Abstract
Atomic resolution crystal structures of the large subunit published since the middle of August 2000 prove that the peptidyl transferase center of the ribosome, which is the site of peptide-bond formation, is composed entirely of RNA; the ribosome is a ribozyme. They also demonstrate that alignment of the CCA ends of ribosome-bound peptidyl tRNA and aminoacyl tRNA in the peptidyl transferase center contributes signficantly to its catalytic power. Several issues remain unresolved. For example, do any components of the site enhance the rate of peptide-bond formation chemically? Do intact ribosomes make peptide bonds the same way as the isolated large subunits that have been the source of all this atomic resolution structural information?
The site in the ribosome where peptide-bond formation occurs, its peptidyl transferase center, catalyzes the synthesis of peptide bonds in vitro using substrates much smaller than the aminoacyl and peptidyl tRNAs that are the ribosome’s normal fare (Traut and Monro 1964; Monro 1967). The lowest-molecular-weight aminoacyl tRNA-like substrate the ribosome will accept is puromycin, which is effectively an adenosine aminoacylated with tyrosine, and molecules as small as CCA aminoacylated with N-formyl methionine will serve in place of peptidyl tRNA. Because the peptidyl transferase center is entirely contained in the large subunit, it should be possible to learn a lot about the mechanism of peptide-bond formation from crystal structures of large subunits with substrates like these bound. Consistent with this expectation, the structures that have been obtained of the large subunit with low-molecular-weight substrates and products bound are highly suggestive regarding the events that occur in the peptidyl transferase center (Nissen et al. 2000; Hansen et al. 2002; Schmeing et al. 2002). Nevertheless, substantial uncertainty remains on two points. First, although the structures provide strong support for the idea that the peptidyl transferase center enhances the rate of peptide-bond formation by aligning its substrates appropriately (Page and Jencks 1971; Nierhaus et al. 1980; Nissen et al. 2000), the extent to which, or even whether the rate of peptide-bond formation is further increased by chemical means remains unclear. Second, it is uncertain how accurately the substrate analog interactions observed in large subunit crystals replicate those that occur in the 70S ribosome during normal protein synthesis. The resolution of these issues is among the most interesting challenges for ribosome research in the post-crystal-structure era.
How peptide-bond formation is catalyzed by the large ribosomal subunit
The reaction that occurs when peptide bonds form on the ribosome is a simple one: the aminolysis of an ester bond. The nucleophilic α-amino group of the amino acid moiety of an aminoacyl tRNA bound to the so-called A site of the peptidyl transferase center attacks the electrophilic carbonyl carbon of the ester bond linking the peptide moiety of a peptidyl tRNA bound to the so-called P site of the peptidyl transferase center. The resulting anionic, tetrahedral carbon intermediate subsequently rearranges to yield a discharged tRNA bound to the P site, and an A-site bound tRNA joined by an ester bond to a peptide that is one amino acid longer than it was before the reaction occurred. There is no evidence for the formation of a covalent intermediate involving ribosomal groups.
Because there are no crystals of 70S ribosomes that diffract to resolutions high enough to enable one to examine the peptidyl transferase center in atomic detail, all of the high-resolution structural information about peptide-bond formation available today derives from structures of the large ribosomal subunit with low-molecular-weight substrates, products, and transition-state analogs bound, and all of that information has been obtained using crystals of the large ribosomal subunit from Haloarcula marismortui (see Table 1 ▶). The pH in these crystals (∼6.0) is far from the optimum for protein synthesis (∼7.5), but nevertheless the large ribosomal subunits they contain bind substrates, products, and transition-state analogs, and will catalyze peptide-bond formation while still in the crystalline state (Schmeing et al. 2002).
TABLE 1.
Structures of large ribosomal subunit with low-molecular-weight substrates and transition-state analogs bound
| Structure | PDB file # | Reference |
| Large subunit with no ligands bound | 1JJ2 | Klein et al. 2001 |
| Large subunit with a transition-state analog (CCdA-p–puro) bound | 1FFZ | Nissen et al. 2000 |
| Large subunit with an acceptor stem in the A site | 1FGO | Nissen et al. 2000 |
| Large subunit with CCA in the P site and CC-puromycin–phe-caproic acid–biotin in the A site | 1KQS | Schmeing et al. 2002 |
| Large subunit with CC-puromycin–phe-caproic acid–biotin stabilized in the P site by sparsomycin | 1M90 | Hansen et al. 2002 |
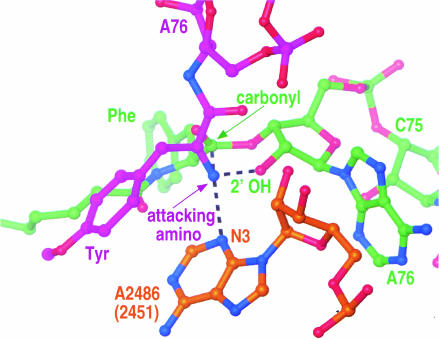
The large ribosomal subunit/substrate structure one would like most to have that is not available at this point is the structure of the large subunit with substrates bound to both its A and P sites. A model for that state of the peptidyl transferase center has been generated by superimposing existing structures for large subunits with A-site and P-site substrates bound separately (Fig. 1 ▶; Hansen et al. 2002). This superposition indicates that the peptidyl transferase center is capable of bringing the nucleophilic α-amino group of an aminoacyl tRNA into van der Waals contact with the electrophilic carbonyl carbon with which it must react during peptide-bond formation. An accurate understanding of the angular orientation of these atoms cannot be obtained from the data now available because of the limitations imposed by the resolution of the structures from which the model is derived and the uncertainties inherent in the superposition process itself, and, indeed, the inferred orientation is not that expected for the two groups at the instant of reaction. Therefore, although it is abundantly clear that the peptidyl transferase center, like the active sites of all other enzymes that catalyze the reaction of pairs of substrates, accelerates the rate of the reaction it catalyzes by using the favorable energy of substrate binding to overcome the entropic cost of aligning substrates (Page and Jencks 1971), significant details remain to be elucidated.
FIGURE 1.
A model for the peptidyl transferase center of the large ribosomal subunit from Haloarcula marismortui with substrates bound to both the A site and its P site. This model was obtained by superimposing the structure of an A-site substrate complex (PDB #1FGO) on the structure of a P-site substrate complex (PDB #1M90; Hansen et al. 2002). The α-amino group of the A-site substrate (purple) is positioned for a pro-S attack on the carbonyl carbon of the ester linking the peptide moiety of the P-site substrate (green). Possible hydrogen-bonding interactions involving the α-amino group and the N3 of A2486 (2451) and the 2‘-OH of A76 are indicated. The 2‘-OH of A2486 (2451) is also close enough so that it could interact.
What chemical resources does the peptidyl transferase center possess for accelerating the rate of peptide-bond formation beyond that achievable by the juxtaposition of substrates? In none of the relevant structures is there any evidence for an active-site metal ion, although until a structure has been obtained with analogs bound in both the A site and the P site, the possibility that metals participate in the reaction cannot be ruled out. There are, however, three groups in the neighborhood of the reactive α-amino group that could conceivably form hydrogen bonds with it, and two are likely to do so: (1) the 2‘-OH of A76 in the P-site-bound peptidyl tRNA, (2) the N3 of A2486 (A2451 in Escherichia coli) of 23S rRNA, and (3) the 2‘-OH of A2486 (2451; Hansen et al. 2002). The hydrogen bonds these groups form with the α-amino group, either singly or collectively, almost certainly help align it, and if any of these groups were to have an elevated pKa, its hydrogen-bonding interaction would facilitate the peptide-bond-forming reaction by enhancing the nucleophilicity of the α-amino group.
There is evidence that the first two of the three groups just mentioned are important for the peptidyl transferase activity of the ribosome, but none regarding the role of the third, the 2‘-OH of A2486. It has long been known that tRNAs that terminate with a 2‘-deoxyadenosine can be aminoacylated, and that they are A-site substrates for the ribosome. The peptidyl tRNAs they form, however, are inactive as P-site substrates; they will not donate their peptide moieties to A-site substrates (Quiggle et al. 1981). Clearly the 2‘-OH of A76 of tRNAs bound in the P site is important for P-site function, but the reason remains to be established. Interactions involving the 2‘-OH of A76 could help position the attacking amino group relative to the ester bond, a hypothesis consistent with the superposition model, but it could also play a more active, chemical role in catalysis, or in fact, do some of both.
Stimulated by the crystal structure of the large subunit with a transition-state analog bound, which indicated that the N3 of A2486 (2451) may interact with the oxyanion intermediate formed during peptide-bond synthesis (Nissen et al. 2000), much work has been done recently on the role of that nucleotide in peptide-bond formation. The role of A2486 in protein synthesis is still not understood, but three important points have been clarified. First, the structural data now available indicate that whatever A2486 may contribute to peptide-bond formation, it is unlikely to be involved in stabilization of the oxyanion intermediate generated during peptide-bond formation. The orientation of substrates in the active site is such that the anionic oxygen of the tetrahedral intermediate is likely to point away from the N3 of A2486 (2451) rather than toward it (Hansen et al. 2002). Second, initial reports notwithstanding (Muth et al. 2000), the chemical reactivity data available do not support the proposal that the pKa of the N3 of A2486 (2451) is anomalously high, as it would have to be if its interaction with the nucleophilic α-amino group were to have a large impact on catalytic rate enhancement, but they do not disprove it either (Bayfield et al. 2001; Muth et al. 2001). Third, mutation of A2486 (2451) reduces the rate of the chemical step of peptide-bond formation in 70S ribosomes by ∼100-fold at basic pHs, which is much larger than the rate changes first reported (Polacek et al. 2001; Thompson et al. 2001; Xiong et al. 2001). This rate effect titrates with a pKa of ∼7.5 (Katunin et al. 2002). These observations would be explained if the N3 of A2486 does, indeed, have a pKa of 7.5 and helps activate the α-amino group as originally proposed (Nissen et al. 2000), but are equally consistent with the hypothesis that the peptidyl transferase center is subject to a pH-dependent conformational change having a pKa of 7.5 that depends on the identity of the base at position 2486 for some other reason. One important fact that is still unknown is whether peptide-bond formation has the same pH-dependence in the 50S subunit, the object for which high-resolution structural information exists, as it does in the 70S ribosome, the object used in most of the recent kinetic studies.
Although the contributions each component in the catalytic site makes to the rate of peptide-bond formation remain to be determined and many other details need to be worked out, the most significant conclusion that has emerged from the structural studies of the large ribosomal subunit and its substrate complexes so far is that the ribosome is a ribozyme. All of the components of the ribosome involved in orienting the CCA ends of tRNA and properly positioning the A-site α-amino group and the carbonyl carbon it attacks are made of RNA, as is everything else in the neighborhood (Nissen et al. 2000). Although proteins do contact ribosome-bound tRNAs and undoubtedly play a role in their orientation, the business of catalysis is conducted entirely by RNA. Thus, in the beginning, there was RNA, at least as far as protein synthesis is concerned. Unlike other known ribozymes, which, for all we know, could have evolved after proteins appeared, the RNA-based catalytic center that is the functional heart of the ribosome must have evolved before gene-encoded proteins did.
Is large-subunit-catalyzed peptide-bond formation a good model for peptide-bond formation catalyzed by the full ribosome?
The studies on the structures of substrate analogs and large subunits summarized above have all been motivated by the belief that they will shed light on the events that occur in the peptidyl transferase center of the 70S ribosome during protein synthesis, and there are many reasons for believing that the reaction of low molecular peptidyl transferase substrates catalyzed by the large ribosomal subunit, that is, the fragment reaction, is relevant. First, both the 70S ribosome and the 50S subunit catalyze the reaction of fragment-reaction substrates. Second, the substrates and products of the fragment reaction are obvious analogs of those processed by the 70S ribosome when it is synthesizing protein normally. Third, the fragment reaction and the normal peptidyl transferase reaction occur at exactly the same site in the large ribosomal subunit. Fourth, both reactions are sensitive to many of the same inhibitors.
At this point, the only reason for questioning the relevance of the large-ribosomal-subunit fragment reaction to the full peptidyl transferase reaction is its rate. In live bacteria, protein is synthesized at a rate of ∼20 amino acids per second (at 37°C; Kjeldgaard and Gaussing 1974), and only a tiny fraction of the ∼50 msec it takes to incorporate the average amino acid is consumed by the chemical step of peptide-bond formation; the rate of that step in the peptide elongation cycle is probably >100 sec−1 (Rodnina et al. 2000). Consistent with this estimate, under saturating conditions, the rate of the reaction of puromycin with peptidyl tRNA bound to mRNA-programmed 70S ribosomes is ∼70 sec−1 (Katunin et al. 2002). The rates that have been reported for the fragment reaction catalyzed by the large subunit, however, are of the order of a few per minute, which is 3 to 4 orders of magnitude slower (Maden et al. 1968). This rate difference is not understood. At worst it could imply that the configuration of the peptidyl transferase center in reaction-ready 70S ribosomes is significantly different from the one seen in the large ribosomal subunit, or even that a mechanistic difference exists between what occurs in 70S ribosomes and isolated large ribosomal subunits. But it need not. A small alteration in the presentation of the nucleophilic α-amino group to the carbonyl group it attacks or in its relationship to surrounding groups could easily result in a change in reaction rate of that magnitude.
Future mechanistic challenges
The ideal structural basis for understanding the mechanism of any enzyme is a knowledge of the atomic positions of all relevant substrate and enzyme atoms from the binding of substrates, through intermediates and transition states to products. Whereas attempts have been made to achieve this level of understanding by time-resolved crystallography, the approach more generally pursued has been to make substrate and intermediate complexes that are trapped at different points along the reaction pathway. Necessarily, every approach to preventing the progression of the reaction has the possibility of altering the reaction pathway and hence of providing a seriously distorted view of what occurs when the enzyme is operating normally. There is little indication this has happened in the past, but it is an important reason to have more than one way to examine each mechanistic issue. The same principles, of course, apply to establishing the source of catalytic power in peptide-bond synthesis.
In the case of the ribosome, it is now very important to obtain a structure that has substrate analogs bound to both the A and P sites. The simultaneous presence of both substrates could induce a change in ribosome structure that neither alone can accomplish, and perhaps reveal why peptidyl tRNA in the P site is not hydrolyzed (in the absence of release factor). It is also possible that some new catalytic element that has not yet been identified in any of the structures obtained to date might be revealed in such a two-substrate structure, a magnesium ion, for example.
It would also be very useful to obtain a structure for a complex with an analog that better represents the tetrahedral carbon intermediate than the CCdA-P–puro analog structure now available, which is now known to present an inaccurate picture of that state of the reaction (Hansen et al. 2002). At the moment, no functional group in the ribosome has been identified that is properly positioned to stabilize the oxyanion that develops during peptide-bond formation the way that the ‘‘oxyanion hole’’ in the serine proteases or the Zn++ ion in the metalloproteases stabilizes similar intermediates during the reverse reaction, the hydrolysis of peptide bonds. The means used by the ribosome to stabilize this intermediate, if any, might be revealed by such a structure.
As with all enzyme mechanistic studies, appropriate mutagenic studies and kinetic analyses sensitive to the chemical step in the reaction are of ongoing importance. A handicap those who study very large RNA enzymes must endure is their inability to make the RNA equivalent of the alanine mutations used by their colleagues who study proteins. It is possible to make base changes in RNAs by mutation, of course, but the four bases are far more similar in their properties than Phe, Asp, His, and Ala, for example. The N3s of A and G are identical, and in significant ways similar to the O2s of U and C. A more powerful approach for getting the information desired would be to insert abasic nucleotides into the RNA of interest, but the execution of such experiments in a system as complex as the ribosome would be very demanding. Finally, prior enzyme mechanistic studies have established that it is of great importance to examine the structure of mutant enzymes captured in the reaction step their mutations are believed to affect. This challenge needs to be addressed with the 50S subunit.
Finally, perhaps the biggest challenge of all is obtaining crystals of the 70S ribosome with aminoacyl- and peptidyl-tRNA substrates bound that diffract to 3 Å resolution, or better. Only when 70S crystals of this quality become available will it be possible to obtain an atomic resolution structure for the entire ribosome with substrates bound that is fully independent of the structures that have been determined for the two subunits separately. Comparisons of that structure with existing structures of 50S subunits with low-molecular-weight substrates bound will reveal whether the conclusions that have been reached about the mechanism of peptide-bond formation from large-subunit structures need to be modified.
Acknowledgments
The authors thank Jeffrey Hansen and Martin Schmeing for preparing the figure presented here. This work was supported by grants to P.B.M. and T.A.S. from NIH (GM22778) and the Agouron Institute.
Article and publication are at http://www.rnajournal.org/cgi/doi/10.1261/rna.2127103.
REFERENCES
- Bayfield, M.A., Dahlberg, A.E., Schulmeister, U., Dorner, S., and Barta, A. 2001. A conformational change in the ribosomal peptidyl transferase center upon active/inactive transition. Proc. Natl. Acad. Sci. 98: 10096–10101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen, J.L., Schmeing, T.M., Moore, P.B., and Steitz, T.A. 2002. Structural insights into peptide bond formation. Proc. Natl. Acad. Sci. 99: 11670–11675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katunin, V.I., Muth, G.W., Strobel, S., Wintermeyer, W., and Rodnina, M.V. 2002. Important contribution to catalysis of peptide bond formation by a single ionizing group within the ribosome. Mol. Cell 10: 339–346. [DOI] [PubMed] [Google Scholar]
- Kjeldgaard, N.O. and Gaussing, K. 1974. Regulation of biosynthesis of ribosomes. In Ribosomes (eds. M. Nomura et al.), pp. 369–392. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- Klein, D.J., Schmeing, T.M., Moore, P.B., and Steitz, T.A. 2001. The kink-turn: A new RNA secondary structure motif. EMBO J. 20: 4214–4221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maden, B.E.H., Traut, R.R., and Monro, R.E. 1968. Ribosome-catalyzed peptidyl transfer: The polyphenylalanine system. J. Mol. Biol. 35: 333–345. [DOI] [PubMed] [Google Scholar]
- Monro, R.E. 1967. Catalysis of peptide bond formation by 50S ribosomal subunits from Escherichia coli. J. Mol. Biol. 26: 147–151. [DOI] [PubMed] [Google Scholar]
- Muth, G.W., Ortoleva-Donnelly, L., and Strobel, S.A. 2000. A single adenosine with a neutral pKa in the ribosomal peptidyl transferase center. Science 289: 947–950. [DOI] [PubMed] [Google Scholar]
- Muth, G.W., Chen, L., Kosek, A., and Strobel, S. 2001. pH-dependent conformational flexibility within the ribosomal peptidyl transferase center. RNA 7: 1403–1415. [PMC free article] [PubMed] [Google Scholar]
- Nierhaus, K.H., Schulze, H., and Cooperman, B.S. 1980. Molecular mechanisms of the ribosomal peptidyl transferase center. Biochem. Internat. 1: 185–192. [Google Scholar]
- Nissen, P., Ban, N., Hansen, J., Moore, P.B., and Steitz, T.A. 2000. The structural basis of ribosome activity in peptide bond synthesis. Science 289: 920–930. [DOI] [PubMed] [Google Scholar]
- Page, M.I. and Jencks, W.P. 1971. Aminolysis of acetylimidazole and rate acceleration caused by intramolecular catalysis. Fed. Proc. 30: 1240. [Google Scholar]
- Polacek, N., Gaynor, M., Yassin, A., and Mankin, A.S. 2001. Ribosomal peptidyl transferase can withstand mutations at the putative catalytic nucleotide. Nature 411: 498–501. [DOI] [PubMed] [Google Scholar]
- Quiggle, K., Kumar, G., Ott, T.W., Ryu, E.K., and Chladek, S. 1981. Donor site of ribosomal peptidyltransferase: Investigation of substrate specificity using 2‘(3′)-O-(N-acylaminoacyl)dinucleoside phosphates as models of the 3′ terminus of N-acylaminoacyl transfer ribonucleic acid. Biochemistry 20: 3480–3485. [DOI] [PubMed] [Google Scholar]
- Rodnina, M.V., Pape, T., Savelsbergh, A., Mohr, D., Matassova, N.B., and Wintermeyer, W. 2000. Mechanisms of partial reaction of the elongation cycle catalyzed by elongation factors Tu and G. In The ribosome: Structure, function, antibiotics and cellular interaction (eds. R.A. Garrett et al.), pp. 301–317. ASM Press, Washington, DC.
- Schmeing, T.M., Seila, A.C., Hansen, J.L., Freeborn, B., Soukup, J.K., Scaringe, S.A., Strobel, S., Moore, P.B., and Stetiz, T.A. 2002. A pretranslocational intermediate in protein synthesis observed in crystals of enzymatically active 50S subunits. Nat. Struct. Biol. 9: 225–230. [DOI] [PubMed] [Google Scholar]
- Thompson, J., Kim, D.F., O’Connor, M., Lieberman, K.R., Bayfield, M.A., Gregory, S.T., Green, R., Noller, H.F., and Dahlberg, A.E. 2001. Analysis of mutations at residues A2451 and G2447 of 23S rRNA in the peptidyltransferase active site of the 50S ribosomal subunit. Proc. Natl. Acad. Sci. 98: 9002–9007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traut, R.R. and Monro, R.E. 1964. The puromycin reaction and its relation to protein synthesis. J. Mol. Biol. 10: 63. [DOI] [PubMed] [Google Scholar]
- Xiong, L., Polacek, N., Sander, P., Boettger, E.G., and Mankin, A.S. 2001. pKa of adenine 2451 in the ribosomal peptidyl transferase center remains elusive. RNA 7: 1365–1369. [PMC free article] [PubMed] [Google Scholar]