Abstract
In higher eukaryotes, intron-containing and intronless versions of otherwise identical genes can exhibit dramatically different expression profiles. Introns and the act of their removal by the spliceosome can affect gene expression at many different levels, including transcription, polyadenylation, mRNA export, translational efficiency, and the rate of mRNA decay. However, the extent to which each of these steps contributes to the overall effect of any one intron on gene expression has not been rigorously tested. Here we report construction and initial characterization of a luciferase-based reporter system for monitoring the effects of individual introns and their position within the gene on protein expression in mammalian cells. Quantitative analysis of constructs containing human TPI intron 6 at two different positions within the Renilla luciferase open reading frame revealed that this intron acts primarily to enhance mRNA accumulation. Spliced mRNAs also exhibited higher translational yields than did intronless transcripts. However, nucleocytoplasmic mRNA distribution and mRNA stability were largely unaffected. These findings were extended to two other introns in a TCR-β minigene.
Keywords: Gene expression, splicing, transcription, translation, luciferase, TCR-β
INTRODUCTION
Most genes in higher eukaryotes are interrupted by at least one intron. Although introns are often considered superfluous junk that must be removed to create a translatable mRNA, in actuality they are both vital facilitators of molecular evolution and crucial regulators of gene expression. In addition to promoting the creation of new genes by exon duplication and allowing for the synthesis of multiple proteins from a single gene by alternative splicing, many studies have shown that constitutively spliced introns are required for optimal gene expression. For example, early experiments revealed that recombinant SV-40 cDNAs produced little or no mRNA, but this deficiency could be corrected by including an intron of either viral or cellular origin (Gruss et al. 1979; Hamer et al. 1979). Later it was demonstrated in several systems, including mammalian tissue culture cells, transgenic mice, insects, and plants, that optimal expression of many endogenous genes similarly requires the presence of one or more introns (Callis et al. 1987; Buchman and Berg 1988; Chiou et al. 1991; Palmiter et al. 1991; Duncker et al. 1997; Bourdon et al. 2001). Although having an intron is not a univeral requirement for eukaryotic gene expression, as evidenced by the existence of naturally intronless genes, protein expression from such genes can nonetheless be boosted by intron addition (Buchman and Berg 1988; B. Cullen, pers. comm.).
When considered as a whole, the extensive literature describing the effects of introns on gene expression presents a rather complicated mosaic. For example, the magnitude of intron-dependent effects can vary tremendously, from almost nothing to more than a 400-fold increase in mRNA levels (Buchman and Berg 1988; Bourdon et al. 2001). Factors affecting this magnitude are intron identity, exon sequence context and intron position within the gene. In some cases, different introns positioned identically within a single gene elicited entirely opposite effects on protein expression (Bourdon et al. 2001). In others, the same intron placed into different exons yielded opposite results (Buchman and Berg 1988; Bourdon et al. 2001). To date, the intronic characteristics that contribute to these differences remain largely undefined. However, some of this variability is likely explained by introns influencing gene expression at multiple steps, with individual introns contributing differentially to each step.
Individual steps in gene expression reported to be affected by introns include transcription, polyadenylation, mRNA export, translational efficiency, and mRNA decay. Promoter proximal introns can augment pre-mRNA synthesis by enhancing both transcription initiation and RNA polymerase II processivity (Fong and Zhou 2001; Furger et al. 2002; Kwek et al. 2002). Further, synergistic interactions between the splicing and polyadenylation machineries contribute to more efficient 3′ end processing of intron-containing transcripts (Proudfoot et al. 2002). When injected into Xenopus oocyte nuclei, some spliced RNAs are exported to the cytoplasm more rapidly than identical RNAs transcribed from cDNAs (Luo and Reed 1999; Zhou et al. 2000; Le Hir et al. 2001). Also in Xenopus oocytes, spliced mRNAs have been reported to have different translational efficiencies than identical mRNAs not generated by splicing (Braddock et al. 1994; Matsumoto et al. 1998). Finally, an intron positioned downstream from an in-frame termination codon can target the spliced mRNA for rapid degradation by the process of nonsense-mediated mRNA decay (NMD; Maquat and Carmichael 2001; Wilusz et al. 2001; Wilkinson and Shyu 2002). The effects of introns on nuclear export and NMD have been attributed to components of the exon junction complex (EJC; Kim et al. 2001; Le Hir et al. 2001; Lykke-Andersen et al. 2001). The EJC is an assemblage of proteins deposited on mRNAs 20–24 nt upstream of exon–exon junctions (Le Hir et al. 2000). Some of these proteins can accompany the spliced mRNA through the nuclear pore and thereby influence cytoplasmic mRNA metabolism (Le Hir et al. 2001; Dreyfuss et al. 2002).
Although numerous effects of introns on various steps of gene expression have been described, to date, each effect has been characterized independently on different genes and in diverse expression systems. Furthermore, many of the early studies (see above) were published before it was realized just how many steps in the gene expression pathway are potentially affected by introns. Consequently, the extent to which each step contributes to the overall effect of any one intron on gene expression is currently unknown for any single gene. Here we describe construction and initial characterization of a luciferase-based reporter system for monitoring the effects of individual introns on gene expression and how these effects vary with intron position. By allowing independent analysis of intron identity and intron position, this system provides a useful tool for dissecting the molecular mechanisms by which individual introns and the act of splicing influence mammalian gene expression.
In this study, we monitored the effects of the sixth intron from the human triose phosphate isomerase (TPI) gene at two different positions inside the open reading frame (ORF) of Renilla luciferase. Both intron positions significantly enhanced gene expression, with the promoter proximal intron having a much more pronounced effect. For both positions, stimulation of protein expression was largely due to increased mRNA accumulation and translational yield. We did not observe any significant splicing-dependent alteration in mRNA stability or nucleocytoplasmic distribution. Curiously, enhanced mRNA accumulation directed by the promoter proximal intron was highly sensitive to the means of 3′ end formation. To determine whether these observations were generalizable to another system, we also examined the consequences of removing one or both introns from a highly expressed TCR-β minigene. Again, we found that the major intron effects were at the levels of mRNA accumulation and translational yield, not nucleocytoplasmic distribution.
RESULTS
Constructs
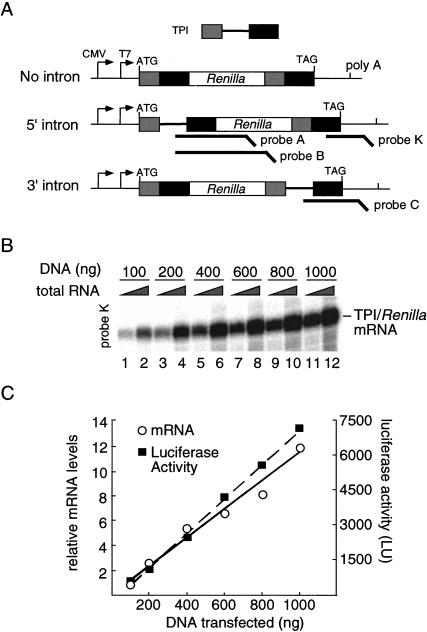
To initiate a systematic analysis of intron effects, we chose the intronless Renilla luciferase cDNA as a reporter system (Fig. 1A ▶). Because optimal excision of an individual intron often requires the context of its naturally flanking exons, we first inserted exons 6 and 7 from human TPI at both ends of the Renilla ORF to generate a fusion protein carrying identical TPI peptide sequences at both the N- and C-termini (no-intron control). To measure the effects of adding a single intron and varying its position within the ORF, derivative constructs were created that contained TPI intron 6 between either the 5′ or 3′ set of TPI exons (5′ intron and 3′ intron). All constructs carried a CMV promoter and the polyadenylation signal from the bovine growth hormone gene for in vivo expression, as well as a T7 promoter for in vitro transcription. To ensure that the observed Renilla luciferase activity accurately reflected in vivo mRNA levels over the range of DNA concentrations we used, we transfected HeLa cells with varying amounts of the 5′ intron construct (which yielded the highest levels of luciferase expression; see below). As expected, both luciferase mRNA and luciferase activity increased linearly over the full range of DNA concentrations tested (Fig. 1B,C).
FIGURE 1.
Schematic representation of TPI/Renilla luciferase reporter constructs. (A) Human triose phosphate isomerase (TPI) exons 6 and 7 were cloned in frame at both ends of the Renilla luciferase ORF (no intron). TPI intron 6 was inserted at either end to generate the 5′ and 3′ intron constructs. (B) Increasing the amount of transfected 5′ intron plasmid elicits a linear increase in mRNA levels. RPA was performed 24 h post-transfection with probe K (Fig. 2C ▶). Two-hundred-fifty nanograms (lanes 1,3,5,7,9,11) or 1 μg (lanes 2,4,6,8,10,12) total RNA was used for RPA. (C) Renilla luciferase activity increases linearly with increasing mRNA levels. mRNA levels are represented relative to 100 ng DNA transfected. (LU) Luminescence units.
Both luciferase activity and mRNA levels are enhanced by introns
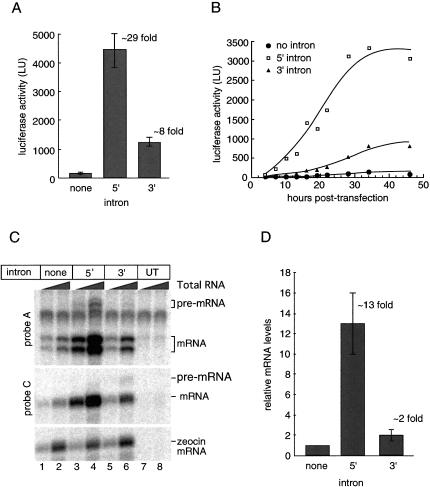
When monitored 42 h post-transfection, both intron-containing constructs yielded significantly more luciferase activity than the no-intron control (Fig. 2A ▶). However, the 5′ intron had a much greater impact (29-fold enhancement) than the 3′ intron (8-fold enhancement). A time course revealed that these intron-dependent differences were detectable as early as 7 h post-transfection, with luciferase activity from all three constructs continuing to increase for at least 21 h thereafter (Fig. 2B ▶).
FIGURE 2.
Introns enhance both luciferase expression and mRNA accumulation in transiently transfected HeLa cells. (A) Cell lysate luciferase activity measured 42 h post-transfection. Activities shown are an average of three independent experiments; error bars represent standard deviation. (B) Time course of luciferase expression in living cells. Transfected HeLa cells were monitored for luciferase activity from 4 to 46 h post-transfection. (C) Analysis of total TPI/Renilla mRNA levels 42 h post-transfection. RPA reactions contained 250 ng or 1 μg total RNA using probe A and probe C (schematized in Fig. 1A ▶). Bands corresponding to pre-mRNA and spliced mRNA are as indicated. Doublet observed with probe A is due to partial homology in the noncomplimentary tail of the probe. Zeocin mRNA, which is transcribed from the same plasmid as TPI/Renilla mRNA was used as a control. (UT) Untransfected. (D) Quantitation of mRNA levels. TPI/Renilla mRNA levels were normalized to zeocin mRNA and plotted relative to the no-intron control. Relative mRNA levels reflect an average of three independent experiments; error bars represent standard deviation.
To determine the extent to which the above intron-dependent enhancements in luciferase activity were due to altered mRNA levels, we next performed ribonuclease protection assays (RPAs) with probes designed to detect both pre-mRNA and mRNA for either intron position (Fig. 2C ▶). Both intron-containing constructs yielded significantly more mRNA than the no-intron control (Fig. 2D ▶), with the 5′ intron giving a greater enhancement (∼13-fold) than the 3′ intron (∼2-fold). Further, both the 5′ and 3′ introns appeared to be efficiently processed (compare relative pre-mRNA:mRNA signals in lanes 3–4 and 5–6), suggesting that splicing of TPI intron 6 is not highly dependent on its distance from either end of the mRNA.
Taken together, the above results confirm that even in the context of a strong CMV promoter, it is possible to detect dramatic enhancement of gene expression by the inclusion of a single intron within the ORF. Furthermore, the level of the response is dependent on intron position, with TPI intron 6 being more stimulatory at the 5′ end than at the 3′ end. In both cases, however, the enhancement in luciferase activity was greater than would be predicted by increases in mRNA levels alone. Whereas the 5′ intron construct exhibited a 29-fold enhancement in luciferase activity over the no-intron control, the level of mRNA only increased 13-fold. Similarly, whereas the 3′ intron construct exhibited an 8-fold increase in luciferase activity over the no intron control, mRNA accumulation was only enhanced 2-fold. Thus both spliced mRNAs exhibited a 2–4 fold increased translational yield (i.e., the amount of protein produced per mRNA) compared to the no-intron control mRNA.
No effect of splicing on mRNA decay
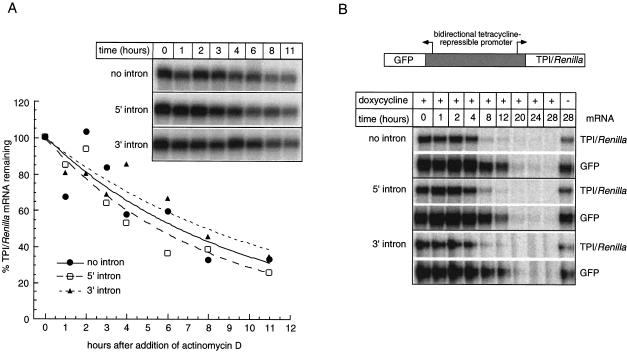
Higher mRNA levels for the intron-containing constructs could result from increased transcriptional efficiency, increased polyadenylation of nascent transcripts, decreased decay of pre-mRNA or mature mRNA, or some combination of these. To examine relative mRNA decay rates, we initially determined half-lives for all three TPI/Renilla mRNAs after inhibition of transcription with actinomycin D (Fig. 3A ▶). Because so much more mRNA was produced by the intron-containing constructs, we wanted to avoid the possibility that the decay machinery might become saturated by the spliced mRNAs. Therefore, to compare similar initial mRNA levels for all three constructs, we used proportionately less of the intron-containing plasmids for transfection, while maintaining a constant amount of total transfected DNA (see Materials and Methods). All three mRNAs exhibited half-lives of ∼7 h, indicating no significant differences in mRNA stability dependent on splicing. To eliminate the possibility that global transcription inhibition led to pleotropic effects on mRNA decay, we also monitored mRNA half-lives by expressing all three constructs from a tetracycline-repressible promoter (Tet-Off™ system, Clontech). This promoter also drove expression of GFP mRNA in the opposite orientation, giving an internal standard for mRNA decay following transcriptional repression. Again, all three TPI/Renilla mRNAs exhibited almost identical half-lives (Fig. 3A,B). We conclude that inclusion of TPI intron 6 at either the 5′ or 3′ end of the TPI/Renilla ORF is of little or no consequence for stability of the mature mRNA.
FIGURE 3.
Splicing does not affect mRNA stability. (A) Time course of TPI/Renilla mRNA decay after treatment of transfected cells with actinomycin D. RPAs were performed on 4 μg total RNA using probe C (Fig. 1A ▶) at times indicated following actinomycin D addition. The graph represents a quantitative analysis of the inset gel. (B) Time course of TPI/Renilla and GFP mRNA decay after transcription repression with 1 μg/mL doxycycline. Constructs expressed both mRNAs in opposing orientations as schematized at top. RPAs were performed on 2 μg total RNA using TPI/Renilla probe B (Fig. 1A ▶) and a GFP-specific probe at times indicated following doxycycline addition. Right-most lane monitors mRNA levels at the last time point in the absence of transcriptional repression.
Effects of ribozyme cleavage on splicing-dependent mRNA accumulation
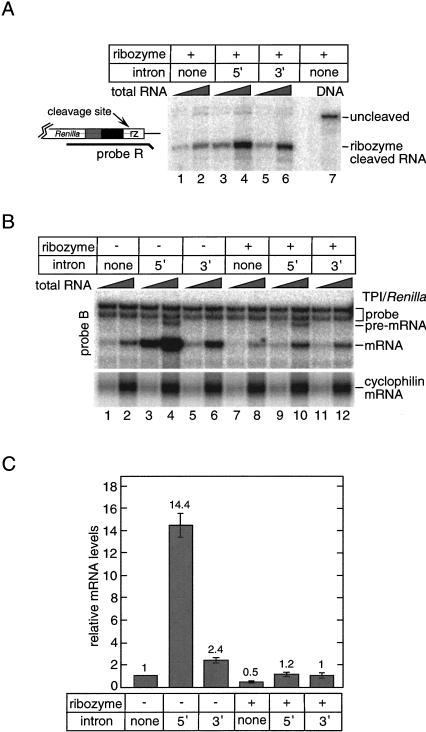
We next constructed variants of the CMV-promoter constructs in which a self-cleaving ribozyme was inserted upstream of the polyadenylation site (Fig. 4 ▶). This hammerhead ribozyme has previously been used to generate poly(A)-minus mRNAs in Saccharomyces cerevisiae with high efficiency (Samarsky et al. 1999). Our rationale for making these constructs was as follows. If some of the intron-dependent enhancement of mRNA accumulation was dependent on poly(A) site cleavage and/or polyadenylation of nascent transcripts, then 3′ end formation with a ribozyme should abrogate this portion of the enhancement.
FIGURE 4.
Uncoupling transcription and 3′ end formation reduces mRNA accumulation from the 5′ intron construct, but has little effect on 3′ intron mRNA. (A) Efficiency of ribozyme (rz) self-cleavage. A probe spanning the ribozyme sequence (probe R) was hybridized to 1.5 μg (lanes 1,3,5) or 6 μg (lanes 2,4,6) total RNA from transfected cells. No intron plasmid DNA was used to obtain protected fragments corresponding to uncleaved mRNA (lane 7). (B) Comparison of mRNA levels from cells transfected with either the parental constructs (Fig. 1A ▶) or their ribozyme-containing equivalents. RPAs were performed with probe B on 1.5 or 6 μg total RNA. Lower panel shows endogenous cyclophilin mRNA levels. (C) Summary of TPI/Renilla mRNA levels for each construct relative to the no-intron/no-ribozyme control. Numbers represent the average of two independent experiments and error bars indicate standard deviations.
When compared to the parental constructs, all three ribozyme constructs expressed lower levels of TPI/Renilla mRNA (Fig. 4 ▶). RPA with a probe spanning the ribozyme sequence showed that in all three cases, most of the TPI/Renilla mRNA did terminate at the ribozyme cleavage site (Fig. 4A ▶). However, given the importance of poly(A) tails for mRNA stability (Caponigro and Parker 1995; Duvel et al. 2002, and references therein), we were somewhat surprised that ribozyme cleavage reduced mRNA levels from the no-intron construct just twofold compared to transcripts terminating at the poly(A) site (Fig. 4B ▶, lanes 2,8, and 4C). This rather small effect on mRNA accumulation has been observed previously in S. cerevisiae, in which 3′ end formation with a similar ribozyme only reduced steady-state mRNA levels by 15%. In that case it was suggested that either the 2‘, 3′ cyclic phosphate terminus left behind by the ribozyme impaired access of 3′ exonucleases to the mRNA, or the absence of a poly(A) tail prevented deadenylation-dependent removal of the 5′ cap structure (Duvel et al. 2002). Finally, inclusion of the ribozyme in the 3′-UTR had no detectable effect on the level of unspliced pre-mRNA from the 5′ intron construct (Fig. 4B ▶, cf. lanes 4 and 10). This suggests that the ribozyme was of little consequence for either transcription rate or pre-mRNA stability.
The decrease in mRNA accumulation upon ribozyme cleavage was much more significant for the 5′ intron construct than for either the 3′ intron or no-intron constructs (Fig. 4B,C). Whereas the 5′ intron construct capable of polyadenylation yielded ∼14-fold more mRNA than its respective no intron control, the ribozyme-containing 5′ intron construct yielded only ∼2-fold more mRNA than its no-intron control. In contrast, the 3′ intron construct exhibited almost no dependence on polyadenylation for enhanced mRNA accumulation, as both the poly(A)-site-containing and ribozyme-containing 3′ intron constructs yielded ∼2-fold more mRNA than their respective no-intron control constructs (Fig. 4B,C). Possible explanations for this phenomenon are presented in the Discussion.
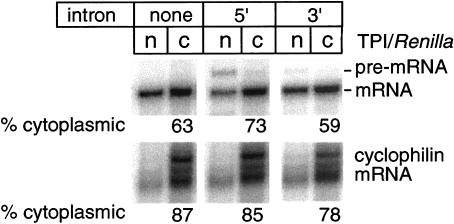
No significant effect of splicing on nucleocytoplasmic mRNA distribution
To test whether splicing affected the nucleocytoplasmic distribution of TPI/Renilla mRNA, we compared the relative amounts of all three mRNAs in the nuclear and cytoplasmic compartments 24 h post-transfection. As with the mRNA decay experiments (see above), we used proportionately less of the intron-containing plasmids for transfection to compare similar in vivo levels of spliced and no-intron mRNAs. All three mRNAs were predominantly cytoplasmic, and there was no significant difference in the nuclear:cytoplasmic mRNA ratio dependent on splicing (Fig. 5 ▶). However, consistent with the known role of the poly(A)-tail in mRNA export (Eckner et al. 1991; Huang and Carmichael 1996), the ribozyme-cleaved transcripts accumulated to a greater extent in the nuclear fraction (data not shown). This confirmed that our assay was capable of detecting differences in nucleocytoplasmic RNA distributions. Therefore, we conclude that the splicing-dependent increase in mRNA translational yield observed in Figure 1 ▶ was not due to enhanced accumulation of mRNA in the cytoplasm.
FIGURE 5.
Splicing does not influence the nuclear/cytoplasmic distribution of TPI/Renilla mRNA. HeLa cells were transfected with varying amounts of the three plasmids in Figure 1A ▶ to achieve similar in vivo mRNA levels, and nuclear and cytoplasmic RNAs were fractionated as described (see Materials and Methods). RPA was performed on equal volumes of isolated RNA with TPI/Renilla probe B (Fig. 1A ▶) or one specific to endogenous cyclophilin mRNA. (n) nuclear; (c) cytoplasmic.
Similar relative effects are observed for TCR-β minigene constructs
Taken together, the above results indicate that for the TPI/Renilla luciferase constructs, the main effects of adding TPI intron 6 within the ORF were at the levels of mRNA synthesis and mRNA translational yield. To determine whether these findings could be extended to another reporter system, we examined the effects of removing either or both introns from a TCR-β minigene (Fig. 6A ▶). However, it should be noted that in this case, the two introns are not identical. All four TCR constructs were driven by the CMV promoter and contained a C-terminal FLAG™ epitope for protein detection.
FIGURE 6.
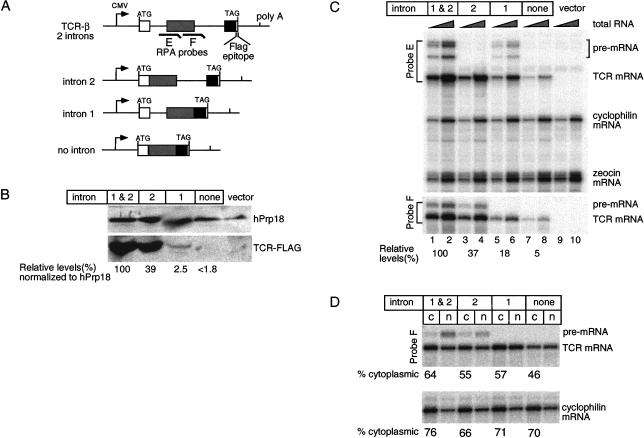
Analysis of overall gene expression, mRNA levels and nucleocytoplasmic distributions for TCR-β minigene constructs. (A) Schematic representation of TCR-β constructs containing two introns, intron 2 alone, intron 1 alone, or no intron. (B) Western blot analysis of total protein extracted from HeLa cells 42 h post-transfection with anti-FLAG™ and anti-hPrp18 antibodies. Note that the relative TCR-β protein level indicated for lane 4 (<1.8) only represents an upper estimate, as no specific Western signal could be detected in this lane. (C) RPA of 500 ng or 2 μg total RNA with probes E and F (schematized in A), and probes specific to endogenous cyclophilin mRNA as well as plasmid-encoded zeocin mRNA (for determination of transfection efficiencies). (D) Nucleocytoplasmic distribution of TCR-β and cyclophilin mRNAs. (n) Nuclear; (c) cytoplasmic.
Similar to the TPI/Renilla constructs, protein expression from the no-intron TCR-β minigene was severely compromised compared to the minigene containing both introns (Fig. 6B ▶). In fact, in transient transfections, we could not detect any protein produced from the no-intron construct, indicating that its protein expression was reduced at least 56-fold (Fig. 6B ▶, 100/1.8) compared to the two-intron gene. Because the no-intron mRNA could direct translation of TCR-β protein in rabbit reticulocyte lysate (data not shown), lack of detectable protein in the transfection experiments was not due to any inherent defect in the construct. Comparison of protein levels from the two-intron and one-intron constructs revealed that having two introns was better than having either single intron alone. However, the effect of each intron alone was highly intron specific. That is, while intron 2 yielded almost half as much protein as having both introns, intron 1 yielded only minimal amounts of protein.
We next examined the relative mRNA levels for each TCR-β construct (Fig. 6C ▶). Compared to the two-intron construct, the no-intron control expressed 20-fold less mRNA. Again, the single-intron constructs exhibited intermediate effects, with intron 2 alone being more advantageous than intron 1 alone. These results are consistent with a previous report that different introns can have quite dissimilar consequences for gene expression even when placed in identical positions (Bourdon et al. 2001). When taken together, the above data indicate that, similar to TPI/Renilla mRNA, spliced TCR-β mRNAs have a >2.7-fold higher translational yield than the no-intron mRNA.
To determine the extent to which the observed differences in protein expression were due to differences in subcellular mRNA localization, we performed biochemical fractionations of nuclear and cytoplasmic compartments (Fig. 6D ▶). As before, these experiments were performed under conditions where in vivo mRNA levels were normalized by titrating the amount of transfected DNA. As with the TPI/Renilla mRNAs, again we saw only minor, if any, differences in the subcellular distributions of TCR-β mRNA dependent on the presence or absence of introns. Notably, removal of both introns reduced the levels of mRNA in the cytoplasm by at most 1.4-fold compared to the two-intron construct. Therefore, differences in the nucleocytoplasmic distribution due to splicing cannot fully explain the increased translational yield from spliced TCR-β mRNA.
DISCUSSION
In this article, we have quantified the effects of three different introns (one from the human TPI gene and two from a mouse TCR-β minigene) on multiple steps of gene expression. We find that even when expression is driven by the strong CMV promoter, all three of these introns significantly enhance protein expression. Their major effect is to increase steady-state mRNA levels. A secondary consequence is an increased translational yield from spliced mRNAs. However, we did not observe any significant effects of introns within ORFs on either mRNA stability or nucleocytoplasmic distribution. Below, we discuss how each of these observations fits with our current understanding of the mechanisms by which introns influence individual steps of gene expression.
Effects of introns on mRNA synthesis
In both systems examined here, the major effect of including one or more introns within the ORF was to increase mRNA abundance. For the TPI/Renilla constructs, we could observe no splicing-dependent alteration in mRNA stability (Fig. 3 ▶). This is consistent with previous data indicating that introns positioned within an ORF do not significantly influence mRNA half-life once the mRNA has been released from the site of transcription (Ryu and Mertz 1989). At present, however, we cannot rule out the possibility that some of the splicing-dependent mRNA accumulation was due to pre-mRNA stabilization. In this regard, the Tollervey lab has described an exosome-dependent pre-mRNA discard pathway in S. cerevisiae (Bousquet-Antonelli et al. 2000), and the exosome was recently shown to associate with RNA pol II at sites of active transcription in Drosophila (Andrulis et al. 2002). It is possible that a higher percentage of intron-containing transcripts than cDNA transcripts escapes surveillance and degradation by nuclear exosomes because they are more efficiently processed and released from the site of transcription than intronless transcripts.
Another mechanism by which introns can influence mRNA synthesis is by modulating the transcriptional machinery. In HeLa cell nuclear extract, introns can enhance transcriptional elongation by recruiting P-TEFb, a RNA pol II CTD kinase, through its interaction with the splicing factor TAT-SF1 (Fong and Zhou 2001). CTD phosphorylation by P-TEFb makes the polymerase more processive, resulting in a greater percentage of full-length transcripts (Price 2000). However, to what extent this intron-dependent increase in processivity contributes to pre-mRNA synthesis in vivo is not yet known. On the other hand, nuclear run-on analysis in both yeast and mammalian tissue culture cells has revealed an intron-dependent enhancement in transcription initiation rates (Furger et al. 2002; Kwek et al. 2002). This effect is most pronounced for a promoter-proximal intron. This is consistent with our finding that the TPI intron had a much greater effect on mRNA accumulation when positioned near the 5′ end of the Renilla luciferase gene than when positioned near its 3′ end.
Insertion of a ribozyme upstream of the poly(A) site allowed us to determine whether any of the enhancement in spliced mRNA levels required normal 3′ end formation (Fig. 4 ▶). This experiment revealed that the increase in mRNA levels promoted by the 3′ intron over the no-intron control was independent of how the 3′ end was generated (aproximately twofold in both cases). In contrast, enhancement of mRNA levels by the 5′ intron exhibited a very strong dependence on the mechanism of 3′ end formation, suggesting that the major effect of the 5′ intron is to enhance polyadenylation efficiency. While the synergistic connection between splicing and polyadenylation has been well established (for review, see Proudfoot et al. 2002), what has not been previously tested is how this link is affected by varying the position of a single intron within an otherwise intronless gene. What our data suggest is that there is a strong relationship between polyadenylation efficiency and intron position, with a promoter proximal intron being more stimulatory than the same intron positioned more distally. Clearly, it will now be of interest to examine this phenomenon in greater detail.
In contrast to the TPI intron in the context of Renilla luciferase, removal of intron 2 from the TCR-β minigene had a greater effect on mRNA accumulation than removal of intron 1. In this case, however, the two introns were not identical and we did not determine the extent to which the accumulation effects were due to changes in mRNA synthesis or stability. Because identically positioned, but distinct introns can have markedly different effects on gene expression (Bourdon et al. 2001), it would be necessary to examine TCR-β introns 1 and 2 individually before one could make any firm conclusions about how they differentially affect mRNA accumulation.
Minimal effects of splicing on nucleocytoplasmic mRNA distribution in mammalian cells
One effect of splicing on downstream mRNA metabolism that has received much attention of late is enhancement of nuclear export (Reed and Magni 2001; Maniatis and Reed 2002; Reed and Hurt 2002). In 1999, Luo and Reed showed that some spliced RNAs are exported more efficiently from Xenopus oocyte nuclei than identical cDNA-derived transcripts (Luo and Reed 1999). Subsequent work demonstrated that a known mRNA export adaptor, REF/Aly, is recruited to mRNAs by splicing as part of the EJC (Le Hir et al. 2000; Zhou et al. 2000). Confirmation that the EJC is the species responsible for positive effects of splicing on RNA export in oocytes came from analysis of spliced RNAs that either did or did not carry an EJC (Le Hir et al. 2001). However, whereas RNAs shorter than ∼300 nt are inefficiently exported unless they carry an EJC, this effect becomes less pronounced as RNA length increases (Le Hir et al. 2001; Rodrigues et al. 2001). This length dependence is most likely explained by the ability of REF/Aly and other mRNA export adaptors to bind to low affinity sites independent of splicing (Rodrigues et al. 2001). Examples of other export adaptors are hnRNP A1, SRp20, 9G8, ASF/SF2, and HuR, all of which can bind mRNA independent of splicing (Michael et al. 1997; Caceres et al. 1998; Gallouzi and Steitz 2001; Huang and Steitz 2001). Thus, as the length of an mRNA increases, so does the likelihood that it will contain binding sites for one or more export adapters. Longer RNAs may therefore be able to establish productive interactions with the export machinery independent of splicing.
The positive effects of splicing on RNA export described above were observed exclusively in Xenopus oocytes. In the present study, we examined the effect of splicing on nuclear:cytoplasmic mRNA ratios in HeLa cells. For the TPI/Renilla constructs, we observed no splicing-dependent increase in cytoplasmic mRNA levels (Fig. 5 ▶), and for the TCR-β transcripts, there was at best a 1.4-fold increase in cytoplasmic mRNA for the two-intron construct compared to the no-intron mRNA (Fig. 6D ▶). Our results are in agreement with several other studies that examined the effects of splicing on cytoplasmic mRNA accumulation in mammalian and insect cells. In 1988, Buchman and Berg reported that despite a 400-fold diminution in the overall level of rabbit β-globin mRNA upon removal of both its natural introns, there was no change in the proportion of mRNA that was cytoplasmic (Buchman and Berg 1988). Similarly, Lu and Cullen recently found no significant export effect in several mammalian and avian cell lines upon removal of introns from a number of indicator genes of various lengths (B. Cullen, pers. comm.). Finally, RNAi knockdown experiments revealed that REF/Aly and other EJC components are dispensable for export of bulk mRNA in Drosophila cells (Gatfield and Izaurralde 2002).
Taken together with our results, the currently available data therefore indicate that splicing and/or EJC deposition is of little or no consequence for the nuclear export of most mRNAs. However, it does remain possible that splicing is an important effector for the export of some individual mRNAs. In this regard, Ryu and Mertz reported that removal of introns from an SV40 late gene resulted in a ∼100-fold reduction in cytoplasmic mRNA levels. They went on to show that the main contributors to this reduction were decreased nuclear mRNA stability and decreased mRNA export (Ryu and Mertz 1989). Another example is provided by human ceruloplasmin, in which transcripts from a cDNA construct were almost entirely retained in the nucleus. In this case, addition of the second intron of rabbit β-globin in the 5′ UTR led to cytoplasmic mRNA accumulation and a 500-fold increase in protein expression (Rafiq et al. 1997).
Effects of splicing on mRNA translational yield
For both the TPI/Renilla and TCR-β constructs, we found that spliced mRNAs yielded two to four times more protein per mRNA molecule than unspliced mRNAs. Theoretically, this increased translational yield could either reflect increased translatability of spliced messages or differential stability of the encoded polypeptide. Our experiments do not distinquish between these possibilities. However they do show that downstream effects of splicing on mRNA metabolism in mammalian cells are not limited to mRNA export and NMD. Splicing-dependent alteration of mRNA translational output has previously been described in Xenopus oocytes (Braddock et al. 1994; Matsumoto et al. 1998). Braddock et al. (1994) reported that when a mature mRNA was injected directly into oocyte nuclei, it was translationally repressed after export to the cytoplasm. This repression could be overcome either by also injecting antibodies against the FRGY2 family of DNA/RNA-binding proteins or by including a spliceable intron in the 3′ UTR. Binding of FRGY2 proteins in the nucleus is known to translationally inactivate or “mask” mRNPs (Bouvet and Wolffe 1994). Therefore, they concluded that splicing can enable an mRNA to escape such masking and actively engage cytoplasmic ribosomes. However, whether these effects are a peculiarity of the oocyte system, which is known to rely heavily on translational regulation of maternal mRNAs, was previously unknown. Our finding that spliced mRNAs also yield more protein in mammalian cells indicates that splicing enhancement of translational yield is a more general phenomenon extendable to other organisms and cell types. In the future, it will be of great interest to determine whether, like export and NMD, the effects of splicing on translational yield are attributable to deposition of an EJC or are due to some other alteration in mRNP structure.
MATERIALS AND METHODS
Plasmids
The TPI/Renilla constructs were created by cloning human triose phosphate isomerase exons 6 and 7 (85 and 38 nt, respectively) plus or minus intron 6 (from pMT-TPI(Norm); Zhang et al. 1998) in frame with the Renilla luciferase gene (from pRL-TK; Promega) in the pcDNA3.1/Zeocin (+) vector (Invitrogen). For the tetracycline repressible constructs, each TPI/Renilla fusion gene was cloned into pBI-EGFP (Clontech). For the ribozyme experiments, a hammerhead ribozyme sequence (Samarsky et al. 1999) was cloned immediately downstream from the stop codon in each construct. T-cell receptor minigenes containing a C-terminal FLAG epitope were derived from Construct A, (Fig. 1 ▶ in Li et al. 1997) and were expressed from the pcDNA3.1 vector. Intron 2, which was greater than 1 kb in Construct A (Li et al. 1997), was reduced to 358 nt in our constructs by restriction digestion and religation. Introns 1 and 2 were deleted individually by sitedirected mutagenesis.
Cell culture and transfections
HeLa cells were cultured in DMEM supplemented with 10% fetal calf serum. HeLa Tet-OffTM cells (Clontech) were cultured in DMEM with tetracycline-tested 10% fetal calf serum (Clontech), supplemented with 100 μg/ml Geneticin. All cells were transfected at 40%–50% confluency using Superfect (Qiagen) according to the manufacturer’s instructions. Unless otherwise indicated, experiments were performed with cells grown in 6-well plates. 500 ng of each TPI/Renilla plasmid was used for experiments shown in Figure 2 ▶ and Figure 4 ▶. For nucleocytoplasmic fractionation (Figure 5 ▶), 5 μg no-intron plasmid, 500 ng 5′ intron plasmid and 2.5 μg 3′ intron TPI/Renilla plasmid were transfected. For TCR-β experiments (Figure 6B ▶, C) 4 μg of each plasmid was used for transfections. For nuclear/cytoplasmic fractionation experiment transfections were done using 400 ng 2-intron, 800 ng intron 2 alone, 1.5 μg intron 1 alone and 4 μg of no intron TCR-β plasmid (Figure 6D ▶). In each set of experiments the total DNA transfected was kept the same by including appropriate amount of empty vector.
Luciferase assays and Western analysis
For luciferase activity measurements, all cells from a single well were lysed in 100 μL 1× Renilla Lysis buffer (Promega) and activity measured by diluting 10% of this lysate fivefold with Renilla Assay buffer (Promega). Light production was monitored in a Turner Designs model TD-20/20 Luminometer with 2-sec delay and 10-sec integration. The remaining lysate was diluted with 900 μL TRI reagent (Molecular Research Corp.) and RNA extracted according to the manufacturer’s protocol. For the time-course experiment, 500 ng of each TPI/Renilla construct were transfected into HeLa cells in a 35-mm dish. At each time point, medium was removed from the dishes and 200 μL Renilla Assay buffer added. The dish was then placed directly in the luminometer and luminescence intergrated over 120 sec after a 10-sec delay. Cells were then washed with 1× PBS and returned to the incubator after addition of fresh medium.
For the TCR minigenes, cells were lysed directly 42 h post-transfection in the six-well plates with TRI reagent, and RNA and protein isolated according the manufacturer’s directions. Extracted proteins were separated on a 15% SDS-polyacrylamide gel and Western blotted with a 1:1000 dilution of anti-FLAG M2 antibody (Sigma), followed by a 1:30,000 dilution of alkaline phosphatase conjugated anti-mouse IgG (Sigma). The same membrane was stripped and probed with anti-hPrp18 (a gift from D. Horowitz, Uniformed Services University of the Health Sciences [USUHS]) as a loading control. Both blots were developed using Enhanced Chemifluorescence reagent (Amersham) and scanned and quantified using a Molecular Dynamics PhosphorImager and associated software.
RNase protection assays (RPA) and subcellular fractionations
For RPA, radiolabeled probes were synthesized by T7 transcription with [α-32P]-UTP from templates generated by PCR. Each probe was designed to span 100–300 nt of the exons, 20–40 nt of the intron, and a 15-nt noncomplimentary sequence to distinguish between undigested probe and protected fragments. Cyclophilin probe was transcribed from pTRI-cyclophilin (Ambion). RPAs were performed with 250 ng to 6 μg of total RNA plus 1 fmole probe essentially as described in Dumas et al. (1996). RNase digestions were performed either with RNases A+T1 or with RNaseOneTM (Promega). To ensure that the probe was not limiting, RPAs were performed with two different amounts of total RNA where indicated. Gels were scanned and quantified using a Molecular Dynamics PhosphorImager and associated software.
Nuclear and cytoplasmic RNA was fractionated as described by Wilkinson (1988), except that after separation of nuclear and cytoplasmic fractions, total RNA was isolated using the TRI reagent. RNA from both fractions was resuspended in equal volume of water and equal volumes were used for RPA.
RNA stability
For the actinomycin D experiment (Fig. 3A ▶), HeLa cells were transfected with 3.5 μg no-intron plasmid, 700 ng 5′ intron plasmid, or 2 μg of 3′ intron plasmid per well. Total DNA was normalized to 4 μg by adding an appropriate amount of pEGFP-C3 vector (Clontech). Actinomycin D (100 μg/mL final) was added to the medium 13 h post-transfection. At times indicated, total RNA was extracted using TRI reagent and analyzed by RPA as above. For the tetracycline repression experiment (Fig. 3B ▶), HeLa Tet-Off cells (Clontech), were transfected with 1 μg no-intron plasmid, 250 ng 5′ intron plasmid, and 600 ng 3′ intron plasmid. Total DNA was normalized to 1 μg using empty pcDNA3.1 vector. Doxycycline was added to culture medium (1 μg/mL final) 20 h post-transfection.
Acknowledgments
We thank Shveta Bagga and Hervé Le Hir for discussion and encouragement, Ken Dower and Annapoorni Rangarajan for technical advice, members of the Moore lab for critical comments on the manuscript, and Bryan Cullen for sharing results prior to publication. M.J.M. is an HHMI associate investigator, and this work was supported, in part, by NIH Grant GM53007.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.rnajournal.org/cgi/doi/10.1261/rna.5250403.
REFERENCES
- Andrulis, E.D., Werner, J., Nazarian, A., Erdjument-Bromage, H., Tempst, P., and Lis, J.T. 2002. The RNA processing exosome is linked to elongating RNA polymerase II in Drosophila. Nature 420: 837–841. [DOI] [PubMed] [Google Scholar]
- Bourdon, V., Harvey, A., and Lonsdale, D.M. 2001. Introns and their positions affect the translational activity of mRNA in plant cells. EMBO Rep. 2: 394–398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bousquet-Antonelli, C., Presutti, C., and Tollervey, D. 2000. Identification of a regulated pathway for nuclear pre-mRNA turnover. Cell 102: 765–775. [DOI] [PubMed] [Google Scholar]
- Bouvet, P. and Wolffe, A.P. 1994. A role for transcription and FRGY2 in masking maternal mRNA within Xenopus oocytes. Cell 77: 931–941. [DOI] [PubMed] [Google Scholar]
- Braddock, M., Muckenthaler, M., White, M.R., Thorburn, A.M., Sommerville, J., Kingsman, A.J., and Kingsman, S.M. 1994. Intron-less RNA injected into the nucleus of Xenopus oocytes accesses a regulated translation control pathway. Nucleic Acids Res. 22: 5255–5264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchman, A.R. and Berg, P. 1988. Comparison of intron-dependent and intron-independent gene expression. Mol. Cell. Biol. 8: 4395–4405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caceres, J.F., Screaton, G.R., and Krainer, A.R. 1998. A specific subset of SR proteins shuttles continuously between the nucleus and the cytoplasm. Genes & Dev. 12: 55–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callis, J., Fromm, M., and Walbot, V. 1987. Introns increase gene expression in cultured maize cells. Genes & Dev. 1: 1183–1200. [DOI] [PubMed] [Google Scholar]
- Caponigro, G. and Parker, R. 1995. Multiple functions for the poly(A)-binding protein in mRNA decapping and deadenylation in yeast. Genes & Dev. 9: 2421–2432. [DOI] [PubMed] [Google Scholar]
- Chiou, H.C., Dabrowski, C., and Alwine, J.C. 1991. Simian virus 40 late mRNA leader sequences involved in augmenting mRNA accumulation via multiple mechanisms, including increased polyadenylation efficiency. J. Virol. 65: 6677–6685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreyfuss, G., Kim, V.N., and Kataoka, N. 2002. Messenger-RNA-binding proteins and the messages they carry. Nat. Rev. Mol. Cell. Biol. 3: 195–205. [DOI] [PubMed] [Google Scholar]
- Dumas, S., Le Hir, H., Bodeau-Pean, S., Hirsch, E., Thermes, C., and Mallet, J. 1996. New species of human tyrosine hydroxylase mRNA are produced in variable amounts in adrenal medulla and are overexpressed in progressive supranuclear palsy. J. Neurochem. 67: 19–25. [DOI] [PubMed] [Google Scholar]
- Duncker, B.P., Davies, P.L., and Walker, V.K. 1997. Introns boost transgene expression in Drosophila melanogaster. Mol. Gen. Genet. 254: 291–296. [DOI] [PubMed] [Google Scholar]
- Duvel, K., Valerius, O., Mangus, D.A., Jacobson, A., and Braus, G.H. 2002. Replacement of the yeast TRP4 3′ untranslated region by a hammerhead ribozyme results in a stable and efficiently exported mRNA that lacks a poly(A) tail. RNA 8: 336–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckner, R., Ellmeier, W., and Birnstiel, M.L. 1991. Mature mRNA 3′ end formation stimulates RNA export from the nucleus. EMBO J. 10: 3513–3522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fong, Y.W. and Zhou, Q. 2001. Stimulatory effect of splicing factors on transcriptional elongation. Nature 414: 929–933. [DOI] [PubMed] [Google Scholar]
- Furger, A., O’Sullivan, J.M., Binnie, A., Lee, B.A., and Proudfoot, N.J. 2002. Promoter proximal splice sites enhance transcription. Genes & Dev. 16: 2792–2799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallouzi, I.E. and Steitz, J.A. 2001. Delineation of mRNA export pathways by the use of cell-permeable peptides. Science 294: 1895–1901. [DOI] [PubMed] [Google Scholar]
- Gatfield, D. and Izaurralde, E. 2002. REF1/Aly and the additional exon junction complex proteins are dispensable for nuclear mRNA export. J. Cell. Biol. 159: 579–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruss, P., Lai, C.J., Dhar, R., and Khoury, G. 1979. Splicing as a requirement for biogenesis of functional 16S mRNA of simian virus 40. Proc. Natl. Acad. Sci. 76: 4317–4321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamer, D.H., Smith, K.D, Boyer, S.H., and Leder, P. 1979. SV40 recombinants carrying rabbit β-globin gene coding sequences. Cell 17: 725–735. [DOI] [PubMed] [Google Scholar]
- Huang, Y. and Carmichael, G.C. 1996. Role of polyadenylation in nucleocytoplasmic transport of mRNA. Mol. Cell. Biol. 16: 1534–1542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, Y. and Steitz, J.A. 2001. Splicing factors SRp20 and 9G8 promote the nucleocytoplasmic export of mRNA. Mol. Cell. 7: 899–905. [DOI] [PubMed] [Google Scholar]
- Kim, V.N., Kataoka, N., and Dreyfuss, G. 2001. Role of the nonsense-mediated decay factor hUpf3 in the splicing-dependent exon–exon junction complex. Science 293: 1832–1836. [DOI] [PubMed] [Google Scholar]
- Kwek, K.Y., Murphy, S., Furger, A., Thomas, B., O’Gorman, W., Kimura, H., Proudfoot, N.J., and Akoulitchev, A. 2002. U1 snRNA associates with TFIIH and regulates transcriptional initiation. Nat. Struct. Biol. 9: 800–805. [DOI] [PubMed] [Google Scholar]
- Le Hir, H., Gatfield, D., Izaurralde, E., and Moore, M.J. 2001. The exon–exon junction complex provides a binding platform for factors involved in mRNA export and nonsense-mediated mRNA decay. EMBO J. 20: 4987–4997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Hir, H., Izaurralde, E., Maquat, L.E., and Moore, M.J. 2000. The spliceosome deposits multiple proteins 20–24 nucleotides upstream of mRNA exon–exon junctions. EMBO J. 19: 6860–6869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, S., Leonard, D., and Wilkinson, M.F. 1997. T cell receptor (TCR) mini-gene mRNA expression regulated by nonsense codons: A nuclear-associated translation-like mechanism. J. Exp. Med. 185: 985–992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo, M.J. and Reed, R. 1999. Splicing is required for rapid and efficient mRNA export in metazoans. Proc. Natl. Acad. Sci. 96: 14937–14942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lykke-Andersen, J., Shu, M.D., and Steitz, JA. 2001. Communication of the position of exon–exon junctions to the mRNA surveillance machinery by the protein RNPS1. Science 293: 1836–1839. [DOI] [PubMed] [Google Scholar]
- Maniatis, T. and Reed, R. 2002. An extensive network of coupling among gene expression machines. Nature 416: 499–506. [DOI] [PubMed] [Google Scholar]
- Maquat, L.E. and Carmichael, G.G. 2001. Quality control of mRNA function. Cell 104: 173–176. [DOI] [PubMed] [Google Scholar]
- Matsumoto, K., Wassarman, K.M., and Wolffe, A.P. 1998. Nuclear history of a pre-mRNA determines the translational activity of cytoplasmic mRNA. EMBO J. 17: 2107–2121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michael, W.M., Eder, P.S., and Dreyfuss, G. 1997. The K nuclear shuttling domain: A novel signal for nuclear import and nuclear export in the hnRNP K protein. EMBO J. 16: 3587–3598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmiter, R.D., Sandgren, E.P., Avarbock, M.R., Allen, D.D., and Brinster, R.L. 1991. Heterologous introns can enhance expression of transgenes in mice. Proc. Natl. Acad. Sci. 88: 478–482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price, D.H. 2000. P-TEFb, a cyclin-dependent kinase controlling elongation by RNA polymerase II. Mol. Cell. Biol. 20: 2629–2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proudfoot, N.J., Furger, A., and Dye, M.J. 2002. Integrating mRNA processing with transcription. Cell 108: 501–512. [DOI] [PubMed] [Google Scholar]
- Rafiq, M., Suen, C.K., Choudhury, N., Joannou, C.L., White, K.N., and Evans, R.W. 1997. Expression of recombinant human ceruloplasmin—An absolute requirement for splicing signals in the expression cassette. FEBS Lett. 407: 132–136. [DOI] [PubMed] [Google Scholar]
- Reed, R. and Hurt, E. 2002. A conserved mRNA export machinery coupled to pre-mRNA splicing. Cell 108: 523–531. [DOI] [PubMed] [Google Scholar]
- Reed, R. and Magni, K. 2001. A new view of mRNA export: Separating the wheat from the chaff. Nat. Cell. Biol. 3: E201–204. [DOI] [PubMed] [Google Scholar]
- Rodrigues, J.P., Rode, M., Gatfield, D., Blencowe, B.J., Carmo-Fonseca, M., and Izaurralde, E. 2001. REF proteins mediate the export of spliced and unspliced mRNAs from the nucleus. Proc. Natl. Acad. Sci. 98: 1030–1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu, W.S. and Mertz, J.E. 1989. Simian virus 40 late transcripts lacking excisable intervening sequences are defective in both stability in the nucleus and transport to the cytoplasm. J. Virol. 63: 4386–4394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samarsky, D.A., Ferbeyre, G., Bertrand, E., Singer, R.H., Cedergren, R., and Fournier, M.J. 1999. A small nucleolar RNA:ribozyme hybrid cleaves a nucleolar RNA target in vivo with near-perfect efficiency. Proc. Natl. Acad. Sci. 96: 6609–6614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson, M. 1988. A rapid and convenient method for isolation of nuclear, cytoplasmic and total cellular RNA. Nucleic Acids Res. 16: 10934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson, M.F. and Shyu, A.B. 2002. RNA surveillance by nuclear scanning? Nat. Cell. Biol. 4: E144–147. [DOI] [PubMed] [Google Scholar]
- Wilusz, C.J., Wang, W., and Peltz, S.W. 2001. Curbing the nonsense: The activation and regulation of mRNA surveillance. Genes & Dev. 15: 2781–2785. [DOI] [PubMed] [Google Scholar]
- Zhang, J., Sun, X., Qian, Y., LaDuca, J.P, and Maquat, LE. 1998. At least one intron is required for the nonsense-mediated decay of triosephosphate isomerase mRNA: A possible link between nuclear splicing and cytoplasmic translation. Mol. Cell. Biol. 18: 5272–5283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, Z., Luo, M.J., Straesser, K., Katahira, J., Hurt, E., and Reed, R. 2000. The protein Aly links pre-messenger-RNA splicing to nuclear export in metazoans. Nature 407: 401–405. [DOI] [PubMed] [Google Scholar]