Abstract
The DA strain of Theiler's murine encephalomyelitis virus persists in the white matter of the spinal cords of susceptible mice. Previous results showed that the difference in susceptibility to viral persistence between the susceptible SJL/J strain and the resistant B10.S strain was due to multiple non-H-2 loci. The respective roles of hematopoietic and nonhematopoietic cells in this difference have been evaluated with bone marrow chimeras. The results show that non-H-2 loci with a major effect on susceptibility are expressed in hematopoietic cells. However, the study of the SJL.B10-D10Mit180-D10Mit74 congenic line suggests that other loci expressed in nonhematopoietic cells also play a role.
The primary demyelinating disease induced by Theiler's murine encephalomyelitis virus is studied as an animal model for multiple sclerosis (13, 16). After intracranial inoculation, the DA strain of Theiler's virus replicates in neurons of the brain and spinal cord in all strains of mice (30). This encephalomyelitis disappears after 2 weeks regardless of the mouse genotype. However, in genetically susceptible mice the virus persists for the lifetime of the animal in the white matter of the spinal cord in oligodendrocytes, macrophages, and possibly astrocytes (3, 15, 18, 25, 26) and induces chronic inflammation and primary demyelination (1, 8, 17, 22). A previous study accounted for the variation of viral RNA level in 17 inbred strains by the interaction of two groups of loci (11). One locus with a major effect was named Tmevp1 for Theiler's murine encephalomyelitis virus persistence locus 1. It is located on chromosome 17 in the H-2D region. Several reports strongly suggest that the same locus controls not only viral persistence but also demyelination (14, 20, 27, 29) and that the H-2Db class I gene plays a major role in resistance to both (4, 19, 28). The existence of non-H-2 susceptibility loci is shown by the fact that the SJL/J strain is more susceptible to viral persistence than the B10.S strain, although both bear the same H-2s haplotype (11). Two of these non-H-2 loci, named Tmevp2 and Tmevp3, have been located on chromosome 10 close to Ifng by studying an F1(SJL/J × B10.S) × B10.S backcross and SJL/J lines congenic for different B10.S genetic intervals of chromosome 10 (7, 10). However, the Ifng gene does not explain the effect of either Tmevp2 or Tmevp3 (7, 24). One of these studies also showed that other susceptibility loci, with minor effects, must contribute to the difference in viral RNA load between these two mouse strains (10).
The non-H-2 loci responsible for the difference in susceptibility between the SJL/J and the B10.S strains could affect the efficiency of the immune response against the virus or the viral life cycle. To distinguish between these possibilities, we measured the viral RNA load in bone marrow chimeras between these two immunocompatible H-2s strains. The SJL.B10-D10Mit180-D10Mit74 congenic line, which has a small B10.S genetic interval containing the Tmevp3 locus (7), was also studied in an attempt to understand the mechanism of action of this locus.
Five- to 6-week-old mice were irradiated with a 137Cs source at 9.5 Gy for B10.S-H2S/Sg McdJ mice and at either 9.5 or 12 Gy for SJL/J mice (Janvier, Saint-Berthevin, France) and SJL.B10-D10Mit180-D10Mit74 congenic mice (rate of delivery, 1.13 to 1.22 Gy per min). Mice were reconstituted with syngeneic or allogeneic bone marrow cells that had been harvested from the tibias and femurs of age- and sex-matched mice. For the reconstitution, we used 4 × 106 to 6 × 106 bone marrow cells from SJL/J or congenic mice and 1.2 ×107 to 1.5 × 107 bone marrow cells from B10.S mice. Reconstituted and 12- to 14-week-old control mice were anesthetized and inoculated intracranially with 104 PFU of the molecularly cloned TMDA1 strain (21, 23) in 40 μl of phosphate-buffered saline. Mice were sacrificed 45 days postinoculation (p.i.). The efficiency of the reconstitution was assessed with peripheral blood lymphocytes at the time of sacrifice for each mouse reconstituted with allogeneic bone marrow cells. The degree of chimerism varied from 72 to 97% regardless of the genotype of the donor and recipient strains (data not shown). Viral RNA load in the spinal cord was quantified by a dot blot assay (11; also see the discussion in reference 1).
Effect of the mouse genotype and bone marrow reconstitution on viral RNA load.
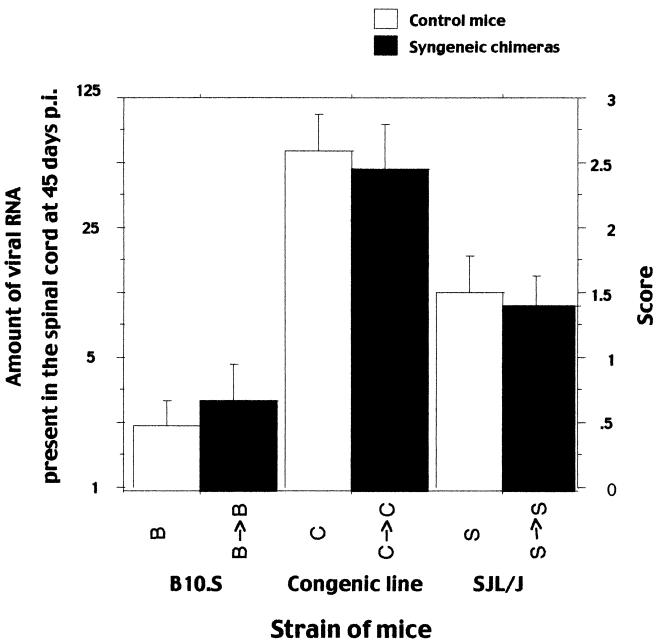
The amount of viral RNA present in the spinal cord at 45 days p.i. was measured for the B10.S, the SJL/J, and the SJL.B10-D10Mit180-D10Mit74 congenic mice and for the same types of mice that had been reconstituted with syngeneic bone marrow cells (Fig. 1; Table 1). The effect of strain origin and reconstitution on viral RNA load was analyzed using a two-way analysis of variance and the Scheffé test. The means of the amount of viral RNA were significantly different among the three strains [F(2; 147) = 19.983; P < 0.0001]. The SJL/J mice were infected at a higher level than the B10.S mice (P = 0.0066). The congenic mice were infected at a higher level than the SJL/J mice (P = 0.0002) and the B10.S mice (P < 0.0001). The mean amount of viral RNA for mice reconstituted with syngeneic bone marrow was similar to that of the nonreconstituted mice [F(1; 147) = 0.006; P = 0.9391]. No interaction between the strain origin and the reconstitution was detected [F(1; 147) = 0.157; P = 0.8551]. Thus, the three strains have different susceptibilities to viral load, and the differences are not affected by the technique used for bone marrow reconstitution.
FIG. 1.
Viral RNA load at 45 days p.i. in the spinal cords of mice reconstituted with syngeneic bone marrow and of control mice of the same genotype. The amount of viral RNA is expressed as the highest RNA dilution which gave a hybridization signal in a dot blot assay. Left ordinate, dilution factor; right ordinate, score (the dilution number in the series). B → B, B10.S mice reconstituted with B10.S bone marrow; C → C, SJL congenic mice reconstituted with SJL congenic bone marrow; S → S, SJL/J mice reconstituted with SJL bone marrow.
TABLE 1.
Viral RNA loada
| Mouse line (no.) | Viral RNA load
|
|
|---|---|---|
| Mean | SEM | |
| B (26) | 0.5 | 0.2 |
| C (28) | 2.6 | 0.3 |
| S (27) | 1.5 | 0.3 |
| B → B (12) | 0.7 | 0.3 |
| C → B (13) | 1.5 | 0.4 |
| S → B (19) | 1.8 | 0.4 |
| B → C (13) | 1.5 | 0.3 |
| C → C (20) | 2.4 | 0.3 |
| S → C (26) | 2.4 | 0.3 |
| B → S (15) | 0.6 | 0.2 |
| C → S (24) | 1.7 | 0.3 |
| S → S (40) | 1.4 | 0.2 |
Reconsitution conditions: B → B, B10.S mice reconstituted with B10.S bone marrow; C → B, B10.S mice reconstituted with SJL congenic bone marrow; S → B, B10.S mice reconstituted with SJL/J bone marrow; B → C, SJL congenic mice reconstituted with B10.S bone marrow; C → C, SJL congenic mice reconstituted with SJL congenic bone marrow; S → C, SJL congenic mice reconstituted with SJL bone marrow; B → S, SJL/J mice reconstituted with B10.S bone marrow; C → S, SJL/J mice reconstituted with SJL congenic bone marrow; S → S, SJL/J mice reconstituted with SJL bone marrow. SEM, standard error of the mean.
Immunological status of the chimeras.
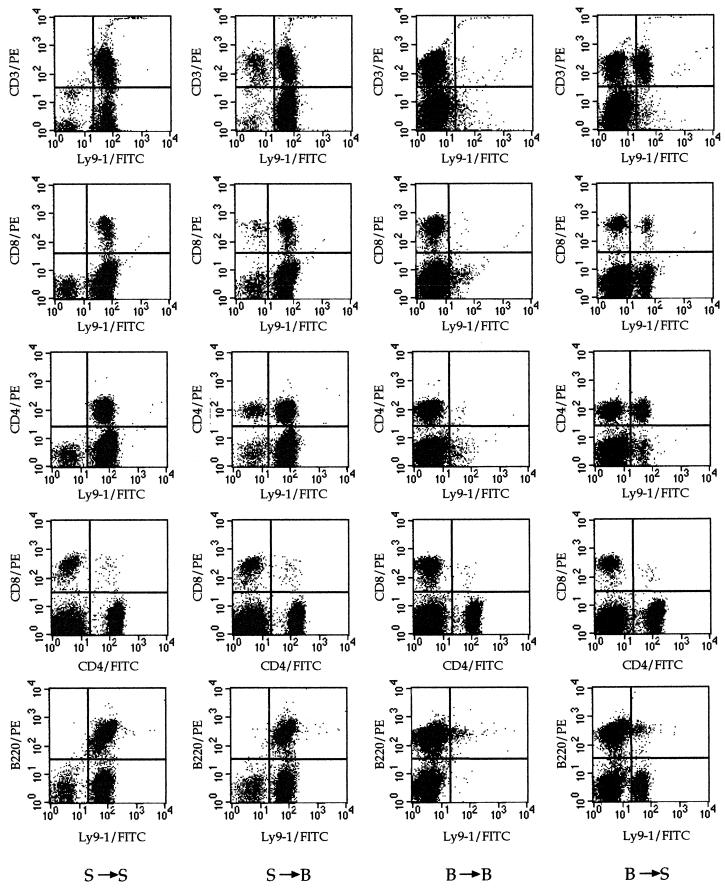
The degree of chimerism for reconstituted SJL/J and B10.S mice was measured at 8 weeks postreconstitution, the time at which the mice were inoculated, by studying the expression of Ly-9.1 on B220+, CD3+, CD4+, and CD8+ spleen cells with FACScan (Fig. 2; Table 2). The efficiency of labeling the four cell populations with the anti-Ly-9.1 monoclonal antibody was always higher than 96.5% for control mice and mice reconstituted with syngeneic bone marrow. The degree of chimerism was close to 100% for B220+ splenocytes. The B10.S mice reconstituted with SJL/J bone marrow showed a high degree of chimerism for CD3+, CD4+, and CD8+ splenocytes (85 to 90%). A lower degree of chimerism (60 to 70%) was detected for CD3+ and CD4+ splenocytes of SJL/J mice reconstituted with B10.S bone marrow.
FIG. 2.
Surface staining of spleen cells from representative mice reconstituted with either syngeneic or allogeneic bone marrow. S → S, SJL/J mice reconstituted with SJL bone marrow; S → B, B10.S mice reconstituted with SJL/J bone marrow; B → B, B10.S mice reconstituted with B10.S bone marrow; B → S, SJL/J mice reconstituted with B10.S bone marrow.
TABLE 2.
Degree of chimerism in the spleen at 8 weeks postreconstitutiona
| Splenocyte | Reconstitution
|
|||||
|---|---|---|---|---|---|---|
| S → B
|
B → S
|
|||||
| No. | DOC | Range | No. | DOC | Range | |
| B220+ | 5 | 99.7 | 99.7-99.8 | 6 | 98.1 | 97.4-98.5 |
| CD3+ | 5 | 88.4 | 85.5-90.6 | 6 | 68.3 | 60.7-87.4 |
| CD4+ | 5 | 89.1 | 85.9-91.8 | 6 | 61.5 | 54.7-82.4 |
| CD8+ | 5 | 85.2 | 83.3-85.7 | 6 | 82.8 | 73.4-92.6 |
DOC, degree of chimerism.
Viral RNA load of chimeras between the two parental strains.
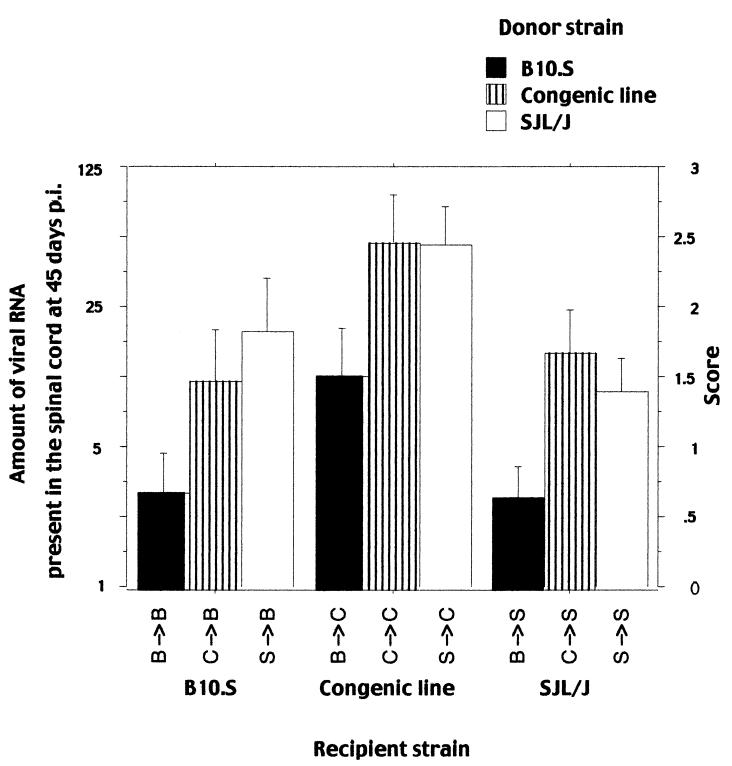
The means of the amount of viral RNA present in the spinal cords of the chimeras 45 days p.i. were compared according to the nature of the recipient and the donor strains, using a two-way analysis of variance and the Scheffé test. As shown in Fig. 3 and Table 1, the means depended on the donor strain [F(1; 82) = 8.580; P = 0.0044] and not on the recipient strain [F(1; 82) = 0.475; P = 0.4927]. No interaction between the origin of the donor and the recipient strain was detected [F(1; 82) = 0.345; P = 0.5588]. These results indicate that the resistance of the B10.S strain is mediated mainly by hematopoietic cells and that the lower degree of chimerism for CD3+ and CD4+ splenocytes of SJL/J mice reconstituted with B10.S bone marrow does not affects their phenotype. Therefore, the non-H-2 loci with a major effect on viral RNA load are expressed in the hematopoietic system, most likely in the immune system. Since the Tmevp1 locus has the characteristics of an H-2 class I gene (2, 5, 6), all loci identified so far with a major effect on Theiler's viral RNA load in the central nervous system of mice seem to act via the immune system. However, we cannot rule out that the non-H-2 loci modify permissiveness of monocytes/macrophages to the virus. It has been noted that susceptibility to demyelination among mouse strains correlates more with viral genome load than with infectious titer (11, 12). Also, Trottier et al. (31) recently reported a striking discrepancy during persistent infection between the viral genome load (109 per spinal cord) and infectious titers (102 to 104 PFU per spinal cord). They explain this discrepancy by a restriction in the viral life cycle and/or by the action of neutralizing antibody. Although our data are consistent with this (9, 11), we offer an alternative hypothesis. Accordingly, the immune system of the resistant strains clears the virus by killing infected cells before viral assembly, whereas that of the susceptible strains kills infected cells at the beginning of virion assembly, allowing low infectivity titers to persist. At the time of killing, virus RNA replication would be already well under way, explaining the high viral RNA load observed.
FIG. 3.
Viral RNA load at 45 days p.i. in the spinal cords of the nine chimeras. The amount of viral RNA is expressed as the highest RNA dilution which gave a hybridization signal in a dot blot assay. Left ordinate, dilution factor; right ordinate, score (the dilution number in the series). B → B, B10.S mice reconstituted with B10.S bone marrow; C → B, B10.S mice reconstituted with SJL congenic bone marrow; S → B, B10.S mice reconstituted with SJL/J bone marrow; B → C, SJL congenic mice reconstituted with B10.S bone marrow; C → C, SJL congenic mice reconstituted with SJL congenic bone marrow; S → C, SJL congenic mice reconstituted with SJL bone marrow; B → S, SJL/J mice reconstituted with B10.S bone marrow; C → S, SJL/J mice reconstituted with SJL congenic bone marrow; S → S, SJL/J mice reconstituted with SJL bone marrow.
Viral RNA load of chimeras between the parental strains and the SJL.B10-D10Mit180-D10Mit74 congenic line.
The viral RNA load was higher for the congenic line than for the SJL/J and B10.S strains (Fig. 1). Immunological chimeras between these three strains were used to test if the high susceptibility of the congenic line was due to its hematopoietic cells (Fig. 3; Table 1). The mean RNA loads were compared according to the nature of the recipient and the donor strain using a two-way analysis of variance and the Scheffé test. These means depended on both the donor strain [F(2; 173) = 13.504; P = 0.0013] and the recipient strain [F(2; 173) = 13.741; P = 0.0012]. Chimeras with a B10.S donor strain were infected at lower levels than chimeras with either an SJL/J (P = 0.0051) or a congenic (P = 0.0042) donor. When the recipient was the congenic line, the chimeras were infected at a higher level than when the recipient was either the SJL/J (P = 0.0012) or the B10.S (P = 0.0121) strain. These results indicate that the susceptibility of the congenic line is mediated in part by nonhematopoietic cells. Since the non-H-2 loci responsible for the difference in viral RNA load between the two parental strains cannot explain this result, other susceptibility loci must exist. Different models were tested to assess the effects of the donor and the recipient on the phenotypes of the nine chimeras obtained with the three parental strains. These models assume that (i) non-H-2 loci are responsible for the difference of susceptibility of the SJL/J and the B10.S strains and that (ii) other non-H-2 loci explain the phenotype of the congenic line and act independently of the first loci. The number and position of the second group of non-H-2 loci vary according to the model. In one model, one such locus is located in the B10.S chromosome 10 interval of the congenic line and the B10.S haplotype is susceptible. In another model, the same locus exists, but it interacts with another locus in the SJL background. This interaction occurs only when the first locus has a B10.S haplotype and the second one has an SJL/J haplotype. Both models were tested by one-way analysis of variance against a null model in which the only loci considered were those that explain the difference between the SJL/J and B10.S strains. The second model was the only one which was not rejected in our test when the phenotype was determined by the genotype of the recipient strain (the P value was 482 times lower than that of the null model). A biological consequence of this model is that the two interacting loci are probably expressed in nonhematopoietic cells. However, we cannot completely rule out that they are expressed in some immune cells, such as microglia, which are radiologically resistant and have a life span longer than the 8 weeks of reconstitution.
In conclusion, non-H-2 loci with a major effect on the susceptibility of the SJL/J and the B10.S strains to Theiler's virus persistence are most probably expressed in the immune system. This result, together with the major effect of H-2 class I genes on persistence, shows that the immune system plays a central role in the control of Theiler's virus RNA load in the central nervous system. Surprisingly, the congenic mouse studied carried a higher viral RNA load than its parents. The study of nine immunological chimeras showed that this could be due to an interaction between two loci. The congenic mouse is presently being studied to clarify the relationship between the interacting locus located in the B10.S chromosome 10 interval and the Tmevp3 locus.
Acknowledgments
We thank M. Gau for secretarial assistance and A. Freitas, D. Gonzalez-Dunia, P. Rohrlich, and S. Vigneau for helpful discussion.
This work was supported by grants from the Institut Pasteur Fondation, the Centre National de la Recherche Scientifique, the Association pour la Recherche sur la Sclérose en Plaques, and the National Multiple Sclerosis Society. S.A. is the recipient of a scholarship from the Ministère de l'Education Nationale de l'Enseignement Supérieur et de la Recherche and from the Fondation pour la Recherche Médicale.
REFERENCES
- 1.Aubagnac, S., M. Brahic, and J.-F. Bureau. 1999. Viral load and a locus on chromosome 11 affect the late clinical disease caused by Theiler's virus. J. Virol. 73:7965-7971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aubagnac, S., M. Brahic, and J.-F. Bureau. 2001. Viral load increases in SJL/J mice persistently infected by Theiler's virus after inactivation of the β2 m gene. J. Virol. 75:7723-7726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aubert, C., M. Chamorro, and M. Brahic. 1987. Identification of Theiler's virus infected cells in the central nervous system of the mouse during demyelinating disease. Microb. Pathog. 3:319-326. [DOI] [PubMed] [Google Scholar]
- 4.Azoulay, A., M. Brahic, and J.-F. Bureau. 1994. FVB mice transgenic for the H-2Db gene become resistant to persistent infection by Theiler's virus. J. Virol. 68:4049-4052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Azoulay-Cayla, A., S. Dethlefs, B. Pérarnau, E. L. Larsson-Sciard, F. A. Lemonnier, M. Brahic, and J.-F. Bureau. 2000. H-2Db−/− mice are susceptible to persistent infection by Theiler's virus. J. Virol. 74:5470-5476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Azoulay-Cayla, A., S. Syan, M. Brahic, and J. F. Bureau. 2001. Roles of the H-2Db and H-Kb genes in resistance to persistent Theiler's murine encephalomyelitis virus infection of the central nervous system. J. Gen. Virol. 82:1043-1047. [DOI] [PubMed] [Google Scholar]
- 7.Bihl, F., M. Brahic, and J.-F. Bureau. 1999. Two loci, Tmevp2 and Tmevp3, located on the telomeric region of chromosome 10, control the persistence of Theiler's virus in the central nervous system. Genetics 152:385-392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brahic, M., and J.-F. Bureau. 1998. Genetics of susceptibility to Theiler's virus infection. Bioessays 20:627-633. [DOI] [PubMed] [Google Scholar]
- 9.Bureau, J.-F., S. Chirinian, S. Ozden, C. Aubert, and M. Brahic. 1990. Isolation of a specific cellular mRNA by subtractive hybridization in Theiler's virus persistent infection. Microb. Pathog. 8:335-341. [DOI] [PubMed] [Google Scholar]
- 10.Bureau, J.-F., X. Montagutelli, F. Bihl, S. Lefebvre, J.-L. Guénet, and M. Brahic. 1993. Mapping loci influencing the persistence of Theiler's virus in the murine central nervous system. Nat. Genet. 5:87-91. [DOI] [PubMed] [Google Scholar]
- 11.Bureau, J.-F., X. Montagutelli, S. Lefebvre, J.-L. Guénet, M. Pla, and M. Brahic. 1992. The interaction of two groups of murine genes determines the persistence of Theiler's virus in the central nervous system. J. Virol. 66:4698-4704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Clatch, R. J., H. L. Lipton, and S. D. Miller. 1987. Class II-restricted T cell responses in Theiler's murine encephalomyelitis virus (TMEV)-induced demyelinating disease. II. Survey of host immune responses and central nervous system virus titers in inbred mouse strains. Microb. Pathog. 3:327-337. [DOI] [PubMed] [Google Scholar]
- 13.Clatch, R. J., R. W. Melvold, M. C. Dal Canto, S. D. Miller, and H. L. Lipton. 1987. The Theiler's murine encephalomyelitis virus (TMEV) model for multiple sclerosis shows a strong influence of the murine equivalents of HLA-A, B, and C. J. Neuroimmunol. 15:121-135. [DOI] [PubMed] [Google Scholar]
- 14.Clatch, R. J., R. W. Melvold, S. D. Miller, and H. L. Lipton. 1985. Theiler's murine encephalomyelitis virus (TMEV)-induced demyelinating disease in mice is influenced by the H-2D region: correlation with TMEV-specific delayed-type hypersensitivity. J. Immunol. 135:1408-1413. [PubMed] [Google Scholar]
- 15.Clatch, R. J., S. D. Miller, R. Metzner, M. C. Dal Canto, and H. L. Lipton. 1990. Monocytes/macrophages isolated from the mouse central nervous system contain infectious Theiler's murine encephalomyelitis virus (TMEV). Virology 176:244-254. [DOI] [PubMed] [Google Scholar]
- 16.Dal Canto, M. C., and H. L. Lipton. 1977. Multiple sclerosis. Animal model: Theiler's virus infection in mice. Am. J. Pathol. 88:497-500. [PMC free article] [PubMed] [Google Scholar]
- 17.Lipton, H. L. 1975. Theiler's virus infection in mice: an unusual biphasic disease process leading to demyelination. Infect. Immun. 11:1147-1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lipton, H. L., J. Kratochvil, P. Sethi, and M. C. Dal Canto. 1984. Theiler's virus antigen detected in mouse spinal cord 2 1/2 years after infection. Neurology 34:1117-1119. [DOI] [PubMed] [Google Scholar]
- 19.Lipton, H. L., R. Melvold, S. D. Miller, and M. C. Dal Canto. 1995. Mutation of a major histocompatibility class I locus, H-2D, leads to an increased virus burden and disease susceptibility in Theiler's virus-induced demyelinating disease. J. Neurovirol. 1:138-144. [DOI] [PubMed] [Google Scholar]
- 20.Lipton, H. L., and R. W. Melvold. 1984. Genetic analysis of susceptibility to Theiler's virus-induced demyelinating disease in mice. J. Immunol. 132:1821-1825. [PubMed] [Google Scholar]
- 21.McAllister, A., F. Tangy, C. Aubert, and M. Brahic. 1989. Molecular cloning of the complete genome of Theiler's virus, strain DA, and production of infectious transcripts. Microb. Pathog. 7:381-388. [DOI] [PubMed] [Google Scholar]
- 22.McAllister, A., F. Tangy, C. Aubert, and M. Brahic. 1990. Genetic mapping of the ability of Theiler's virus to persist and demyelinate. J. Virol. 64:4252-4257. (Author's correction, 67:2427, 1993.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Michiels, T., V. Dejong, R. Rodrigus, and C. Shaw-Jackson. 1997. Protein 2A is not required for Theiler's virus replication. J. Virol. 71:9549-9556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Monteyne, P., F. Bihl, F. Levillayer, M. Brahic, and J.-F. Bureau. 1999. The Th1/Th2 balance does not account for the difference of susceptibility of mouse strains to Theiler's virus persistent infection. J. Immunol. 162:7330-7334. [PubMed] [Google Scholar]
- 25.Njenga, M. K., K. Asakura, S. F. Hunter, P. Wettstein, L. R. Pease, and M. Rodriguez. 1997. The immune system preferentially clears Theiler's virus from the gray matter of the central nervous system. J. Virol. 71:8592-8601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pena Rossi, C., M. Delcroix, I. Huitinga, A. McAllister, N. van Rooijen, E. Claassen, and M. Brahic. 1997. Role of macrophages during Theiler's virus infection. J. Virol. 71:3336-3340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rodriguez, M., and C. S. David. 1985. Demyelination induced by Theiler's virus: influence of the H-2 haplotype. J. Immunol. 135:2145-2148. [PubMed] [Google Scholar]
- 28.Rodriguez, M., and C. S. David. 1995. H-2Dd transgene suppresses Theiler's virus-induced demyelination in susceptible strains of mice. J. Neurovirol. 1:111-117. [DOI] [PubMed] [Google Scholar]
- 29.Rodriguez, M., J. L. Leibowitz, and C. S. David. 1986. Susceptibility to Theiler's virus-induced demyelination. Mapping of the gene within the H-2D region. J. Exp. Med. 163:620-631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stroop, W. G., J. R. Baringer, and M. Brahic. 1981. Detection of Theiler's virus RNA in mouse central nervous system by in situ hybridization. Lab. Investig. 45:504-509. [PubMed] [Google Scholar]
- 31.Trottier, M., P. Kallio, W. Wang, and H. L. Lipton. 2001. High numbers of viral RNA copies in the central nervous system of mice during persistent infection with Theiler's virus. J. Virol. 75:7420-7428. [DOI] [PMC free article] [PubMed] [Google Scholar]