Abstract
Alternative splicing of mammalian transcripts, which yields many diverse protein products from one gene, is the rule and not the exception. Although the mechanisms that govern alternative splicing are being unraveled, little is known about the evolution of this critical engine of proteome diversity. Here we present a phylogenetic analysis from a sea urchin to humans of the alternative splicing unit encoding the third Ig domain of fibroblast growth factor receptors. The remarkable conservation of intronic control elements, both in structure and function, indicates that the mechanisms that regulate this alternative splicing unit evolved over 600 million years ago.
Keywords: Exon, intron, alternative splicing, evolution
INTRODUCTION
Proteomic diversity is a hallmark of complex eukaryotic organisms. Although this complexity has been attributed to expansion in gene number, the human genome is predicted to contain <50,000 protein-coding genes versus ∼19,100 in the nematode Caenorhabditis elegans (Lander et al. 2001; Venter et al. 2001). Perhaps a more powerful engine of proteome diversity is alternative splicing of primary transcripts. Although alternative splicing occurs in C. elegans and in Drosophila melanogaster, it has reached unprecedented frequency in mammals, where as many as 60% of primary transcripts may be alternatively spliced (Modrek and Lee 2002). It is not clear at which point in evolution alternative splicing became the rule rather than the exception and little is known about the evolution of the mechanisms that regulate this splicing.
Alternative splicing and gene duplication increase the diversity of fibroblast growth factor receptors (FGF-R), particularly in the extracellular Ig domains that determine ligand-binding specificity (McKeehan et al. 1998). Two isoforms of FGF-R2, FGF-R2(IIIb) and FGF-R2(IIIc), contain different versions of the third Ig domain (IgIII) and bind a different subset of FGFs (McKeehan et al. 1998). These two isoforms are produced by the regulated and mutually exclusive splicing of one of two exons, exon IIIb or exon IIIc (Fig. 1A ▶) (Carstens et al. 1998). The cell-type specific inclusion of exon IIIb is controlled in large part by two regulatory elements (IAS2 and ISAR) that activate the constitutionally weak exon IIIb and repress exon IIIc, whereas the cell-type specific inclusion of exon IIIc is regulated by elements that silence exon IIIb (UISS, ESS, and DISS) and lead to the exclusive use of the strong exon IIIc (Carstens et al. 1998,2000; Del Gatto and Breathnach 1995; Del Gatto et al. 1997; Del Gatto-Konczak et al. 2000; Wagner and Garcia-Blanco 2001,2002). In this report, we examine the evolution of the alternative splicing unit that regulates IgIII isoform choice and we show that critical aspects of this regulation have been conserved since the divergence of echinoderms and vertebrates.
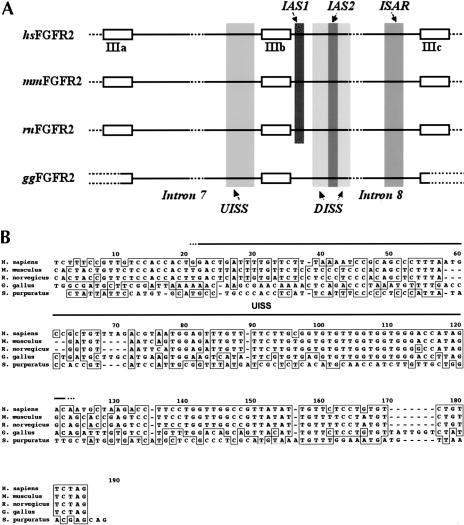
FIGURE 1.
The IgIII alternative splicing unit in vertebrate FGF-R genes is highly conserved. (A) A schematic diagram of the genomic region spanning the IgIII alternative splicing unit is shown for human (hsFGF-R2), mouse (mmFGF-R2), rat (rnFGF-R2), and chicken (ggFGF-R2) FGF-R2 genes. Exons IIIa, IIIb, and IIIc, and introns 7 and 8 are indicated, and the nomenclature is based on the structure of the gene in mammals. ClustalW alignments revealed high conservation of sequence in the regions indicated in the figure: the upstream intronic splicing silencer (UISS), the downstream intronic splicing silencer (DISS), the intronic activation sequence-2 (IAS2), and the intronic splicing activator and repressor (ISAR)(also known as IAS3). Broken lines in exons IIIa and IIIc of ggFGF-R2 represent regions where sequence has not been rigorously confirmed. (B) A ClustalW alignment of the 3′ end of intron 7 from vertebrate FGF-R2 and spFGF-R genes revealed conservation of UISS among vertebrates. (C) A ClustalW alignment of intron 8 from vertebrate FGF-R2 and spFGF-R genes shows the remarkable conservation of cis-acting elements, most notably IAS2 and ISAR. Divergent sequences in the middle of the intron are not shown. (D) Sequence alignment revealed high conservation within the IAS2 and ISAR regulatory elements among the four vertebrate species.
RESULTS AND DISCUSSION
Mining the human and mouse genome databases and our own data for the rat FGF-R2 genomic sequence, we concluded that the IgIII alternative splicing unit for the FGF-R2 gene, which is the region between and including exons IIIa and IIIc, was highly conserved among placental mammals (Fig. 1 ▶). Not surprisingly, high homology was found among the three exons of the IgIII unit and among the cis-acting elements that have critical roles in IgIII splicing regulation. To probe the conservation of structure of the IgIII alternative splicing unit among nonmammalian vertebrates, we cloned and sequenced this genomic region in the chicken (Gallus gallus) FGF-R2 gene. Even though mammals diverged from the avian/reptilian lineage more than 300 million years ago (Hedges and Poling 1999), we found that the overall structure of the IgIII region in the chicken FGF-R2 gene is almost identical to that seen in mammals. In addition to the conservation of exonic sequences, the noncanonical sequences of the splice sites that border exon IIIb were found to be conserved: the 5′ splice site sequence (5′-GTAACA-3′) is identical to that in mammals, and the 3′ splice site includes a weak polypyrimidine tract as is the case in the three mammalian FGF-R2 genes (Fig. 1B,C ▶). Whereas the majority of intronic regions were divergent (see, for example, sequences 330–390 and 1110–1170 in Fig. 1C ▶ and data not shown), segments of the chicken gene that flank exon IIIb were homologous to the mammalian splicing silencers UISS (Fig. 1B ▶) and DISS (Fig. 1C ▶). More remarkable was the conservation of the IAS2 and ISAR activating elements, for example the chicken IAS2 (ggIAS2) differs by only one transversion when compared with the 18-nucleotide mammalian IAS2 element (Fig. 1D ▶). The mammalian IAS2 and ISAR elements are partially complementary and can base-pair to form a secondary structure (Del Gatto et al. 1997; Jones et al. 2001); consequently, it was not surprising that ggIAS2 and ggISAR could form a comparable secondary structure (see Fig. 3B ▶ below). Not all regulatory elements were conserved, however, a Tia-1 binding site (IAS1), which has an ancillary role in activating exon IIIb (Del Gatto-Konczak et al. 2000), was not present in the chicken FGF-R2 gene (Fig. 1A,C ▶). These data indicate that the general architecture of the IgIII region of the FGF-R2 gene and the sequence of the critical cis-elements that control IIIb versus IIIc choice evolved before the divergence of mammals and birds. These data also suggest that chicken FGF-R2 transcripts will use exons IIIb and IIIc in a mutually exclusive fashion.
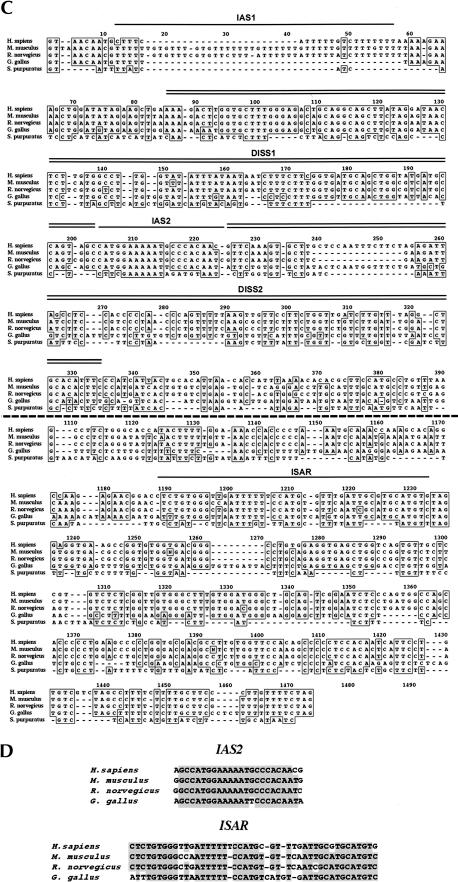
FIGURE 3.
Conservation of IAS2–ISAR structure and function among FGF-R genes from sea urchins to humans. (A) A ClustalW alignment of intron sequences from H. sapiens, G. gallus and S. purpuratus downstream of exon IIIb revealed the conservation of several cis-acting elements. The 5′ splice site that borders exon IIIb and the conserved IAS2 and ISAR elements are shown for hsFGF-R2, ggFGF-R2, and spFGF-R genes. The position of the regulatory elements relative to the 5′ splice site (+1) is also indicated in each case. (B) Remarkable conservation of structure is apparent when comparing the potential for IAS2 and ISAR to form a secondary structure composed of two stems separated by a bulge (proximal and distal stems are indicated). (C) The spIAS2 and spISAR stem can partially substitute for the mammalian stem in an alternative splicing functional assay in rat DT3 cells (Carstens et al. 1998). The percentage of exon IIIb (with standard deviation) included is shown for a rat minigene construct were the rnIAS2 and rnISAR sequences were replaced with pBluescript sequences (Blue/Blue) as a negative control or replaced with the stem-forming portions of IAS2 and ISAR from either R. norvegicus or S. purpuratus. (D) An 89-nucleotide RNA, which included the spIAS2 and spISAR sequences separated by six nucleotides was synthesized by T7 RNA polymerase (see Materials and Methods). The structure of this RNA was probed with RNase A and RNase T1. Strong RNase A cleavage sites are indicated by large black arrowheads and weak sites are indicated by small black arrowheads. Weak RNase T1 cleavage sites are indicated by small gray arrowheads. The cleavage pattern shown is consistent with the specificity of the nucleases and the structure shown except for the RNase A cleavage 5′ of U30.
To study the splicing of FGF-R2 transcripts in G. gallus, we dissected a female chicken and isolated RNA from seven organs. These organs are presumed to have both epithelial and stromal components in different ratios. These ratios should dictate the levels of FGF-R2(IIIb) or FGF-R2(IIIc), assuming that the regulation of ggFGF-R2 is cell-type specific. RT-PCR analysis revealed wide variation among organs in the relative levels of FGF-R2(IIIb) versus FGF-R2(IIIc) (Fig. 2A ▶). Interestingly, in the GI tract, which was primarily intestinal tissue, we find almost exclusive use of exon IIIb. This is consistent with the preponderance of epithelial cells in these tissues (Bloom and Fawcett 1975). In the heart, however, we detected mostly FGF-R2(IIIc), which was expected for an organ composed of endothelium, myocardium and mesothelium (Bloom and Fawcett 1975). Other tissues revealed a mixture of FGF-R2(IIIb) and FGF-R2(IIIc) as is the case in mammalian tissues. Interestingly, the gall bladder contained almost exclusively FGF-R2(IIIc), which may be attributable to the preponderance of muscle and connective tissue in this organ (Bloom and Fawcett 1975). The identity of FGF-R2(IIIc) in the gall bladder and FGF-R2(IIIb) in intestine was confirmed by cloning and sequencing of the PCR products (data not shown). It is important to note that we did not observe RNA products with exon IIIb and exon IIIc double inclusion. This strongly suggests that the regulation of the alternative splicing of chicken FGF-R2 transcripts parallels the regulation observed in mammals.
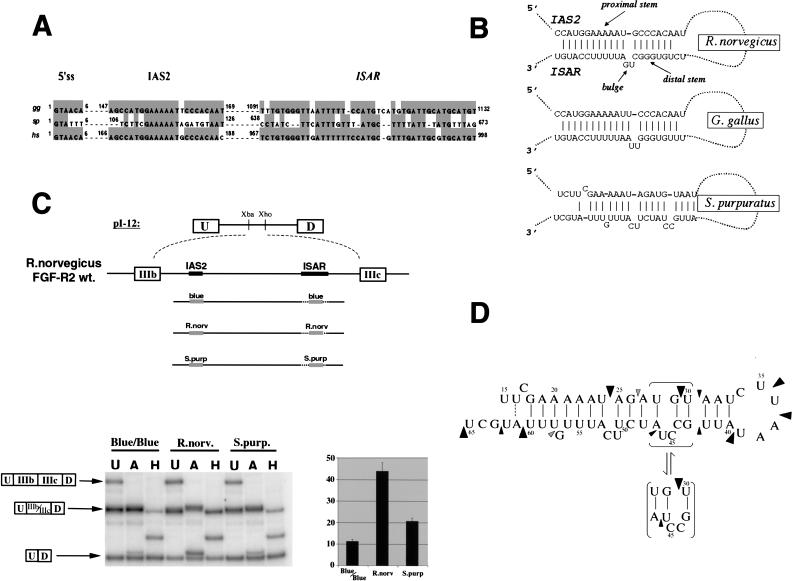
FIGURE 2.
Chicken FGF-R2 transcripts are differentially spliced to IIIb or IIIc isoforms. (A) A schematic of the RT-PCR assay used to detect FGF-R2 transcripts and to distinguish between inclusion of IIIb and IIIc. Products of RT-PCR were separated on agarose gels and visualized by EtBr staining. RT-PCR products (U) were incubated with either HaeIII (H), which cleaves within exon IIIb, or EcoRV (E), which cleaves within exon IIIc. The middle panel shows RT-PCR analysis for chicken organs and right panel shows RT-PCR analysis for cell lines. Cell lines UM and SL-29 have a fibroblast morphology and cell line LMH has an epithelial morphology. (B) Schematic of four minigenes spanning the IgIII region of the rat FGF-R2 gene indicates the presence or absence of the IAS2 and ISAR (left panel). The level of exon IIIb inclusion from these minigenes transfected into LMH or UM cells is shown in the middle and right panels.
To address the cell-type specificity further, we also used three chicken cell lines: the fibroblast lines, UM and SL-29, and the epithelial-like line LMH. As expected, the UM and SL-29 fibroblast cell lines include only the IIIc exon, whereas the epithelial-like LMH cell line used predominantly the IIIb exon (Fig. 2A ▶). Inclusion of exon IIIb in mammalian epithelial cells requires the activating elements IAS2 and ISAR ( Del Gatto et al. 1997; Carstens et al. 1998;). The importance of these elements in the chicken LMH cells was tested by transfecting minigene constructs, which span the Ig-III region of the rat FGF-R2 gene. The minigene constructs included exon IIIb and, as in mammals, this exon inclusion required IAS2 and ISAR (Fig. 2B ▶). The pattern of exon IIIb versus exon IIIc use and the dependence on IAS2 and ISAR elements in chicken cells strongly suggests a conservation of function as well as structure in FGF-R2 alternative splicing.
To unravel the origins of the IgIII alternative splicing unit, we studied the only identified FGF-R gene in the echinoderm Strongylocentrotus purpuratus. spFGF-R encodes two IgIII isoforms, which have been justifiably presumed to be the result of alternative splicing (McCoon et al. 1996, 1998). The isoforms, IgIIIS (short) and IgIIIL (long), encode IgIII domains of different length with IgIIIL having an additional 34 amino acids presumably because of the differential inclusion of a 102-nucleotide exon (McCoon et al. 1996). We cloned and sequenced the S. purpuratus IgIII region, which was noted to include three exons, spIIIa, spIIIb, and spIIIc, and confirmed that the IgIIIL isoform was the result of the inclusion of exon spIIIb. Alternative inclusion of exon spIIIb is regulated during development of S. purpuratus and is tissue-specific in the adult, for instance intestine has a predominance of the IgIIIL isoform, whereas the lantern muscle expresses exclusively the IgIIIS isoform (McCoon et al. 1996,1998). If the mechanism of this regulation were shared between S. purpuratus and vertebrates, one would expect conservation of regulatory cis-acting elements.
An alignment of the sequences of the intron downstream of exon IIIb revealed remarkable conservation of IAS2, of proximal ISAR sequences, and of the relative position of IAS2 and ISAR within the intron (Figs. 1C and 3A ▶ ▶). Perhaps most importantly, spIAS2 and spISAR are predicted to form a structure similar to that proposed in vertebrates (Fig. 3B ▶). It should be noted that the functional importance for the mammalian stem structure has been confirmed using mutations to disrupt the stem and abrogate function, and compensatory mutations that rescue both stem formation and function (Del Gatto et al. 1997; Jones et al. 2001). In S. purpuratus, the ISAR sequences that form the distal stem were noted to diverge; however, compensatory changes in IAS2 recreate a distal stem (Fig. 3B ▶), a phylogenetic conservation of structure that strongly suggests conservation of function. In fact, S. purpuratus IAS2–ISAR sequences could partially substitute for the Rattus norvegicus IAS2–ISAR stem-forming sequences in activating the inclusion of the rat IIIb exon in FGF-R2 minigenes (Fig. 3C ▶). Establishing the in vivo structure of RNA is very difficult, especially in S. purpuratus, and therefore we asked whether a model RNA containing spIAS2 and spISAR could form a stem structure in vitro. Structure probing of this model RNA with RNase A and RNase T1 revealed that spIAS2 and spISAR form a stem (Fig. 3D ▶). The specific conservation of cis-acting elements that regulate IgIII alternative splicing in FGF-R genes indicates that the basic mechanism of exon IIIb inclusion probably evolved more than 600 million years ago.
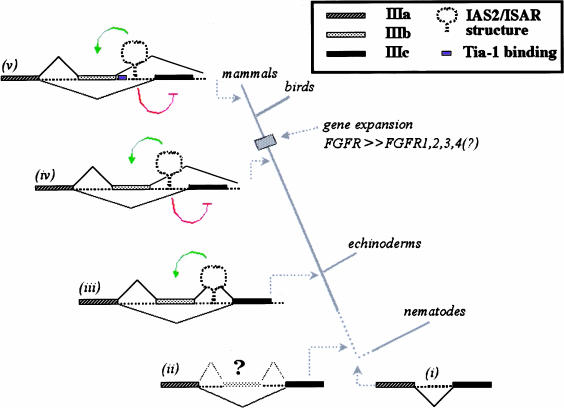
We propose that the common ancestor of present day FGF-R genes had an intron that bisected the IgIII-coding region (in Fig. 4 ▶, i); the location of an intron relative to the FGF-R ORF is conserved in C. elegans and humans (DeVore et al. 1995). An incomplete duplication of the exon encoding the carboxy-terminal half of IgIII led to the formation of a very weak internal exon (IIIb proto-exon), which was rarely, if ever, included in FGF-R transcripts and therefore resulted in low levels of this larger and likely inactive FGF-R (Fig. 4 ▶, ii). We posit that the IAS2–ISAR regulatory elements evolved in a common ancestor of echinoderms and vertebrates providing these organisms with the ability to exert spatiotemporal regulation over the inclusion of this proto-exon IIIb (Fig. 4 ▶, iii). In S. purpuratus, exon spIIIb inclusion appears to inactivate FGF-R and therefore downregulate its expression during development, whereas exon spIIIb skipping leads to functional receptor expression in adult lantern muscle tissue (McCoon et al. 1998). The regulated IIIb proto-exon evolved the potential to encode a Ig carboxy-terminal half, possibly by gene conversion, and the IAS2–ISAR elements developed the capacity to promote mutually exclusive use of exons IIIb or IIIc. The regulated alternative inclusion of IIIb or IIIc led to the tissue-specific expression of different FGF-R2 isoforms in vertebrates (Fig. 4 ▶, iv) (Sato et al. 1992; Yan et al. 1993; Poulin and Chiu 1994; Shi et al. 1994). The FGF-R2 IgIII alternative splicing unit appears to have undergone further evolution after the divergence of mammals and birds. An apomorphy in the mammalian FGF-R2 genes is the aforementioned Tia-1 binding site (IAS1), an activating element that is absent in the chicken FGF-R2 gene and in the other mammalian FGF-R genes (Fig. 4 ▶, v). This continued refinement of the FGF-R IgIII alternative splicing unit indicates the importance of FGF-R isoform diversity. Of the two mechanisms that generate protein variance among the FGF-R genes—gene duplication and alternative splicing—the latter was the original mechanism for generating isoform diversity (MacLean et al. 1997).
FIGURE 4.
The evolution of the FGF-R IgIII alternative splicing unit. Three FGF-R genes from protostomes had been sequenced previously, the Dfr1 (X74030) and Dfr2 (X74031) in the fruit fly Drosophila melanogaster and the egl-15 gene in the nematode C. elegans (U39761). The D. melanogaster FGF-R Ig domains are all encoded within one exon. The IgIII domain is encoded by two exons in egl-15 and the position of the intron relative to the protein domain has been precisely conserved from vertebrates to C. elegans (i) (De Vore et al. 1995). The amino-terminal half of the domain is encoded by exon 8 (ceIIIa), which is homologous to vertebrate exon IIIa. The carboxy-terminal half of the IgIII domain is encoded by exon 9 (ceIIIc), which is more homologous to IIIc exon of vertebrates than to the IIIb exon. Intron 8 is only 46 nucleotides in length and does not have sequences that resemble IAS2 and ISAR. Given the conserved position of the intron in the C. elegans FGF-R gene and in deuterostomes, we suggest that the ancestral FGF-R gene contained an intron splitting the IgIII domain (i). A weak proto-exon IIIb evolved and was sometimes included in FGF-R transcripts (ii). Our data indicate that the common ancestor of echinoderms and vertebrates evolved the regulation of this proto-exon IIIb and developed an FGF-R IgIII alternative splicing unit with IAS2 and ISAR elements, which activate the proto-exon (indicated in green) (iii). We believe that before the evolution of vertebrates, an organism evolved the mutually exclusive use of exon IIIb or exon IIIc (iv). This unit must have evolved the capacity to repress exon IIIc inclusion (indicated in red). The IgIII alternative splicing unit of FGF-R2 has evolved further and among mammals we observed the IAS1 element immediately downstream of exon IIIb (indicated in blue)(v).
MATERIALS AND METHODS
Cloning and sequencing of genomic fragments
Chicken genomic DNA was obtained from Novagen (catalog no. 69233–3) and sea urchin genomic DNA was obtained from soft tissues of one Strongylocentrotus purpuratus organism (kindly provided by the Marine Resources Division of the Marine Biological Laboratory, Woods Hole, MA) and purified using the Qiagen DNeasy Tissue kit (catalog no. 69504) according to the manufacturer’s protocol. Genomic DNA samples were treated with the Ambion RNase cocktail (catalog no. 2286) according to the manufacturer’s protocol. Taq PCR reactions (200 μM dNTPs, 1 μg template, 200 nM forward primer, 200 nM reverse primer, 100 units/mL Taq DNA polymerase) were used to amplify genomic DNA fragments containing introns and exons of interest. The nomenclature above (e.g., intron 8) assumes that overall structure of the chicken and sea urchin gene is similar to that of mammalian FGF-R genes. PCR products were gel-purified using the Qiagen Qiaquick Gel Extraction kit (catalog no. 28704) and subsequently cloned into the pGEM-T Easy vector using the Promega T-Easy Vector System I (catalog no. A1360). PGEM-T Easy clones containing PCR product inserts were initially sequenced with pUC M13 Forward and Reverse primers and subsequently sequenced with product-specific primers. Sequences were assembled using Sequencher v3.1.1 (Gene Codes Corp.) and aligned using MacVector 6.5.3 (Oxford Molecular Group). Oligonucleotide sequences will be provided on request. Sequence for these regions can be found as follows: GenBank accession no. NT_027097 for hsFGF-R2, GenBank accession no. MGI:95523 for mmFGF-R2, GenBank accession no. AF456422 for rnFGF-R2, and GenBank accession no. AF456421 and no. AF456423 for ggFGF-R2 sequences. The sequence of the spFGF-R2 IgIII alternative splicing unit was deposited in GenBank (accession no. AF456424).
Alignment of nucleic acid sequences
Alignment parameters for intron 7 were as follows: Slow pairwise alignment with an open gap penalty of 50.0 and an extended gap penalty of 10.0 (Higgins et al. 1996). Multiple alignment with an open gap penalty of 5.0, extend gap penalty of 0.5, delay divergence of 30%, and weighted transitions. Alignment of intron 8 employed a gap penalty of 3.7 and an extended gap penalty of 0.5.
Analysis of ggFGF-R2 isoforms
One female Leghorn chicken was sacrificed and the indicated organs (Fig. 2B ▶) were collected and immediately chilled in 8 mL of RNA Later (Ambion catalog no. 7020). Total RNA was extracted using the Qiagen RNeasy mini kit (catalog no. 74104) or Trizol reagent (Invitrogen, catalog no. 15596–026). The ggFGF-R2 IIIb versus IIIc isoforms were detected by RT-PCR. The PCR primers used and the diagnostic restriction digests are schematically shown in Figure 2A ▶. Three chicken cell lines were also used to analyze ggFGF-R2 isoforms. Cell line LMH (ATCC catalog no. CRL-2117) is derived from a hepatocellular carcinoma and has epithelial morphology. Cell line UMNSAH/DF-1 (UM) (ATCC catalog no. CRL-12203) is derived from a primary embryo and has fibroblast morphology. Cell line SL-29 (ATCC catalog no. CRL-1590) is derived from a whole embryo and has fibroblast morphology. The RNA extraction and RT-PCR analysis for the cell lines were as described above for organs.
Functional analysis of cis-acting elements
UM and LMH cells were transfected with 2 μg of rat FGF-R2 minigene constructs (derived from minigene constructs described in Carstens et al. 1998) in FuGene reagent. Stable transfectants were selected with G418 and the fraction of RNAs including exon IIIb was determined and quantified as described before (Carstens et al. 1998). Minigene reporters were transfected into DT3 cells, stable transfectants selected with G418, and the fraction of RNAs including exon IIIb was determined and quantified as described previously (Carstens et al. 1998).
RNA structure probing
An RNA that included spIAS2 and spISAR sequences (bold) separated by a six nucleotide loop (underlined) (5′-GGGAGAAGAGA AUUUUCGAAAAAUAGAUGUAAUCUUAAUAUUGCCUAUC UUCAUUUGUUUAUGCUAGAGUCGACCUGCAGGCAUGCA UA-3′) was synthesized using T7 RNA polymerase as described previously (Colvin and Garcia-Blanco 1992). Structure probing by limiting digestion with RNase A and RNase T1 was also essentially as described by Colvin and Garcia-Blanco (1992) except that primer extension was carried out with Superscript RT.
Acknowledgments
The authors thank M. Abou Donia for providing chicken tissues; Cliff Cunningham for interesting discussions on phylogeny; and Andrew Baraniak for comments on the manuscript. The authors thank Annette Kennett for her help in the preparation of the manuscript and members of the Garcia-Blanco laboratory for encouragement and advice. This work was funded by grants to M.A.G-B. from the National Institutes of Health and the American Heart Association, and by the generous support of the Josiah Macy, Jr. Research Foundation for work at the Marine Biological Laboratory. E.J.W. acknowledges the support of a Department of Defense predoctoral fellowship. M.A.G-B. was a scholar of the Raymond and Beverly Sackler Foundation.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.rnajournal.org/cgi/doi/10.1261/rna.2470903.
REFERENCES
- Bloom, W. and Fawcett, D.W. 1975. A Textbook of histology. W.B. Saunders Co., Philadelphia, PA.
- Carstens, R.P., McKeehan, W.L., and Garcia-Blanco, M.A. 1998. An intronic sequence element mediates both activation and repression of rat fibroblast growth factor receptor 2 pre-mRNA splicing. Mol. Cell. Biol. 18: 2205–2217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carstens, R.P., Wagner, E.J., and Garcia-Blanco, M.A. 2000. An intronic splicing silencer causes skipping of the IIIb exon of fibroblast growth factor receptor 2 through involvement of polypyrimidine tract binding protein. Mol. Cell. Biol. 20: 7388–7400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colvin, R.A. and Garcia-Blanco, M.A. 1992. Unusual structure of the human immunodeficiency virus type 1 trans-activation response element. J. Virol. 66: 930–935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Gatto, F. and Breathnach, R. 1995. Exon and intron sequences, respectively, repress and activate splicing of a fibroblast growth factor receptor 2 alternative exon. Mol. Cell. Biol. 15: 4825–4834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Gatto, F., Plet, A., Gesnel, M.C., Fort, C., and Breathnach, R. 1997. Multiple interdependent sequence elements control splicing of a fibroblast growth factor receptor 2 alternative exon. Mol. Cell. Biol. 17: 5106–5116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Gatto-Konczak, F., Bourgeois, C.F., Le Guiner, C., Kister, L., Gesnel, M.C., Stevenin, J., and Breathnach, R. 2000. The RNA-binding protein TIA-1 is a novel mammalian splicing regulator acting through intron sequences adjacent to a 5′ splice site. Mol. Cell. Biol. 20: 6287–6299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeVore, D.L., Horvitz, H.R., and Stern, M.J. 1995. An FGF receptor signaling pathway is required for the normal cell migrations of the sex myoblasts in C. elegans hermaphrodites. Cell 83: 611–620. [DOI] [PubMed] [Google Scholar]
- Hedges, S.B. and Poling, L.L. 1999. A molecular phylogeny of reptiles. Science 283: 998–1001. [DOI] [PubMed] [Google Scholar]
- Higgins, D.G., Thompson, J.D., and Gibson, T.J. 1996. Using CLUSTAL for multiple sequence alignments. Methods Enzymol. 266: 383–402. [DOI] [PubMed] [Google Scholar]
- Jones, R.B., Carstens, R.P., Luo, Y., and McKeehan, W.L. 2001. 5′- and 3′-terminal nucleotides in the FGFR2 ISAR splicing element core have overlapping roles in exon IIIb activation and exon IIIc repression. Nucleic Acids Res. 29: 3557–3565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander, E.S., Linton, L.M., Birren, B., Nusbaum, C., Zody, M.C., Baldwin, J., Devon, K., Dewar, K., Doyle, M., FitzHugh, W., et al. 2001. Initial sequencing and analysis of the human genome. Nature 409: 860–921. [DOI] [PubMed] [Google Scholar]
- MacLean, D.W., Meedel, T.H., and Hastings, K.E. 1997. Tissue-specific alternative splicing of ascidian troponin I isoforms. Redesign of a protein isoform-generating mechanism during chordate evolution. J. Biol. Chem. 272: 32115–32120. [DOI] [PubMed] [Google Scholar]
- McCoon, P.E., Angerer, R.C., and Angerer, L.M. 1996. SpFGFR, a new member of the fibroblast growth factor receptor family, is developmentally regulated during early sea urchin development. J. Biol. Chem. 271: 20119–20125. [DOI] [PubMed] [Google Scholar]
- McCoon, P.E., Blackstone, E., Angerer, R.C., and Angerer, L.M. 1998. Sea urchin FGFR muscle-specific expression: Posttranscriptional regulation in embryos and adults. Dev. Biol. 200: 171–181. [DOI] [PubMed] [Google Scholar]
- McKeehan, W.L., Wang, F., and Kan, M. 1998. The heparan sulfate-fibroblast growth factor family: Diversity of structure and function. Prog. Nucleic Acid Res. Mol. Biol. 59: 135–176. [DOI] [PubMed] [Google Scholar]
- Modrek, B. and Lee, C. 2002. A genomic view of alternative splicing. Nat. Genet. 30: 13–19. [DOI] [PubMed] [Google Scholar]
- Poulin, M.L. and Chiu, I.M. 1994. Nucleotide sequences of two newt (Notophthalmus viridescens) fibroblast growth factor receptor-2 variants. Biochim. Biophys. Acta 1220: 209–211. [DOI] [PubMed] [Google Scholar]
- Sato, M., Kitazawa, T., Katsumata, A., Mukamoto, M., Okada, T., and Takeya, T. 1992. Tissue-specific expression of two isoforms of chicken fibroblast growth factor receptor, bek and Cek3. Cell Growth Differ. 3: 355–361. [PubMed] [Google Scholar]
- Shi, D.L., Launay, C., Fromentoux, V., Feige, J.J., and Boucaut, J.C. 1994. Expression of fibroblast growth factor receptor-2 splice variants is developmentally and tissue-specifically regulated in the amphibian embryo. Dev. Biol. 164: 173–182. [DOI] [PubMed] [Google Scholar]
- Venter, J.C., Adams, M.D., Myers, E.W., Li, P.W., Mural, R.J., Sutton, G.G., Smith, H.O., Yandell, M., Evans, C.A., Holt, et al. 2001. The sequence of the human genome. Science 291: 1304–1351. [DOI] [PubMed] [Google Scholar]
- Wagner, E.J. and Garcia-Blanco, M.A. 2001. Polypyrimidine tract binding protein antagonizes exon definition. Mol. Cell. Biol. 21: 3281–3288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ———. 2002. RNAi mediated PTB depletion leads to enhanced exon definition. Mol. Cell 10: 943–949. [DOI] [PubMed] [Google Scholar]
- Yan, G., Fukabori, Y., McBride, G., Nikolaropolous, S., and McKeehan, W.L. 1993. Exon switching and activation of stromal and embryonic fibroblast growth factor (FGF)-FGF receptor genes in prostate epithelial cells accompany stromal independence and malignancy. Mol. Cell. Biol. 13: 4513–4522. [DOI] [PMC free article] [PubMed] [Google Scholar]