasonication, and cellular debris was removed by centrifugation. The resulting supernatant was boiled for 10 min and centrifuged to remove non-heat-stable proteins. The cleared supernatant was passed over a poly(A)-sepharose column, washed with a high salt buffer (50 mM Tris at pH 7.5, 1 mM EDTA, 1 M NH4Cl, 5% glycerol) and then eluted with urea (8 M urea, 50 mM Tris at pH 7.5, 1 mM EDTA, 1 M NH4Cl, 5% glycerol). Purified Hfq was dialyzed against a storage buffer (50 mM Tris-HCl at pH 7.5, 1 mM EDTA, 250 mM NH4Cl, 10% glycerol). Hfq concentration was assayed at A280/A260 by the Warburg–Christian method (Stoscheck 1990).
Gel mobility shift assays
Binding assays were performed with 0.17 pmole of DsrA and 3 μM purified Hfq in binding buffer (10 mM Tris-HCl at pH 8.0, 50 mM NaCl, 50 mM KCl, 10 mM MgCl2) in the presence and absence of varying concentrations of competitor in a 10-μL reaction mixture. The competitor RNAs DsrADI, DsrADII, and DsrADIII were added to full-length DsrA at a 5- to 100-fold molar ratio; MDII was added at a 10- to 30-fold molar ratio; and poly(A)27 was added at a molar ratio of 0.01–5. Polydisperse poly(U) RNA was added at 5 ng–5 μg and polydisperse poly(A) at 0.1 ng–5 μg. DsrA was incubated with competing RNAs at 25°C for 5 min, followed by the addition of Hfq and incubation at 25°C for 5 min. Samples were then mixed with 2 μL of Hi-Density TBE sample buffer (Invitrogen), analyzed on an 8% native polyacrylamide gel in 1× TBE (Invitrogen) at 100 V for 2 h, dried, and exposed to a PhosphorImager screen (Molecular Dynamics). A PhosphorImager and ImageQuant software (Molecular Dynamics) were used to view the gel.
Nuclease cleavage assays
Labeled DsrA (0.1 pmole) in the absence and presence of varying concentrations of Hfq protein was incubated at 25°C for 10 min. The binding step was followed by cleavage with 0.05 U of RNase I (Promega) at 25°C for 2 min or 0.007 U of RNase V1 (USB) at 25°C for 10 min. DsrA was precipitated and resuspended in Gel Loading Buffer II (Ambion). Samples were analyzed on 12% polyacrylamide–7 M urea gels. A limited alkaline digestion of DsrA and 5′-end-labeled DsrADI (nucleotides 1–25) were added for size comparison. The gel was dried and exposed to a PhosphorImager screen. A PhosphorImager and ImageQuant software were used to view the gel.
Minimal binding assay of DsrA–Hfq complex
Our experiment outlined below is based on experiments previously described (Ziehler and Engelke 2000; Zhang et al. 2002). 5′-end-labeled DsrA (0.1 pmole/μL and 5 × 104 cpm) was ethanol-precipitated and resuspended in 1.2 μL of Na2CO3/EDTA solution (50 mM Na2CO3, 1 mM EDTA). The reaction was incubated for 90 sec and snap-cooled on ice for 1 min. A 2.8-μL volume of CU buffer (25 mM citrate, 10.25 mL of 0.1 M citric acid, 10 M urea, and 30 mL of water; the pH was adjusted to 5.0 with HCl) was then added, and the solution was mixed well. The hydrolyzed DsrA was incubated in the absence or presence of Hfq protein at 25°C for 10 min and separated on an 8% native polyacrylamide gel in 1× TBE. Hydrolyzed DsrA fragments that were bound to Hfq or unbound were visualized by autoradiography. The set of bands corresponding to the bound fractions and unbound fractions were excised, and samples were eluted into Elution Buffer as described above. The RNA was extracted with Trizol, chloroform, and Phase-Lock Gel Tubes (Eppendorf) using the manufacturer’s instructions. Samples were resuspended in 5 μL of the formamide loading dye, Gel Loading Buffer II (Ambion). Samples were analyzed on a 12% polyacrylamide gel in 1× TBE. The samples were viewed using a PhosphorImager screen and accompanying software as described above.
Circular dichroism and thermal melt profile
Samples for CD studies were prepared by dilution of DsrA RNA into buffer containing all components of the final sample except Hfq protein. These samples were annealed by heating at 90°C for 2 min, followed by slow cooling to room temperature (∼25°C). Hfq was added after cooling. CD spectra and CD thermal melt profiles were collected in either 1- or 10-mm pathlength cells depending on the sample composition on a Jasco J715 CD spectrometer equipped with a Peltier temperature-control device. Melt data were fit to a unimolecular two-state folding model for the ΔH° and Tm of the transition using Kaleidagraph software (Bloomfield et al. 2000). Gel shift assays were used to ensure that complete binding was maintained throughout the temperature range of the CD experiments. Samples of 5′-[32P]DsrA were prepared as above and then incubated at 37°C or 65°C for 10 min in the presence of 0, 3, or 7 M urea and 4.5 μM Hfq. These samples were loaded while still hot onto prewarmed 5% PAGE/7 M urea gels running in 1× TBE and visualized by phosphorimagery.
Acknowledgments
We thank B. Uyesugi for preparation of the pBAU-001 construct, J. Fitzgerald for purification of Hfq, and G. Storz for kindly providing the expression clone for Hfq. We also thank R. Blumenthal and R. Buchanan for helpful comments. This work was supported by Indiana University (A.L.F.), I.U. Dept. of Chemistry (A.L.F.), and the National Institutes of Health (RO1-GM56448 to D.D.S. and T32-GM07757 to I.U./P.M.).
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.rnajournal.org/cgi/doi/10.1261/rna.2570803.
REFERENCES
- Arluison, V., Derreumaux, P., Allemand, F., Folichon, M., Hajnsdorf, E., and Regnier, P. 2002. Structural modelling of the Sm-like Protein Hfq from Escherichia coli. J. Mol. Biol. 320: 705–712. [DOI] [PubMed] [Google Scholar]
- Azam, T.A., Iwata, A., Nishimura, A., Ueda, S., and Ishihama, A. 1999. Growth phase-dependent variation in protein composition of the Escherichia coli nucleoid. J. Bacteriol. 181: 6361–6370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloomfield, V.A., Crothers, D.M., and Tinoco, J.I. 2000. Nucleic acids: Structures, properties and function. University Science Books, Sausalito, CA.
- Collins, B.M., Harrop, S.J., Kornfeld, G.D., Dawes, I.W., Curmi, P.M., and Mabbutt, B.C. 2001. Crystal structure of a heptameric Sm-like protein complex from archaea: Implications for the structure and evolution of snRNPs. J. Mol. Biol. 309: 915–923. [DOI] [PubMed] [Google Scholar]
- de Haseth, P.L. and Uhlenbeck, O.C. 1980a. Interaction of Escherichia coli host factor protein with oligoriboadenylates. Biochemistry 19: 6138–6146. [DOI] [PubMed] [Google Scholar]
- ———.1980b. Interaction of Escherichia coli host factor protein with Q β ribonucleic acid. Biochemistry 19: 6146–6151. [DOI] [PubMed] [Google Scholar]
- Fasman, G.D. 1996. Circular dichroism and the conformational analysis of biomolecules. Plenum, New York.
- Fernandez, M.T.F.d., Hayward, W.S., and August, J.T. 1972. Bacterial proteins required for replication of phage Qβ ribonucleic acid. Purification and properties of host factor I, a ribonucleic acid-binding protein. J. Biol. Chem. 247: 824–831. [PubMed] [Google Scholar]
- Hajnsdorf, E. and Regnier, P. 2000. Host factor Hfq of Escherichia coli stimulates elongation of poly (A) tails by poly (A) polymerase I. Proc. Natl. Acad. Sci. 97: 1501–1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lease, R.A. and Belfort, M. 2000. A trans-acting RNA as a control switch in Escherichia coli: DsrA modulates function by forming alternative structures. Proc. Natl. Acad. Sci. 97: 9919–9924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lease, R.A., Cusick, M.E., and Belfort, M. 1998. Riboregulation in Escherichia coli: DsrA RNA acts by RNA:RNA interactions at multiple loci. Proc. Natl. Acad. Sci. 95: 12456–12461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lockard, R.E. and Kumar, A. 1981. Mapping tRNA structure in solution using double-strand-specific ribonuclease V1 from cobra venom. Nucleic Acids Res. 9: 5125–5140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowman, H.B. and Draper, D.E. 1986. On the recognition of helical RNA by cobra venom V1 nuclease. J. Biol. Chem. 261: 5396–5403. [PubMed] [Google Scholar]
- Majdalani, N., Cunning, C., Sledjeski, D., Elliott, T., and Gottesman, S. 1998. DsrA RNA regulates translation of RpoS message by an anti-antisense mechanism, independent of its action as an antisilencer of transcription. Proc. Natl. Acad. Sci. 95: 12462–12467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majdalani, N., Chen, S., Murrow, J., John, K.S., and Gottesman, S. 2001. Regulation of RpoS by a novel small RNA: The characterization of RprA. Mol. Microbiol. 39: 1382–1394. [DOI] [PubMed] [Google Scholar]
- Moller, T., Franch, T., Hojrup, P., Keene, D.R., Bachinger, H.P., Brennan, R.G., and Valentin-Hansen, P. 2002. Hfq. A bacterial Sm-like protein that mediates RNA–RNA interaction. Mol. Cell 9: 23–30. [DOI] [PubMed] [Google Scholar]
- Muffler, A., Traulsen, D.D., Fischer, D., Lange, R., and Hengge-Aronis, R. 1997. The RNA-binding protein HF-I plays a global regulatory role which is largely, but not exclusively, due to its role in expression of the σS subunit of RNA polymerase in Escherichia coli. J. Bacteriol. 179: 297–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mura, C., Cascio, D., Sawaya, M.R., and Eisenberg, D.S. 2001. The crystal structure of a heptameric archaeal Sm protein: Implications for the eukaryotic snRNP core. Proc. Natl. Acad. Sci. 98: 5532–5537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabczenko, A. and Shugar, D. 1971. Studies on the conformation of nucleosides, dinucleoside monophosphates and homopolynucleotides containing uracil or thymine base residues, and ribose, deoxyribose or 2′-O-methylribose. Acta Biochim. Pol. 18: 387–402. [PubMed] [Google Scholar]
- Sambrook, J., Fritsch, E., and Maniatis, T. 1989. Molecular cloning: A laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Schumacher, M.A., Pearson, R.F., Moller, T., Valentin-Hansen, P., and Brennan, R.G. 2002. Structures of the pleiotropic translational regulator Hfq and an Hfq–RNA complex: A bacterial Sm-like protein. EMBO J. 21: 3546–3556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuppli, D., Miranda, G., Tsui, H.C., Winkler, M.E., Sogo, J.M., and Weber, H. 1997. Altered 3′-terminal RNA structure in phage Qβ adapted to host factor-less Escherichia coli. Proc. Natl. Acad. Sci. 94: 10239–10242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senear, A.W. and Steitz, J.A. 1976. Site-specific interaction of Qβ host factor and ribosomal protein S1 with Qβ and R17 bacteriophage RNAs. J. Biol. Chem. 251: 1902–1912. [PubMed] [Google Scholar]
- Shelton, V.M., Sosnick, T.R., and Pan, T. 1999. Applicability of urea in the thermodynamic analysis of secondary and tertiary RNA folding. Biochemistry 38: 16831–16839. [DOI] [PubMed] [Google Scholar]
- Sledjeski, D. and Gottesman, S. 1995. A small RNA acts as an antisilencer of the H-NS-silenced rcsA gene of Escherichia coli. Proc. Natl. Acad. Sci. 92: 2003–2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sledjeski, D.D., Gupta, A., and Gottesman, S. 1996. The small RNA, DsrA, is essential for the low temperature expression of RpoS during exponential growth in Escherichia coli. EMBO J. 15: 3993–4000. [PMC free article] [PubMed] [Google Scholar]
- Sledjeski, D.D., Whitman, C., and Zhang, A. 2001. Hfq is necessary for regulation by the untranslated RNA DsrA. J. Bacteriol. 183: 1997–2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sosnick, T.R., Fang, X., and Shelton, V.M. 2000. Application of circular dichroism to study RNA folding transitions. Meth. Enzymol. 317: 393–409. [DOI] [PubMed] [Google Scholar]
- Stoscheck, C.M. 1990. Quantitation of protein. Meth. Enzymol. 182: 50–68. [DOI] [PubMed] [Google Scholar]
- Sun, X., Zhulin, I., and Wartell, R.M. 2002. Predicted structure and phyletic distribution of the RNA binding protein Hfq. Nucleic Acids Res. 30: 3662–3671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toro, I., Thore, S., Mayer, C., Basquin, J., Seraphin, B., and Suck, D. 2001. RNA binding in an Sm core domain: X-Ray structure and functional analysis of an archaeal Sm protein complex. EMBO J. 20: 2293–2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsui, H.C., Feng, G., and Winkler, M.E. 1997. Negative regulation of mutS and mutH repair gene expression by the Hfq and RpoS global regulators of Escherichia coli K-12. J. Bacteriol. 179: 7476– 7487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vytvytska, O., Moll, I., Kaberdin, V.R., Gabain, A.V., and Blasi, U. 2000. Hfq (HF1) stimulates ompA mRNA decay by interfering with ribosome binding. Genes & Dev. 14: 1109–1118. [PMC free article] [PubMed] [Google Scholar]
- Wassarman, K.M., Repoila, F., Rosenow, C., Storz, G., and Gottesman, S. 2001. Identification of novel small RNAs using comparative genomics and microarrays. Genes & Dev. 15: 1637–1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young, P.R. and Kallenbach, N.R. 1978. Secondary structure in polyuridylic acid. Non-classical hydrogen bonding and the function of the ribose 2′-hydroxyl group. J. Mol. Biol. 126: 467–479. [DOI] [PubMed] [Google Scholar]
- Zhang, A., Altuvia, S., Tiwari, A., Argaman, L., Hengge-Aronis, R., and Storz, G. 1998. The OxyS regulatory RNA represses rpoS translation and binds the Hfq (HF-I) protein. EMBO J. 17: 6061–6068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, A., Wassarman, K.M., Ortega, J., Steven, A.C., and Storz, G. 2002. The Sm-like Hfq protein increases OxyS RNA interaction with target mRNAs. Mol. Cell 9: 11–22. [DOI] [PubMed] [Google Scholar]
- Ziehler, W.A. and Engelke, D.R. 2000. Probing RNA structure with chemical reagents and enzymes. In Current protocols in nucleic acid chemistry (eds. S.L. Beaucage et al.), pp. 6.1.1–6.1.21. Wiley, New York, NY. [DOI] [PMC free article] [PubMed]