Abstract
Protein–RNA recognition between aminoacyl-tRNA synthetases and tRNA is highly specific and essential for cell viability. We investigated the structure–function relationships involved in the interaction of the Escherichia coli tRNAAsp acceptor stem with aspartyl-tRNA synthetase. The goal was to isolate functionally active mutants and interpret them in terms of the crystal structure of the synthetase-tRNAAsp complex. Mutants were derived from Saccharomyces cerevisiae tRNAAsp, which is inactive with E. coli aspartyl-tRNA synthetase, allowing a genetic selection of active tRNAs in a tRNAAsp knockout strain of E. coli. The mutants were obtained by directed mutagenesis or library selections that targeted the acceptor stem of the yeast tRNAAsp gene. The mutants provide a rich source of tRNAAsp sequences, which show that the sequence of the acceptor stem can be extensively altered while allowing the tRNA to retain substantial aminoacylation and cell-growth functions. The predominance of tRNA backbone-mediated interactions observed between the synthetase and the acceptor stem of the tRNA in the crystal and the mutability of the acceptor stem suggest that many of the corresponding wild-type bases are replaceable by alternative sequences, so long as they preserve the initial backbone structure of the tRNA. Backbone interactions emerge as an important functional component of the tRNA-synthetase interaction.
Keywords: Protein–RNA recognition, knockout cell, RNA identity, GU, genetic selection, library construction
INTRODUCTION
Protein–RNA recognition between aminoacyl-tRNA synthetases (aaRSs) and tRNA is highly specific and essential for cell viability, due to the requirement for accurate translation of the genetic code into protein sequences. Moreover, tRNA molecules must interact with many other macromolecules such as precursor tRNA processing and modification enzymes and the protein translation machinery. Because of these characteristics, tRNA recognition is a paradigm for the study of the mechanisms of protein–RNA interactions. A rich repertoire of crystal structures is available of complexes between tRNAs and their aaRSs (Arnez and Moras 1997; Cusack et al. 1998; Steitz 1990). Although we know how many proteins and their respective RNAs fit together structurally, it is important to emphasize how little we understand regarding the molecular basis of recognition specificity. For example, are nucleotides in tRNA recognized individually and independently of one another or are they recognized as groups of nucleotides that function as a unit in the interaction with an aaRS? Also, what is the functional relevance of the many tRNA backbone-mediated interactions with aaRSs observed in crystal structures? Do backbone-mediated interactions with tRNA cause local shifts in the geometry of the pendent bases that also frequently interact with the aaRS? These are some of the questions that will be addressed in future research on protein–RNA interactions.
Here we report the results of our investigation of the structure–function relationship of the acceptor stem of Escherichia coli tRNAAsp in its interaction with AspRS. Our goal is to identify mutants of tRNAAsp that are functionally active and interpret them in terms of the crystal structure of the AspRS-tRNAAsp complex. The mutants are derived from Saccharomyces cerevisiae (yeast) tRNAAsp, which is inactive with E. coli AspRS, so that a genetic selection in a tRNAAsp knockout strain of E. coli can be used to isolate strains producing active tRNAs. The 2.4 Å crystal structure of the E. coli tRNAAsp-AspRS complex shows that the enzyme interacts with the anticodon, the D-stem, and the acceptor stem of the tRNA (Eiler et al. 1999). A series of backbone interactions occurs with the 3′ members of the six contiguous nucleotide pairs in the acceptor stem, the focal point of the present research, which provides a high density of structural information (Fig. 1 ▶).
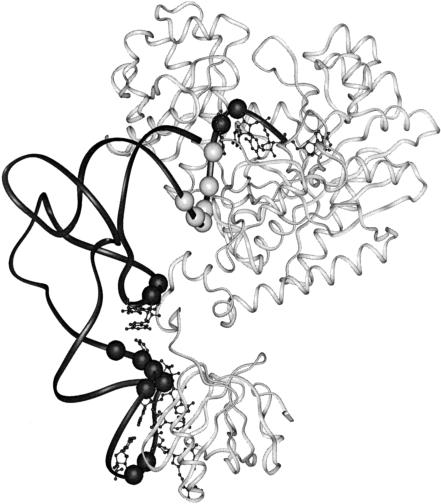
FIGURE 1.
Ribbon representation of one monomer of the dimeric E. coli AspRS-tRNAAsp complex showing the tRNA in black, with spheres representing phosphate sugar backbone residues that interact with AspRS; bases that interact with the protein are also shown. Note the contiguous series of six interacting phosphate sugar residues (light gray) in the tRNA acceptor stem (residues 67–71, upper right). The AspRS protein is shown in off-white (Eiler et al. 1999).
RESULTS AND DISCUSSION
Overview
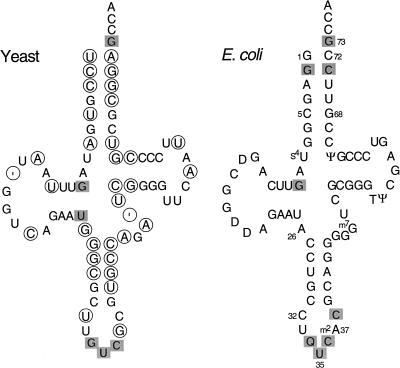
Yeast tRNAAsp is a poor substrate for aminoacylation by E. coli AspRS, and the tRNA does not support growth of E. coli tRNAAsp knockout cells (Moulinier et al. 2001). A position-by-position comparison of yeast and E. coli tRNAAsp molecules reveals variation throughout their sequences, including 10 differences in the amino acid acceptor stem (Fig. 2 ▶). We previously observed that redesigning the end of the acceptor stem of yeast tRNAAsp with the corresponding E. coli bases confers activity on the modified tRNA, as determined by enzyme kinetics and in vivo growth and aminoacylation measurements (Moulinier et al. 2001). However, the tRNA structure–aminoacylation function relationship that emerged was based on just a few mutants, and only the end the acceptor stem was probed. Clearly, a more comprehensive view of this structure–function relationship would emerge from analysis of additional tRNA acceptor-stem mutants, and we therefore constructed and analyzed more. The new mutants were obtained in two ways: directed mutagenesis and randomized library construction. Mutants obtained both ways were functionally tested in tRNAAsp knockout cells. In both approaches, mutant genes were constructed or selected from randomized oligonucleotides so that the transcribed tRNA contained nucleotide changes in the appropriate positions of the acceptor stem. In addition, all yeast tRNAAsp genes were designed so that the tRNA contains the E. coli C32 and A37 nucleotides in the anticodon loop, to optimize the tRNA’s translation properties in E. coli. Finally, the tRNA genes were flanked by restriction sites complementary to sites in the expression plasmids; the latter sites were bracketed by promoter and terminator sequences to drive synthesis of the inserted tRNA gene.
FIGURE 2.
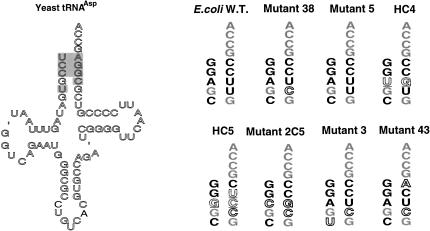
Cloverleaf structure of tRNAAsp from yeast and E. coli. The circled bases in yeast tRNAAsp differ from the corresponding base in E. coli tRNAAsp. The shaded bases are important for in vitro aspartylation of tRNAAsp (Pütz et al. 1991; Nameki et al. 1992). The ‘ symbol is an alignment gap. Standard position numbers are indicated. Q is a modified derivative of G. Other modified bases in E. coli tRNAAsp are indicated.
The tRNAAsp knockout cells are maintained by wild-type E. coli tRNAAsp derived from plasmid pSAD with a regulatable araBAD promoter, which is activated by arabinose and repressed by glucose. The synthetic tRNA genes of mutants were ligated into plasmid pGFIB that constitutively produces tRNA to a steady-state level threefold in excess of total chromosomal tRNAAsp in wild-type E. coli (McClain and Gabriel 2001). Two systems were used to study the functional capacity of the mutant tRNA in knockout cells. First, in the two-plasmid system, wild-type E. coli tRNAAsp produced from pSAD was repressed by glucose so that cell viability depends on the function of the mutant tRNA expressed constitutively from the second plasmid. This two-plasmid system allows an initial functional characterization of the mutant tRNA. Second, the one-plasmid system involves a plasmid switch in which plasmid pGFIB producing a mutant tRNA is switched for the resident plasmid pSAD producing wild-type tRNAAsp. A switch is possible only when the function of the mutant tRNA can support the growth of knockout cells. Mutant tRNAs that support knockout cell growth must be predominantly charged with aspartic acid; otherwise, cells would not grow, because this amino acid is encoded at many sites in cellular proteins. Once the mutants exist in the one-plasmid state, they can be functionally assessed by quantitative growth rate measurements and steady-state levels of aminoacyl-tRNA.
Yeast tRNAAsp acceptor-stem mutants
The acceptor stem of tRNA contains seven base pairs (bp; the standard position numbers range from 1–72 to 7–66) and the adjacent single-stranded position 73 (Fig. 3 ▶, left). Six of the base pairs, but not position 73, differ between E. coli and yeast tRNAAsp molecules. Previous work focused primarily on the first two base pairs, 1–72 and 2–71. Here we performed a systematic mutational analysis of the acceptor stem of yeast tRNAAsp. To this end, we generated tRNAAsp mutants by directed mutagenesis and by selection from randomized libraries to gain a better understanding of how the acceptor stem contributes to function.
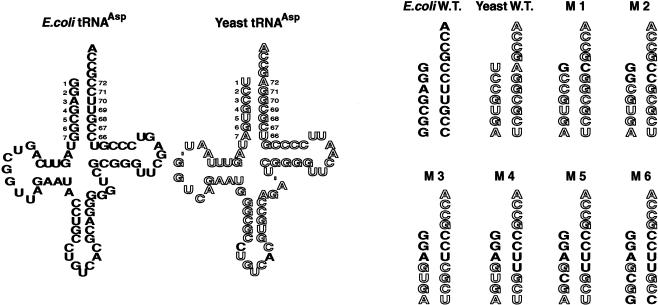
FIGURE 3.
Design of the six acceptor-stem mutants. The mutants are abbreviated as mutant 1, M1, etc. E. coli-specific bases are in solid font, and yeast-specific bases are in outline. Note that all yeast tRNAAsp genes were designed so that the tRNA contains the E. coli C32 and A37 nucleotides (see text). Modified bases in the cloverleaf structures are not indicated.
First, we rationally constructed a series of yeast tRNAAsp mutants in which the acceptor stem was incrementally modified to contain more and more of the corresponding E. coli tRNAAsp nucleotide pairs. We began by substituting the first base pair (giving mutant 1), then the first and second base pairs (mutant 2), and continued in this way until all of the base pairs up to and including the seventh base pair were included in a multiple mutant. This procedure gave six mutants of yeast tRNAAsp (Fig. 3 ▶). These six mutants were initially characterized in the two-plasmid state by streaking cultures across an arabinose agar plate containing a glucose-saturated paper strip in the middle of the plate (Gabriel and McClain 2001). All cultures grow in the arabinose sector of the plate, where the araBAD promoter produces wild-type tRNAAsp from the maintenance plasmid. However, only active mutants grow near the glucose strip, where the araBAD promoter is repressed, so that cells contain little wild-type tRNAAsp and are growth-dependent on the mutant tRNA produced from plasmid pGFIB. This test revealed that only mutants 3, 5, and 6 grew in the presence of glucose (data not shown). A subsequent Northern analysis using a DNA probe that specifically hybridized to the anticodon of yeast tRNAAsp in vast preference to that of E. coli tRNAAsp revealed that all rationally constructed mutants made tRNAAsp-sized molecules, except mutant 4, which showed little if any mature tRNAAsp (data not shown). Thus, the functional properties of mutant 4 tRNAAsp are indeterminate. We surmise that mutant 4 tRNA and/or its precursor is degraded by cellular RNases because the molecule contains a specific tandem wobble pair, G4-U69 and U5-G68.
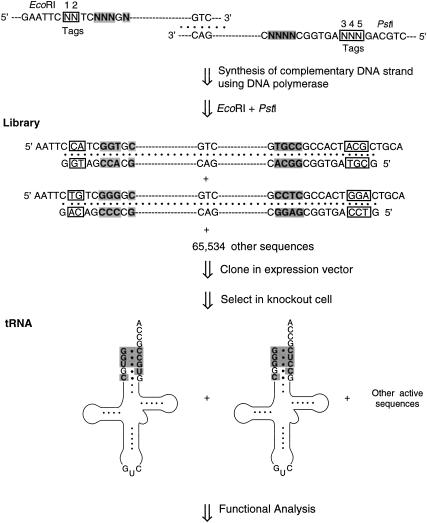
In order to examine a larger spectrum of sequence combinations, we also randomized the first five base pairs in the acceptor stem of the gene for yeast tRNAAsp and selected active clones in tRNAAsp knockout cells (Fig. 4 ▶). By design (Choi et al. 2002a), two oligonucleotides containing the sequence of yeast tRNAAsp overlap just in the anticodon stem/loop and carry degenerate nucleotide mixes in the single-stranded five-base-pair-target sites in the acceptor stem where yeast and E. coli tRNAAsp sequences differ. Only eight nucleotides were randomized at target sites, because two positions in the respective tRNAs contain identical nucleotides. The oligonucleotides were annealed at the anticodon region, and the 3′-OHs were extended by Taq DNA polymerase into duplex DNA, which generated a randomized library of individual mutant genes. The library was cloned into plasmid pGFIB and electroporated into knockout cells. Cells producing active mutant tRNAAsp were selected by growth on plates containing glucose, because expression of the wild-type tRNAAsp gene on maintenance plasmid pSAD is repressed under these conditions.
FIGURE 4.
Method used to construct the randomized library of yeast tRNAAsp mutants. Two synthetic oligonucleotides were annealed, one round of complementary strands was synthesized by Taq DNA polymerase, and the product was cleaved by restriction enzymes. The shaded bases are the targets of mutagenesis, and the boxed residues at positions 1–5 are the sequence tags.
Two-way mix libraries were designed to carry degenerate nucleotide mixes (50% yeast and 50% E. coli bases) in the target sites in the acceptor stem. Thus, the theoretical complexity of the library is 28 or 256 distinct sequences. In the first two-way mix library, 30 active mutants were selected from 15,000 clones plated. Fifteen of these were mutant 5, 14 were mutant 38, and one was mutant 3. Mutant 38 represents a new sequence which contains the changes in mutant 3 plus a U5 to C5 mutation changing the U5-G68 wobble pair of yeast tRNAAsp to a C5-G68 base pair of E. coli; the 4–69 base pair remained that of yeast (Fig. 5 ▶). Because mutant 3 was underrepresented, in a second library we incorporated sequence tags (Fig. 4 ▶) in the initial oligonucleotides in order to assess the chemical randomness of the library. The sequence tags are designed to contain a random mix of four nucleotides sandwiched between the tRNA gene and the cloning restriction sites. Our rationale was that if the sequences of the tags are random, then the mixed positions within the tRNA gene are also random. We analyzed the sequences of the tRNA genes and sequence tags of 80 active clones. The active clones included 46 isolates of mutant 5 and 34 isolates of mutant 38. Statistical analysis of the tags indicated that the nucleotides were random at tag positions 1, 3, and 4 (P-values ≥0.05), whereas those at tag positions 2 and 5 were biased (P-values <0.05). The P-value represents a χ-square analysis that the observed distribution is random at the 95% confidence interval. The lack of randomness at positions 2 and 5 may reflect bias against transcripts with particular bases that impair precursor RNA processing. Although the results at the five tag positions are not uniform, tag positions 1, 3, and 4 are random, and therefore the underrepresentation of mutant 3 may not be due to library bias; rather, it may be due to its poor growth properties (see below).
FIGURE 5.
The acceptor-stem sequences of active mutants selected from libraries. The shaded bases in the cloverleaf of yeast tRNAAsp were randomized. E. coli-specific bases are in solid black font, yeast-specific bases are in outline, new bases are underlined in dotted outline, and common bases are gray. All yeast tRNAs were designed to contain the E. coli C32 and A37 nucleotides (see text). Modified bases in the cloverleaf structure are not indicated.
Mutant 2C5 (Fig. 5 ▶) that had been constructed in earlier work and was shown to be active (Moulinier et al. 2001) was not isolated from this library, suggesting that the selection method may be excluding active mutants. The selection method was therefore modified by adding small amounts of arabinose (from 0.001% w/v to 0.02%) to the selection plates to induce low-level expression of wild-type tRNAAsp. The arabinose was subsequently removed along with the maintenance plasmid during the conversion of the mutants to the one-plasmid system. These modified conditions gave 74 active clones, of which 61 were sequenced. We obtained two isolates of mutant 2C5 and four isolates of a new mutant, mutant 43 (Fig. 5 ▶). (The selection containing arabinose also gave repeats of other mutants found in previous selections, i.e., 31 isolates of mutant 5, 20 isolates of mutant 38, and four isolates of mutant 3.) Mutant 43 is similar to mutant 38 except that the G residue at position 1 is derived from E. coli and that A72 is from yeast tRNAAsp, which creates a G1-A72 wobble pair in the active mutant tRNAAsp.
Finally, to increase the sequence complexity in the five base pairs of yeast tRNAAsp under study, we employed four-way (i.e., 25% of each base) rather than two-way mixes in the library construction of tRNAAsp genes. This gave a theoretical tRNA gene complexity of 48 or 65,536 distinct sequences. Twelve thousand clones were plated, and five active clones were obtained. Because active clones were isolated at a frequency of 1/2,400 clones plated, most sequence combinations were inactive. The active clones included two isolates of mutant 5, one of mutant 38, and one isolate each of two new mutants, HC4 and HC5 (Fig. 5 ▶). These new mutants differ substantially from the others and contain different sequence combinations, a tandem wobble pair (HC4) or a wobble pair at a new position in the acceptor helix (HC5).
Aminoacylation and growth properties of tRNA mutants
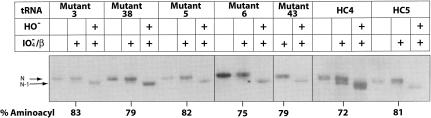
All active clones were converted to the one-plasmid system by switching the mutant tRNA on plasmid pGFIB for the maintenance plasmid. The mutant tRNAs could then be functionally characterized for aminoacylation and growth properties. The steady-state level of aspartyl-tRNAAsp was determined using tRNA preparations that had been isolated under acidic conditions that preserve the aminoacyl bond. The preparations were subjected to periodate oxidation and β-elimination, which cleaves one residue from the 3′ end of uncharged tRNA (Chang et al. 1999). Because the latter tRNA lacks one residue, it can be separated from full-length tRNA, which is derived from aminoacyl-tRNA, on a denaturing polyacrylamide gel. The steady-state level of aspartyl-tRNAAsp for all mutants approached that of wild-type E. coli tRNAAsp (Fig. 6 ▶). The tandem wobble pair in mutant 4 described above resulted in little mature tRNA, in contrast to the tRNA production by mutant HC4; the two mutants are associated with different locations and orientations of the tandem wobble pair.
FIGURE 6.
Steady-state aminoacylation levels of tRNAAsp mutants in the tRNAAsp knockout strain. The aminoacylation level of wild-type E. coli tRNAAsp ranged from 83% to 89% in four independent experiments; that of mutant 2C5 was 71% (Moulinier et al. 2001; data not shown).
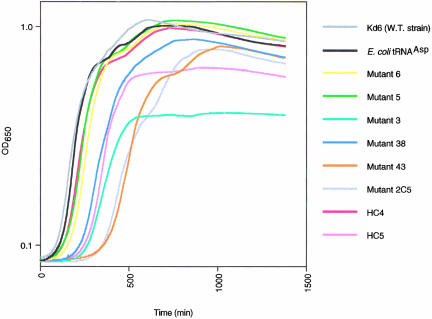
Growth curves of knockout cells containing the mutant tRNAs were determined, which showed that mutants 6, 5, and HC4 were comparable to wild-type E. coli tRNAAsp (Fig. 7 ▶). The growth curves for mutants 38 and HC5 were also similar to wild-type, but the lag phase before entering exponential growth was longer, and the cultures entered stationary phase at a somewhat lower cell density. Mutants 43 and 2C5 had a much longer lag phase, and mutant 3 entered stationary phase at a low cell density (Fig. 7 ▶). The reason for the low cell density at stationary phase for mutant 3 despite its apparent high level of Asp-tRNAAsp is unknown, but may reflect a deficiency in the molecule’s performance in translation subsequent to aminoacylation or in some other physiological process. Based on their growth properties, we anticipate that mutants 3, 43, and 2C5 would be at a growth disadvantage relative to mutants 5 and 38, and this may be why they appear less frequently than the other two mutants in selections for active tRNAAsp clones from randomized libraries. We have not probed the differences in these mutants further.
FIGURE 7.
Growth curves of the tRNAAsp knockout strain producing different tRNAAsp molecules. All mutants except mutant 43 were measured in a single experiment.
Concluding remarks
The E. coli AspRS-tRNAAsp complex is a homodimer with each protein subunit binding one tRNA molecule (Eiler et al. 1999). In the heterologous complex between E. coli AspRS and yeast tRNAAsp, AspRS monomer 1 binds the tRNA in a similar fashion (A76 is out of place) as in the homologous E. coli complex, but the interactions between monomer 2 and the acceptor stem are completely missing (Moulinier et al. 2001). The present study was intended to identify structural features in the acceptor stem of the tRNA that promote functional interaction between tRNAAsp and AspRS.
The present group of mutants obtained by a combination of directed mutagenesis and library selection provides a rich source of tRNAAsp sequences that are functionally capable of supporting growth of tRNAAsp knockout cells and of sustaining high steady-state levels of aspartyl-tRNA. The mutants and the crystal structure of complexes between tRNAAsp and AspRS offer the possibility of an analysis of the molecular basis of the specificity of the interaction between the two macromolecules. The 2.4 Å crystal structure of the homologous complex between E. coli tRNAAsp and AspRS (Eiler et al. 1999) shows that the enzyme makes direct contacts to the bases in the anticodon (G34, U35, C36, and C38), the D stem (G10 and U11), and the single-stranded acceptor end (G73, C74, and C75; Fig. 1 ▶). In contrast, the AspRS enzyme makes phosphate-sugar backbone interactions with six of the 3′ nucleotides of the double-stranded acceptor stem (Fig. 1 ▶). The acceptor stem is not involved in any direct base contacts with the enzyme. We can use the structure of the homologous E. coli complex to interpret our mutant data because most of the same acceptor-stem interactions are observed in monomer 1 of the heterologous complex between wild-type yeast tRNAAsp and E. coli AspRS. The two monomers are in functional communication with respect to the bound tRNA substrate such that aminoacylation occurs only when both tRNAs are properly and fully bound (Moulinier et al. 2001). The heterologous complex is an inactive state because the terminal A76 accepting nucleotide of yeast tRNAAsp is not in the enzyme active site in monomer 1 and none of the acceptor stem of yeast tRNAAsp in monomer 2 is bound, highlighting the importance of acceptor-stem interactions for aminoacylation. These structural aspects of the heterologous complex confirm that yeast tRNAAsp is not a substrate for E. coli AspRS.
The mutants of tRNAAsp described here show clearly that the sequence of the acceptor stem can be extensively altered while allowing the associated tRNAAsp molecules to retain substantial aminoacylation and cell-growth functions. Only particular nucleotide combinations are active, and in fact, most combinations are inactive. The predominance of tRNA backbone-mediated interactions observed between AspRS and the acceptor stem in the E. coli homologous complex suggests that many of the corresponding wild-type bases should be replaceable by alternative sequences so long as they preserve the initial backbone structure of the tRNA. A corollary of structure-based recognition is that the smallest unit of function is a group of nucleotides rather than a single nucleotide or base pair. An aminoacylation specificity that depends on molecular structure rather than base sequence can explain why tRNAAsp with divergent acceptor-stem sequences (Fig. 5 ▶) can be functionally active.
It would be incorrect to link the functional activity of any mutant tRNA to an individual base pair or wobble pair that it contains, because the sequence context in which that element exists undoubtedly contributes to the molecule’s functional activity. Clearly, the amino acid acceptor stem as a whole is being recognized. The twist angles and overall pitch determine acceptor-stem backbone geometry, which is critical for the A76 accepting residue to fit into the enzyme active site. In the heterologous tRNA-AspRS complex, the conformation of the acceptor stem is such that the backbone is not efficiently bound by the protein. The most extreme example of this occurs in monomer 2, where none of the acceptor stem is bound by the protein. We infer that in active mutants of yeast tRNA, the acceptor-stem backbone geometry approximates that of the E. coli tRNA, which permits more efficient interaction with the protein. Thus, one can understand how a different combination of bases in wild-type and mutant tRNAs can lead to a similar overall acceptor stem geometry and molecular function. Structure-based and/or backbone recognition of tRNAs have been observed previously (McClain et al. 1998; Moulinier et al. 2001; Choi et al. 2002b). It will be interesting to learn the extent to which RNA-protein recognition systems in general use the type of structure-base recognition described here.
MATERIALS AND METHODS
Strains, plasmids, and microbiological procedures
The E. coli strains UVT (tRNAAsp knockout; McClain and Gabriel 2001) and plasmids pGFIB (Masson and Miller 1986) and pSAD (Gabriel and McClain 2001) have been described. Cells were grown on LB, SOC broth (Heery et al. 1989) or MinA minimal media. MinA minimal medium was supplemented with MgSO4 (1 mM), B1 (4 μg/mL), arginine (40 μg/mL), tryptophan (40 μg/mL), glycerol (0.2%, v/v), casamino acids (0.2%), and either glucose (0.2%) or arabinose (0.2%). Cultures were incubated at 37°C. Plasmids were selected with ampicillin and/or chloramphenicol at concentrations of 100 μg/mL and 20 μg/mL, respectively.
Library construction
The oligonucleotides DYE5 and DYE3 were ordered from the University of Wisconsin Biotechnology Center (Madison, WI) and oligonucleotides IDT1–5, IDT1–3, IDT2–5, and IDT2–3 were ordered from Integrated DNA Technology (Coralville, IA). The nucleotide sequences were as follows:
DYE5 5′-CCGCAAGCTTGAATTCKSMGYGRTAGTTTAATGGT CAGAATGGGCG CCTGTACGTGCCAG-3′ 61 MER;
DYE3 5′-GGATCCTGCAGTGGCKSMRCGRCGGGGAATTGAA CCCCGATCTGGC ACGTGACAGGCGCCC-3′ 61 MER;
IDT1–5 5′-AAGCTTGAATTCNNTCKSMGYGATAGTTTAATG GTCAGAATGGGC GCCTGTCACGTGCCAG-3′ 61 MER;
IDT1–3 5′-GGATCCTGCAGNNNAGTGGCKSMRCGACGGGG AATTGAACCCCG ATCTGGCACGTGACAGGCGCCC-3′ 66 MER;
IDT2–5 5′-AAGCTTGAATTCNNTCNNNGNGATAGTTTAATG GTCAGAATGGGC GCCTGTCACGTGCCAG-3′ 61 MER;
IDT2–3 5′-GGATCCTGCAGNNNAGTGGCNNNNCGACGGGG AATTGAACCCCG ATCTGGCACGTGACAGGCGCCC-3′ 66 MER.
DYE5 and DYE3 were used for the tRNAAsp acceptor-stem library without sequence tags. The oligonucleotides IDT1–5 and IDT1–3 were used to construct the two-way mix tRNAAsp acceptor-stem library with sequence tags, and IDT2–5 and IDT2–3 were used for the four-way mix tRNAAsp acceptor-stem library with sequence tags. The oligonucleotides were purified by PAGE, and the library construction was performed using Taq DNA polymerase. The degenerate oligonucleotides contained the yeast tRNAAsp sequence, which was followed by the sequence tag and restriction site. The degenerate oligonucleotides were denatured by heating at 94°C for 1 min and annealed by lowering the temperature to 57°C for 30 sec. The complementary strands of the annealed complex were synthesized by Taq DNA polymerase at 72°C for 1 min. The library DNA was digested with the restriction endonucleases EcoRI and PstI and was purified on a 2% agarose gel. The product with appropriate mobility was cloned into pGFIB by ligation with T4 DNA ligase in a 20 μL reaction volume at 14°C for 18 h.
Selection of tRNAAsp mutants
Active mutants were selected in tRNAAsp knockout cells (strain UVT). UVT cells with pSADaspwt were grown overnight at 37°C in MinA with arabinose, chloramphenicol, and casamino acids. The cells were pelleted, washed three times with an equal volume of cold sterile water, and resuspended in 1/20 volume of cold sterile water. The library ligation mixture (1 μL, ∼10 pmole) in plasmid pGFIB was introduced into the competent UVT cells by electroporation at 2.5 kV (GIBCO BRL E. coli Pulser), and the cells were incubated with shaking in 1 mL SOC for 1 h at 37°C. The cells were washed twice with MinA and resuspended in 1 mL MinA. An aliquot (100 μL) was plated on MinA agar containing ampicillin to determine the total number of transformants. The remaining cells were plated in 100-μL aliquots onto LB agar (a glucose-containing medium) containing ampicillin to select for the pGFIB plasmid. Colonies containing active clones appeared after incubation for 12–48 h at 37°C. The pSADaspwt plasmid was cured from individual colonies by serial transfer (2×–7×) in LB liquid medium containing ampicillin. The cultures were then streaked onto LB agar plates containing ampicillin, and after incubation at 37°C for 16 h, the colonies were screened for ability to grow on LB agar containing ampicillin and LB agar containing chloramphenicol. Colonies that grew only on LB agar containing ampicillin had lost the pSADaspwt maintenance plasmid. The absence of wild-type E. coli tRNAAsp was confirmed by Northern blot analysis. Plasmid DNA was obtained from individual colonies lacking the maintenance plasmid, and the sequence of the tRNA gene was determined. The tRNA gene was recloned into fresh plasmid after EcoRI and PstI digestions.
Microtiter growth curves
UVT strains containing wild-type or mutant tRNAAsp in plasmid pGFIB were grown overnight at 37°C in LB medium containing ampicillin. In a sterile microtiter plate, 1 μL of each culture was diluted into 150 μL of LB medium containing ampicillin, and the microtiter plate was incubated in a Molecular Devices SpectraMAX Plus microplate reader for 20 h at 37°C. Optical density readings at 650 nm were taken at 2-min intervals, and the cultures were shaken for 100 sec between OD measurements. The data were collected using the Softmax, Pro 3.1.1 program, and CA-Cricket Graph III 1.5.2 was used to generate the growth curves. The growth curves are the average of two independent measurements for each mutant.
Analysis of aminoacyl-tRNA
A 10-mL culture of 2X YT ampicillin medium was inoculated with newly transformed clones expressing the indicated tRNA gene from plasmid pGFIB and incubated with vigorous shaking at 37°C until the density reached approximately 3 × 108 cells/mL. The following operations were at 4°C. The cells were harvested by centrifugation at 5000 rpm for five min, resuspended in 0.5 mL of 0.3 M sodium acetate (pH 5.2), 1 mM EDTA, transferred to a 2 mL microcentrifuge tube, and mixed with 0.5 mL acid phenol saturated with 0.3 M sodium acetate (pH 5.2). The mixture was vortexed vigorously, allowed to stand for 10 min, and phase-separated in a microcentrifuge for 10 min. The aqueous phase was precipitated with 2.5 volumes (1.25–1.5 mL) of ethanol. After 20–30 min on dry ice, the RNA was collected by centrifugation and washed with 0.5 mL of 70% (v/v) ethanol and 10 mM sodium acetate (pH 5.2). The RNA was stored in 40 μL of 10 mM sodium acetate (pH 5.2) and 1 mM EDTA at −80°C. Half of an RNA aliquot was left untreated and the other half was deacylated by mild alkaline hydrolysis with Tris base (0.5 M Tris, pH 9.0, 30 min, 37°C). Both samples were subjected to periodate oxidation and β elimination (40 mM NaIO4, 90 min, 0°C then 330 mM rhamnose, 30 min, 0°C followed by ethanol precipitation and resuspension in 1M lysine, 60 min, 45°C followed by ethanol precipitation) to remove the terminal nucleoside of deacyl-tRNA. RNA samples (0.05 OD260) were fractionated on an 8% polyacrylamide urea DNA sequencing gel (pH 8.3) at ∼80°C and subjected to a Northern blot analysis. The gel was electroblotted to Nytran Plus membrane (Schleicher and Schuell) in 40 mM Tris-acetate (pH 4.2) and 2 mM EDTA at 20 V for 60 min at 4°C. The membrane was cross-linked to RNA by 254 nm irradiation (Ultra Violet Products no. UVS-54) at 15 cm for 3 min and incubated in hybridization buffer (prehybridized in 6X SSC, 10X DEN, 0.2% (w/v) SDS and 1 mM EDTA, then hybridized in 6X SSC and 5X DEN) at 37°C with the indicated probe. Figures showing Northern blots and gels were obtained by scanning X-ray film images into Photoshop 5.02 and labeled using Illustrator 8.0. Synthetic DNAs to hybridize Northern membranes were obtained from the University of Wisconsin Biotechnology Center. Probes were labeled according to the manufacturer’s directions using phage T4 polynucleotide kinase (New England Biolabs) and 30 μCi of adenosine (5′-γ-32P)-triphosphate (Amersham P32 redivue, 5,000 Ci/mM). The Northern blots of gels were quantified using a Molecular Dynamics Storm 860 instrument and ImageQuaNT 4.1 software. The range of values obtained in individual experiments was <102-fold, which is well within the dynamic range of the instrument.
Acknowledgments
This work was supported by grant GM42123 from the National Institute of General Medical Sciences (NIGMS).
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.rnajournal.org/cgi/doi/10.1261/rna.2139703.
REFERENCES
- Arnez, J.G. and Moras, D. 1997. Structural and functional considerations of the aminoacylation reaction. Trends Biochem. Sci. 22: 211–216. [DOI] [PubMed] [Google Scholar]
- Chang, K.-Y., Varani, G., Bhattacharya, S., Choi, H., and McClain, W.H. 1999. Correlation of deformability at a tRNA recognition site and aminoacylation specificity. Proc. Natl. Acad. Sci. 96: 11764–11769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi, H., Otten, S., and McClain, W.H. 2002a. Isolation of novel tRNAAla mutants by library selection in a tRNAAla knockout strain. Biochimie 84: 705–711. [DOI] [PubMed] [Google Scholar]
- Choi, H., Otten, S., Schneider, J., and McClain, W.H. 2002b. Genetic perturbations of RNA reveal structure-based recognition in protein-RNA interaction. J. Mol. Biol. 324: 573–576. [DOI] [PubMed] [Google Scholar]
- Cusack, S., Yaremchuk, Y., and Tukalo, M. 1998. tRNA recognition by aminoacyl-tRNA synthetases. The many faces of RNA (eds. D.S. Eggleston et al.), pp. 55–65, London: Academic Press, Ltd.
- Eiler, S., Dock-Bregeon, A.-C., Moulinier, L., Thierry, J.-C., and Moras, D. 1999. Synthesis of aspartyl-tRNAAsp in Escherichia coli—A snapshot of the second step. EMBO J. 18: 6532–6541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabriel, K. and McClain, W.H. 2001. Plasmid systems to study RNA function in Escherichia coli. J. Mol. Biol. 310: 543–548. [DOI] [PubMed] [Google Scholar]
- Heery, D.M., Powell, R., Gannon, F., and Dunican, L.K. 1989. Curing a plasmid from E. coli using high-voltage electroporation. Nucleic Acids Res. 17: 10131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masson, J.-M., and Miller, J.H. 1986. Expression of synthetic suppressor tRNA genes under the control of a synthetic promoter. Gene 47: 179–183. [DOI] [PubMed] [Google Scholar]
- McClain, W.H. and Gabriel, K. 2001. Construction of an Escherichia coli knockout strain for functional analysis of tRNAAsp. J. Mol. Biol. 310: 537–542. [DOI] [PubMed] [Google Scholar]
- McClain, W.H., Schneider, J., Bhattacharya, S., and Gabriel, K. 1998. The importance of tRNA backbone-mediated interactions with synthetase for aminoacylation. Proc. Nat. Acad. Sci. 95: 460–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moulinier, L., Eiler, S., Eriani, G., Gangloff, J., Thierry, J.-C., Gabriel, K., McClain, W.H., and Moras, D. 2001. The structure of an AspRS-tRNAAsp complex reveals a tRNA-dependent control mechanism. EMBO J. 20: 5290–5301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nameki, N., Tamura, K., Himeno, H., Asahara, H., Hasegawa, T., and Shimizu, M. 1992. E. coli tRNAAsp recognition mechanism differing from that of the yeast system. Biochem. Biophys. Res. Commun. 189: 856–862. [DOI] [PubMed] [Google Scholar]
- Pütz, J., Puglisi, J.D., Florentz, C., and Giegé, R. 1991. Identity elements for specific aminoacylation of yeast tRNAAsp by cognate aspartyl-tRNA synthetase. Science 252: 1696–1699. [DOI] [PubMed] [Google Scholar]
- Steitz, T.A. 1990. Structural studies of protein-nucleic acid interaction: The sources of sequence-specific binding. Q. Rev. Biophys. 23: 205–280. [DOI] [PubMed] [Google Scholar]