Abstract
Bacterial translation initiation factor IF2 is a GTP-binding protein that catalyzes binding of initiator fMet-tRNA in the ribosomal P site. The topographical localization of IF2 on the ribosomal subunits, a prerequisite for understanding the mechanism of initiation complex formation, has remained elusive. Here, we present a model for the positioning of IF2 in the 70S initiation complex as determined by cleavage of rRNA by the chemical nucleases Cu(II):1,10-orthophenanthroline and Fe(II):EDTA tethered to cysteine residues introduced into IF2. Two specific amino acids in the GII domain of IF2 are in proximity to helices H3, H4, H17, and H18 of 16S rRNA. Furthermore, the junction of the C-1 and C-2 domains is in proximity to H89 and the thiostrepton region of 23S rRNA. The docking is further constrained by the requisite proximity of the C-2 domain with P-site-bound tRNA and by the conserved GI domain of the IF2 with the large subunit’s factor-binding center. Comparison of our present findings with previous data further suggests that the IF2 orientation on the 30S subunit changes during the transition from the 30S to 70S initiation complex.
Keywords: BABE, fMet-tRNA, IF2, phenanthroline, ribosome topography, translation initiation, tethered nuclease
INTRODUCTION
Translation initiation factor 2 (IF2) is an essential GTP/GDP-binding protein whose main recognized function is to interact specifically with initiator fMet-tRNA and to position it correctly in the ribosomal P site, thereby increasing the rate and fidelity of translation initiation (for recent reviews, see Gualerzi et al. 2000, 2001; Boelens and Gualerzi 2002). In the last few years, a wealth of high-resolution crystallographic and electron microscopy data has shed light on structural intricacies of the translational apparatus. The high-resolution x-ray crystal structures of 30S and 50S subunits have been determined by several groups (Ban et al. 2000; Schluenzen et al. 2000; Wimberly et al. 2000) and the structure of the 70S ribosome with tRNAs was determined at 5.5 Å resolution (Yusupov et al. 2001). In addition, high-resolution structures of translation initiation factors IF1 (Sette et al. 1997), IF3 (Biou et al. 1995; Garcia et al. 1995a,b), the ribosome recycling factor (RRF; Selmer et al. 1999), and translation termination factor RF2 (Vestergaard et al. 2001) have been added recently to an impressive list of solved structures, including the elongation factors EF-Tu and EF-G in various functional states (Kjeldgaard and Nyborg 1992; Berchtold et al. 1993; Kjeldgaard et al. 1993; Ævarsson et al. 1994; Czworkowski et al. 1994; Nissen et al. 1995, 1999; Abel et al. 1996; al-Karadaghi et al. 1996; Polekhina et al. 1996). Furthermore, structural models of complexes between factors EF-Tu/EF-Ts (Kawashima et al. 1996) and between ribosomes or ribosomal subunits and IF1, IF3, EF-Tu, and EF-G have elucidated the binding sites of these factors and have suggested mechanisms for their functions (Stark et al. 1997, 2000; Agrawal et al. 1999; McCutcheon et al. 1999; Carter et al. 2001; Dallas and Noller 2001; Pioletti et al. 2001).
The structure of the eubacterial IF2 fMet-tRNA-binding domain has been determined at high resolution (Meunier et al. 2000), whereas the crystallographic structure of the complete archaeal IF2/eIF5B revealed an unusual chalice shape (Roll-Mecak et al. 2000). More recently, the structure of the first 157 amino acids of the amino-terminal domain of Escherichia coli was characterized by NMR (Laursen et al. 2003). Although it is likely that the overall three-dimensional structure of IF2/eIF5B is very similar to that of bacterial IF2, the archaeal protein lacks a large polypeptide segment at its amino terminus (as do a few eubacterial IF2s) and contains two additional α-helices at its carboxyl terminus. Furthermore, at least some of the functions performed by bacterial IF2 are different from those of IF2/eIF5B.
Compared with the other translation factors, much less is known about IF2’s localization on the ribosome and the mechanism by which this factor interacts with and is ejected from the ribosome. In fact, not only is a high-resolution structure of an IF2:ribosome complex still lacking, but even the more traditional biochemical data concerning the binding site of IF2 on the ribosome have been equivocal. Protein–protein cross-linking studies identified a diffuse pattern of IF2 neighbors on the 50S subunit (Heimark et al. 1976), the 30S subunit (Bollen et al. 1975), and IF1 (Boileau et al. 1983). Likewise, protein–rRNA cross-linking and in situ chemical probing with ethylnitrosourea showed that the structure of several regions of the 30S subunit were found to be affected by IF2 binding (Wakao et al. 1990, 1991). On the other hand, no changes in reactivity toward base-specific probes directly attributable to IF2 were observed in 16S rRNA upon IF2 binding to the 30S subunit (Moazed et al. 1995) or to a 70S initiation complex (La Teana et al. 2001). Thus, rather than identifying and precisely localizing IF2 on the 30S subunit, these results were taken variously to indicate the occurrence of a global IF2-induced conformational change of the subunit (Wakao et al. 1990, 1991), that IF2 may interact with the 30S subunit primarily via protein–protein interactions (Moazed et al. 1995), or that it does not establish stable interactions that affect base pairing in 16S rRNA, but could still interact with the sugar-phosphate backbone of the rRNA (La Teana et al. 2001).
The first successful IF2 mapping on the 50S ribosomal subunit came with the identification of specific reactivity increases/decreases of bases within the α-sarcin-ricin loop (G2655, A2660, G2661, and A2665) and helix 89 (A2476 and A2478) of 23S rRNA upon binding of the factor (La Teana et al. 2001). These findings, together with the observation that IF2 could be cross-linked to L7/L12 (Heimark et al. 1976), and the recent finding that IF2 can inhibit EF-G binding to the ribosome (Cameron et al. 2002), confirmed at least a partial overlap of the IF2-binding site with that of the elongation factors EF-Tu and EF-G (Moazed et al. 1988).
Here, we have investigated the proximity between specific positions of IF2 and regions of the 16S and 23S rRNA using two different tethered chemical nucleases, Fe(II)-EDTA and Cu(II):1,10-orthophenanthroline (Cu:oP). The tethered nuclease method has been used successfully by Noller’s group and collaborators to map sites of interaction of several ribosomal proteins and translation factors with the ribosome (Heilek et al. 1995; Wilson and Noller 1998; Lieberman et al. 2000; Dallas and Noller 2001; Lancaster et al. 2002). The present results provide the first complete localization of IF2 on the 70S initiation complex.
RESULTS
Experimental strategy
Bacterial translation initiation factor IF2 consists of five structural domains (Gualerzi et al. 1991; Roll-Mecak et al. 2000; Spurio et al. 2000). The amino-terminal domain (N-domain), which in Bacillus stearothermophilus comprises the first 227 residues, is very rich in alanine as well as in both positively and negatively charged amino acids. Overall, this domain is less conserved than the rest of the molecule, its susceptibility to proteolysis suggests it is weakly structured, and it is apparently dispensable for all basic translational functions of IF2 (Cenatiempo et al. 1987; Gualerzi et al. 1991). The next domain, GI, spans residues 228 to 412, is highly conserved, and contains all of the structural motifs characteristic of the family of GTP/GDP-binding proteins (Cenatiempo et al. 1987; Roll-Mecak et al. 2000). Domain GII consists of 107 highly conserved amino acids (residues 413–520), and is predicted in bacteria, and found in archaea to be a β-barrel module (al-Karadaghi et al. 1996; Roll-Mecak et al. 2000). This module is structurally homologous to domain II of EF-G and EF-Tu (Nissen et al. 1995; Brock et al. 1998) and to the carboxy-terminal-most domain of IF2 itself, called IF2 C-2 (Meunier et al. 2000). Following the GII domain, is C-1, a rather sturdy domain rich in helical structures (Misselwitz et al. 1997; Krafft et al. 2000; Spurio et al. 2000). The last 110 amino acids of the protein (from Glu 632 to Ala 741) constitute the C-2 domain, which is responsible for the recognition and binding of fMet-tRNA (Guenneugues et al. 2000; Krafft et al. 2000; Spurio et al. 2000).
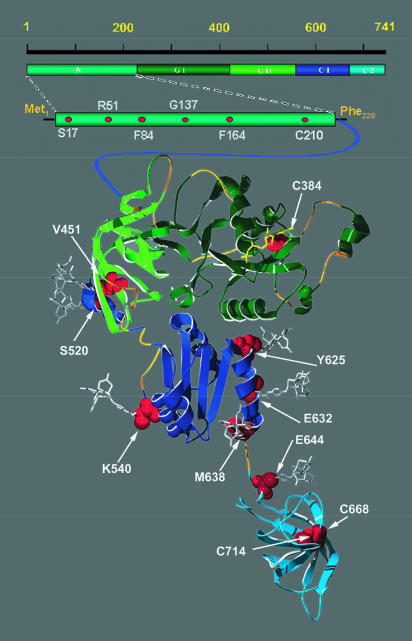
The domains of B. stearothermophilus IF2 are presented in Figure 1 ▶. A structural alignment of B. stearothermophilus IF2 with the corresponding regions of archaeal IF2/eIF5B, EF-Tu, and EF-G (Swiss-PDBViewer, Guex and Peitsch 1997) yielded a three-dimensional homology map of good quality. This alignment enabled us to present, with reasonable confidence, the probable three-dimensional structure of GI, GII, C-1, and the orientation of C-2 of B. stearothermophilus IF2 (Fig. 1 ▶). In this figure, the structure of C-2 is derived from NMR spectroscopy (Meunier et al. 2000), and the regions of IF2 for which this structural alignment is less clear are highlighted in ochre.
FIGURE 1.
Domain structure of B. stearothermophilus IF2 displaying the location of the cysteine residues (natural and introduced by mutagenesis). The five domains of B. stearothermophilus IF2 are depicted schematically and color coded at the top of the figure. The three-dimensional structure of the protein as predicted by homology modeling (Guex and Peitsch 1997) with the archaeal IF2/eIF5B three-dimensional structure (Roll-Mecak et al. 2000) is presented at the bottom of the figure, following the same color scheme (the three-dimensional structure of the C-2 domain was taken from pdb file 1D1NA) (Guennegues et al. 2000). The regions of likely structural discrepancy between bacterial and archaeal proteins, according to the structural alignment and energy minimization, are shown in ochre. Bound GDP is shown in yellow. All Cys residues, including those naturally present in IF2 and those introduced by site-directed mutagenesis, are depicted as space-filled red residues within the three-dimensional structure model of IF2 and within the bar representing the amino-terminal portion of IF2.
IF2 binds both subunits of the ribosome. Of the above-mentioned domains, GII is the best candidate for interacting with the 30S subunit, as suggested by its homology with domain II of EF-G (Moreno et al. 2000; Roll-Mecak et al. 2001) and by its localization in the molecule on the side opposite GI, which is likely involved in establishing the contact with the 50S subunit, ultimately triggering GTP hydrolysis. Additional contacts between IF2 and the 30S subunit may also involve parts of the amino-terminal domain of the factor (Moreno et al. 1999).
Using these structural and functional data as a starting point for directed chemical probing, we introduced Cys residues in the IF2 protein sequence to provide reactive side chains on which to tether chemical nucleases in the amino-terminal and GII domains, and at the junction of domains C-1 and C-2. The two chemical nucleases we used here are Fe(II)-EDTA and Cu(II)-phenanthroline, whose use as structural probes of nucleic acid structure was largely pioneered by Dervan and by Sigman. Fe(II)-EDTA generates diffusible hydroxyl radicals that have the potential to cleave rRNA even at considerable distance (Moser and Dervan 1987; Hall and Fox 1999), whereas Cu(II):1,10-orthophenanthroline (Cu:oP) interacts preferentially with bulged or stacked single-stranded regions of rRNA and generates more localized (Pope and Sigman 1984; Johnson and Nazhat 1987; Mazumder et al. 1992), and generally less-robust RNA cleavages compared with Fe-EDTA (Bowen et al. 2001).
Cysteine substitutions in IF2 and derivatization with chemical nucleases
In this study, we used B. stearothermophilus IF2 (82 kD), which is functionally interchangeable with E. coli IF2 (Brombach et al. 1986). It is able to form complexes with E. coli ribosomes and ribosomal subunits that are somewhat more stable than those formed by the homologous factor (La Teana et al. 2001). Thus, we constructed 12 active mutants, in which amino acids at specific positions of the molecule were individually replaced by a Cys residue. Five of these substitutions (at positions S17, R51, F84, F164, and G137) were introduced in the N-domain of IF2, two were introduced at positions V451 and S520, located in the middle of a β-strand (S13) and at the carboxy-terminal edge of domain GII, respectively, K540 was substituted in the first helix of domain C-1, and four were introduced in the region spanning from C-1 to C-2 (Y625, E632, M638, and E644; Fig. 1 ▶). Formation of a thioether bond between the bromo- or iodo-acetamido linker attached to the chemical nucleases and the reduced thiol of the cysteine residue tethered the chemical nucleases to these residues of the IF2 molecule. Both iron conjugated to the chemical reagent bromoacetamidobenzyl-EDTA (BABE) and copper conjugated to 5-iodoacetamido-1,10-phenanthroline are designed to yield cleavages originating from a tether length of about 13Å (Rana and Meares 1991; Chen et al. 1993; Perrin et al. 1994).
The efficiency of derivatization of the IF2 mutant proteins was assayed by the colorimetric change resulting from the accessibility of the free thiol groups to the Ellman’s reagent, 5,5′-dithiobis, 2-nitrobenzoic acid (DTNB) as described in Materials and Methods. The biological activity of the derivatized IF2 variants was tested in 70S initiation-complex formation by filter-binding assay as well as in an IF2-dependent in vitro translation assay, which indicated that the IF2 conjugates were able to promote translation to levels comparable with wild-type IF2 (see Table 1 ▶). In addition to the Cys residues introduced by site-directed mutagenesis, wild-type B. stearothermophilus IF2 contains four naturally encoded Cys residues. However, only two of these cysteines (positions 210 and 384) could theoretically be accessible to chemical modification under nondenaturing conditions, as an earlier Raman spectroscopy study showed that the two Cys residues (C668 and C714) present in the fMet-tRNA-binding domain (IF2C-2) of IF2 are not reactive (Misselwitz et al. 1999). Of the two potentially reactive and naturally occurring cysteines, one (C210) is located in the N-domain and the second (C384) in the GI domain. Our results indicate that none of these is in fact reactive with DTNB, and because no cleavage was seen from the nucleases tethered to wild-type IF2, we did not investigate this issue in greater depth.
TABLE 1.
Initiation complex formation activity of IF2 mutants expressed as a percentage of wild type activity
| Attached probes | ||||
| Domain | IF2 mutants | None | Fe-EDTA | Cu:oP |
| wt | 100 | 96.7 ± 10.4 | 84.5 ± 11.3 | |
| N | S17 | 76.7 ± 14.7 | 74.3 ± 8.7 | 100.8 ± 29.6 |
| R51 | 86.5 ± 9.3 | 73.7 ± 4.3 | 79.0 ± 8.4 | |
| F84 | 83.4 ± 12.2 | 88.0 ± 7.9 | 78.8 ± 2.0 | |
| G137 | 89.3 ± 9.2 | 100.7 ± 3.7 | 64.5 ± 9.8 | |
| F164 | 68.4 ± 10.7 | 65.7 ± 19.2 | 133.2 ± 3.0 | |
| GII | V451 | 99.9 ± 21.9 | 80.5 ± 16.0 | 96.0 ± 0.1 |
| S520 | 109.8 ± 13.9 | 94.7 ± 21.3 | 96.2 ± 2.2 | |
| C-1 | K540 | 138.1 ± 2.3 | 132.7 ± 17.3 | n.d. |
| Y625 | 144.9 ± 3.0 | 110.0 ± 2.1 | n.d. | |
| E632 | 124.2 ± 4.9 | 97.0 ± 1.9 | n.d. | |
| M638 | 85.8 ± 14.6 | 79.7 ± 1.4 | n.d. | |
| C-1/C-2 | E644 | 103.4 ± 12.5 | 98.3 ± 1.0 | n.d. |
The ability of wild-type and mutant derivatized and underivatized IF2 proteins to stimulate the binding of 35SfMet-tRNA to the 70S ribosome in the presence of IF1, IF3, and 022 mRNA was assayed by a filter-binding assay as described in Materials and Methods. The first column indicates the domains of IF2 in which the cysteine substitution was introduced. The second column specifies the position of the mutagenized amino acids. The third column shows the activity of the underivatized IF2 mutants as a percentage of the wild-type underivatized IF2. The fourth and fifth columns show activity of the Cu-oP or Fe-EDTA-derivatized proteins as a percentage of the underivatized wild-type IF2 activity.
Cleavage of rRNA by tethered nucleases
To identify the rRNA regions proximal to the bound IF2 in 30S and 70S initiation complexes, complexes were formed with derivatized IF2, and cleavage reactions were initiated by addition of reducing agent. The Fe-BABE reaction was initiated with ascorbate and hydrogen peroxide and produces diffusible hydroxyl radicals that cleave predominantly within about a 10Å radius of the iron center (Imlay and Linn 1988; Heilek et al. 1995; Heilek and Noller 1996), whereas the cleavage by the Cu:oP-derivatized proteins was initiated by addition of 3-mercaptopropionic acid (MPA) and, as mentioned above, does not cleave significantly beyond the tether length. Following the reaction, the cleavage sites were mapped by primer extension analysis using primers complementary to multiple sites along the 16S and 23S rRNA.
When wild-type IF2 or the individual mutagenized proteins were bound to 30S ribosomal subunits or to a complete 30S initiation complex, no cleavage of the 16S rRNA was detected, even though IF2 is capable of binding to both free 30S ribosomal subunits (Pon et al. 1985), and with even greater affinity, to a complete 30S initiation complex (Tomsic 2002). On the other hand, cleavage of the 16S rRNA was obtained when derivatized IF2 was bound to a 70S initiation complex.
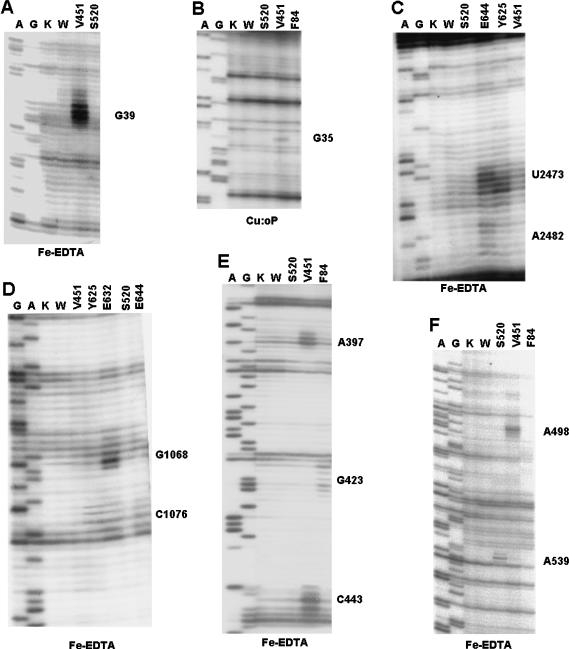
The 30S cleavages obtained are shown in the autoradiograms in Figure 2 ▶ and are summarized in the secondary structure map in Figure 3 ▶. The cleavage locations are superimposed on the structure of the small subunit of Thermus thermophilus in Figure 4 ▶ (Carter et al. 2001). The main cleavage sites from IF2 position V451C (the near GII domain) tethered with Fe-BABE were centered around positions G38-G39-C40 and A498 (Fig. 2A,F ▶), whereas the Cu:oP derivative at position 451 produced a cleavage at nucleotides G35–C36 (Fig. 2B ▶). This was the only instance in this study in which the observed Cu:oP cleavage was different from the homologous Fe:BABE cleavage. These cleavage sites are located in helices H3 and H4 and at the base of H17 (Fig. 3E ▶) and are close to each other in the three-dimensional structure of the 30S subunit (Fig. 4 ▶).
FIGURE 2.
Analysis of the cleavage sites in 16S and 23S rRNA produced by chemical nucleases tethered to IF2. Denaturing electrophoretic analysis of the primer extension products of the 16S (A,B,E,F) and 23S (C,D) rRNAs that were subjected to in situ cleavages by the IF2-tethered chemical nucleases Fe:EDTA (A,C,D–F) or Cu:oP (B). The nucleases were tethered to Cys residues naturally present or introduced by site-directed mutagenesis at the positions indicated at the tops of the lanes. (Lanes K,W) Control reaction mixtures that contained either nonderivatized wild-type IF2 (K) or the cleavage reaction carried out with derivatized wild-type IF2 (W). (Lanes A,G) Dideoxy sequencing lanes that refer to the sequence of 16S rRNA. The hydroxyl radical cleavages are seen as additional bands produced specifically by the chemical nucleases. See text for further details.
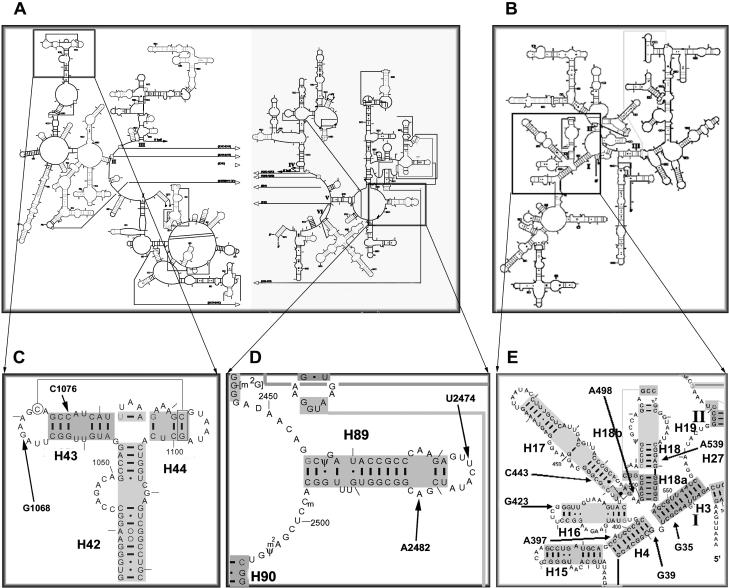
FIGURE 3.
Secondary structure locations of cleavage sites produced by IF2-tethered chemical nucleases. The regions in which the IF2-tethered nucleases produce their cleavages are indicated on secondary structure maps of 23S (A,C,D) and 16S (B,E) rRNA. Cleavages were concentrated in the thiostrepton region (C) and H89 (D) of 23S, and in the 5′ domain (E) of 16S rRNA.
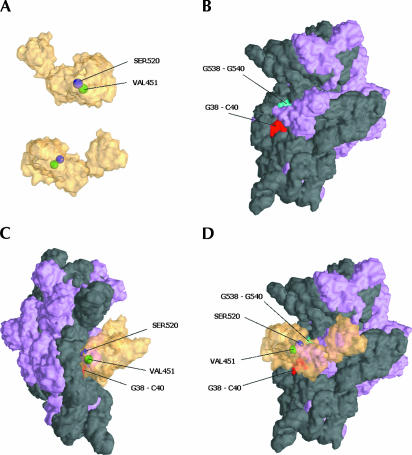
FIGURE 4.
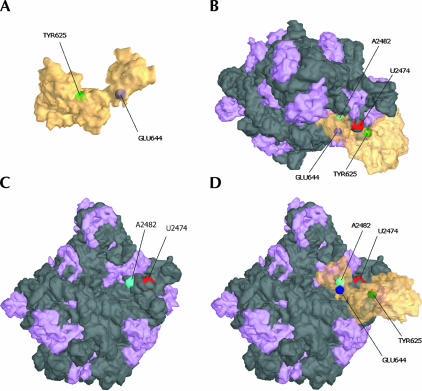
Location of cleavage sites on 16S rRNA superimposed on the three-dimensional structure of the 30S subunit. (A) Two views of IF2 (transparent yellow) with the derivatized amino acid positions S520 (purple) and V451 (green) that produced cleavage on the 16S rRNA are shown. (B) A surface representation of the interface side of the 30S subunit with rRNA shown in dark gray and ribosomal protein colored light purple (Carter et al. 2000). The major rRNA cleavage sites G38-C40 (red) and G538-G540 (green) are indicated. (C,D) Side and interface views of the 30S:IF2 docking.
Fe-BABE-derivatized F84C and S520C of IF2 also cleaved specific sites of the 16S rRNA. Fe-BABE tethered to F84C in the N-domain produced cleavages around G423 (Fig. 2E ▶) located in the apical loop of helix H16 (Fig. 3B,E ▶) in the shoulder of 30S, whereas Fe-BABE at position 520 in domain GII cleaved at nucleotides G538-A539-G540 (Fig. 2F ▶), close to the position to which IF1 has been localized (Carter et al. 2001) (Fig. 4 ▶).
On the 50S subunit, we obtained several characteristic cleavages from Fe-BABE tethered to the C-1/C-2 region. In helix 89 of 23S rRNA, cleavage was observed at positions U2474 from IF2 E644C (and weakly from Y625C) and A2482 from position E644C (Figs. 2C ▶, 3D ▶, 5 ▶). These cleavages are in agreement with previously reported IF2-dependent changes in chemical reactivity in H89 toward base-specific probes (La Teana et al. 2001). In addition, several nucleotides in the thiostrepton/L11-binding region were cleaved. G1068 was cleaved only by BABE-derivatized position E632C, whereas C1076 was cleaved by derivatized positions E632C and E644C, and weakly by position Y625C.
FIGURE 5.
Location of cleavage sites on 23S rRNA superimposed on the three-dimensional structure of the 50S subunit. (A) IF2 shown in transparent yellow with the locations of Y625 and E644 indicated in green and purple, respectively. (B, top) View of the 50S:IF2 complex. (C) A surface representation of the interface side of the 50S subunit with rRNA shown in dark gray and ribosomal protein colored light purple (Ban et al. 2000). The locations the landmark rRNA cleavage sites U2474 (red) and A2482 (green) are indicated. (D) Interface view of the 50S:IF2 interaction. Note that IF2 is transparent; amino acids shown are buried against the 50S interface.
Computer modeling of the IF2:70S initiation complex
After orienting IF2 on the 30S subunit with respect to the cleavage data, we docked IF2 on the 70S ribosome containing a P-site-bound tRNA. The docking was accomplished by first orienting the factor exactly as it was on the 30S subunit alone, then adjusting its position subtly to minimize steric clashes when the 50S subunit was added to the 30S:IF2 complex model. A natural fit was obtained by bringing the C2 domain of IF2 into proximity to the P-site tRNA on the 50S side [the tRNA was prepositioned in the P site exactly as seen in the 70S ribosome backbone crystal structure (Yusupov et al. 2001)]. This docking, based first on the 16S cleavage data, then on steric considerations at the 30S:50S interface, brought the C-1:C-2 junction in proximity to helix 89, the site of our 23S cleavage. This fit also placed IF2’s GI domain in proximity of the α-sarcin loop, in agreement with chemical structure-probing data described above (La Teana et al. 2001). The simultaneous proximity of the cleavage sites on the 16S and 23S rRNA and the protection sites on the 23S strongly supports our docking of IF2 on the 70S ribosome. The proximity of the C2 domain with the -CCA terminus of a P-site-bound tRNA provided yet another solid docking constraint. Significantly, the only residual steric clash remaining in the model concerns a small region of the carboxyl terminus of ribosomal protein S12 and the junction of the IF2 GII and C1 domains. Because a degree of conformational flexibility is likely for both IF2, and perhaps also for this external segment of S12, we do not believe this steric conflict represents a serious challenge to our placement of IF2.
DISCUSSION
To better understand the mechanism of action of IF2 during initiation of protein synthesis, we investigated the binding site(s) of this factor on the ribosome by tethered nuclease probing from IF2. The results obtained represent the first unambiguous mapping of IF2 on the 30S ribosomal subunit. The position of IF2, and more precisely, of its GII domain on the small ribosomal subunit is very similar to that of fusidic acid-stalled EF-G (Agrawal et al. 1998), a similarity further supported by the extensive overlap between the nucleotides cleaved by Fe-EDTA tethered to EF-G and IF2. In fact, the same 16S rRNA cleavage is obtained from nucleases tethered to Cys residues replacing Ser 520 and Val 451 of IF2 and Gly 301 and Glu 314 of EF-G (Wilson and Noller 1998), which are located in almost identical positions of domain II of the two factors (Brock et al. 1998).
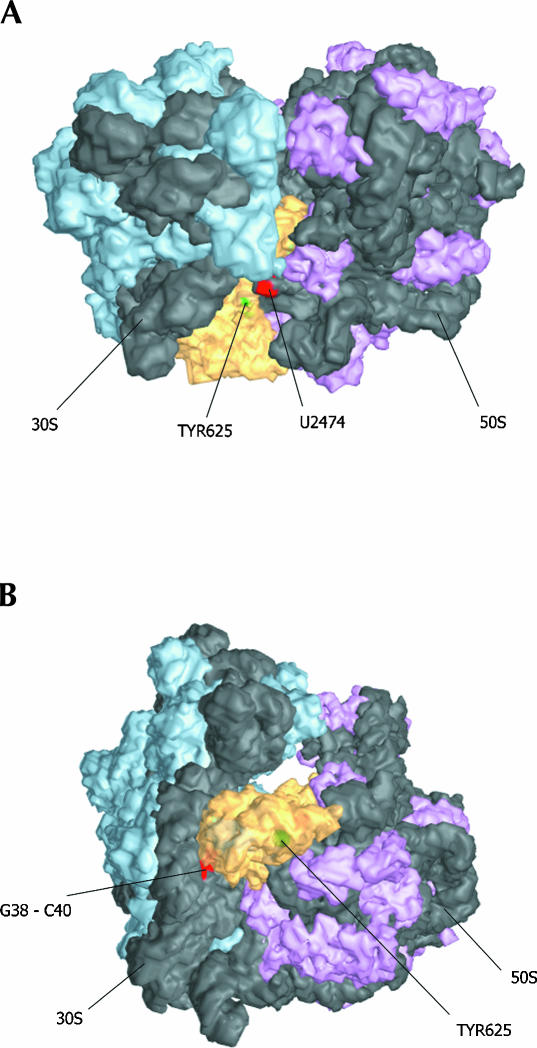
The docking of IF2 here (Fig. 6 ▶) pertains specifically to the position of IF2 within a 70S initiation complex, because, as mentioned above, the 16S rRNA was not cleaved by IF2-tethered nucleases within a 30S initiation complex. The rRNA positions cleaved by the IF2-tethered chemical nucleases highlighted in Figures 4 ▶ and 5 ▶ are in close proximity to the derivatized positions of IF2. In addition to the cleavage data, we used other constraints to validate the docking. We have placed IF2 such that the C-2 domain is very close to the acceptor end of a P-site-bound tRNA (Yusupov et al. 2001), as the molecular determinants for the recognition and binding of fMet-tRNA are located in C-2 (Spurio et al. 2000), and, almost exclusively, within the acceptor end of initiator tRNA (Guenneugues et al. 2000). Thus, in Figure 4 ▶, IF2 is oriented on the 30S subunit so that an approaching 50S subunit would contact IF2 C-2 with its peptidyl transferase center and the sarcin-ricin loop of 23S would be shielded by the GI domain of IF2, in agreement with published chemical protection data (La Teana et al. 2001). The proposed localization of IF2 on the 50S subunit is shown in Figure 5 ▶. It is notable that this docking of IF2 on the 70S ribosome is reminiscent of a similar docking of EF-G on the 50S subunit (Ban et al. 1999). Furthermore, Dahlberg and coworkers recently demonstrated the ability of IF2 to compete with EF-G for binding on the ribosome, suggesting an overlapping binding site (Cameron et al. 2002).
FIGURE 6.
Docking of IF2 on the 70S ribosome. (A) View from the top of the ribosome with several sites of cleavage indicated. Large subunit ribosomal protein density is colored purple, small subunit protein is shown in cyan; rRNA is dark gray. (B) View from the leading edge of the ribosome, facing the subunit interface. This 70S model was assembled from the 30S, 50S, and 70S crystal structures (Ban et al. 2000; Wimberly et al. 2000; Carter et al. 2001; Yusupov et al. 2001) as described in Materials and Methods.
Although the precise structure of the amino-terminal domain of B. stearothermophilus IF2 is not known, the cleavage of 16S rRNA residues in the 421–425 region places it at the shoulder of the leading edge of the 30S subunit. Because this IF2 region is dispensable for translational functions in E. coli, the significance of this interaction is still unclear.
Additional proximity relationships between IF2 and fMet-tRNA that are suggested by footprinting, cross-linking, and proteolysis data, were useful in the development of our model. For example, parts of the elbow region of the tRNA and part of the anticodon arm were found to be shielded by IF2 within an IF-2/fMet-tRNA complex and within a 30S initiation complex, whereas the anticodon could be cross-linked to the N-domain of IF2. Furthermore, residues within the GII peptide Asn 611–Arg 645 of E. coli IF2 (corresponding to Asn 464–Glu 498 of B. stearothermophilus) were found to be close to the elbow region of the tRNA (Wakao et al. 1989; Yusupova et al. 1996). Finally, binding of fMet-tRNA to B. stearothermophilus IF2 was found to protect the Arg 308–Ala 309 bond, located in the GTP-binding domain GI, and, more weakly, the Arg 519–Ser 520 bond in the GII domain from trypsin digestion (Severini et al. 1992).
Thus, whereas the present data indicate that IF2 GII is localized on the 30S subunit within a 70S initiation complex, the cross-linking data place this domain close to the elbow of the initiator tRNA in both IF2/fMet-tRNA binary complex and in 30S initiation complexes. Because the distance between the cleavage sites originating from IF2 GII in the 16S rRNA and in the elbow of P-site-bound fMet-tRNA appears to be too large to allow GII to establish both contacts and/or proximity relationships with both sites at the same time, we must draw the conclusion that the ribosomal localization of the IF2–fMet-tRNA complex changes during the transition from 30S to 70S initiation complex. Drawing from fast kinetic analysis of the late events of translation initiation (Tomsic et al. 2000), as well as our failure here to detect any cleavage of the 16S rRNA in the IF2:30S complex, it appears that a conformational change involving the ribosomal subunits and IF2 accompanies the transition from 30S to 70S initiation complex.
These considerations suggest that, when bound within a 30S initiation complex, IF2 occupies a position somewhat different from that depicted in Figures 4 ▶ and 6 ▶. Moving the GII domain closer to the elbow of the tRNA, and thus more centrally located on the interface side of the 30S subunit compared with Figure 4 ▶, could account for at least some of the earlier protein–protein cross-linking data such as IF2-IF1 (Boileau et al. 1983), IF2-S12, and possibly IF2-S13 and IF2-S19 (Bollen et al. 1975).
In conclusion, we suggest the following general scenario. In solution and during initial interactions with the 30S subunit, IF2 contacts fMet-tRNA through an interaction between its C-2 domain and fMet-ACCAAC, allowing contacts/proximity between GII domain and the T stem and between the N-domain and the anticodon stem. On the 30S subunit, IF2 is near or in direct contact with ribosomal protein S12 and IF1 (which strengthens the affinity of IF2 for the ribosome and occludes the A site). Upon joining of the 50S subunit with the 30S initiation complex, the GII of IF2 moves toward the edge of the 30S ribosomal subunit, away from the elbow of fMet-tRNA, losing some contacts with the IF1-binding region and favoring the ejection of IF1 from the 30S subunit, whereas its GI domain binds in the vicinity of the factor-binding center of the 50S subunit before leaving the 70S initiation complex in a state suitable for the subsequent binding of the EF-Tu–aminoacyl-tRNA–GTP ternary complex.
MATERIALS AND METHODS
Preparation of IF2 mutants
Individual Cys residues were introduced in IF2 by oligonucleotide-directed mutagenesis of B. stearothermophilus infB as described previously (Spurio et al. 2000). After confirming the mutations by DNA sequencing, the mutagenized genes were subcloned into pPLc 2833, transformed in E. coli UT5600. The overexpressed proteins were purified essentially as described previously (Spurio et al. 2000). Protein purity as judged by SDS-PAGE was ≥95%. Protein concentration was determined by the Bradford colorimetric assay (Bradford 1976). The purified proteins were kept at −80°C in storage buffer [20mM Tris-HCl (pH 7.1), 200 mM NH4Cl, 6 mM β-mercaptoethanol, 0.1 mM EDTA, 10% glycerol).
Preparation of mRNA and fMet-tRNA
Model 022 mRNA was transcribed in vitro by T7 RNA polymerase and purified as described by La Teana et al. (1993). fMet-tRNA and 35S-labeled fMet-tRNA were prepared and purified as described previously (Rodnina et al. 1994).
IF2-dependent in vitro translation
IF2-dependent in vitro translation of 022 mRNA by E. coli MRE600 high-salt washed 70S ribosomes was performed essentially as described (La Teana et al. 1993; Grill et al. 2000), except that no DTT was added when derivatized proteins were tested.
Preparation of Cu(II):phenanthroline and Fe(II)-BABE-derivatized IF2
Conjugation of Fe(II)-BABE to IF2 was performed essentially as described in Culver et al. (1999). A total of 2 μL 1 M DTT were added to 1 mL of IF2 solution (1 μg/μL in storage buffer). After a 15-min incubation at 4°C, the buffer was exchanged with prefiltered modification buffer [30 mM Tris-HCl (pH 7.1), 800 mM KCl] using Microcon YM30 (Millipore) speed filter tubes, and the IF2 proteins were concentrated threefold before being derivatized with either Cu(II):phenanthroline or Fe(II)-BABE, following different derivatization procedures (below).
Fe(II)-BABE derivatization
A total of 20 μL of freshly made 50 mM Fe(NH4)2(SO4)2 were mixed with 100 μL 11 mM BABE (Dojindo Molecular Technologies, Inc.) in 100 mM NaOAc, and incubated for 40 min at 22°C before addition of 6 μL 50 mM EDTA (pH 7.1); the resulting Fe(II)-BABE solution was incubated for an additional 20 min at 22°C, and then kept on ice until use. An equal volume (120 μL) of Fe(II)-BABE and of reduced IF2 solution (~3 μg/μL) were mixed with 760 μL Modification buffer containing 0.01% Triton X-100, and protein derivatization was allowed to continue for 30 min at 22°C.
Phenanthroline derivatization
A total of 100 μL of phenanthroline (Molecular Probes) solution (10 mM in 100% DMSO) were mixed with 120 μL of reduced IF2 (~3 μg/μL) and with 780 μL of Modification buffer containing 0.01 % Triton X-100. Complete reaction of the thiol groups was achieved after a 2-h incubation in the dark at 22° C.
Sample preparation and DTNB assay
To remove the free reagents from the modified protein, the samples derivatized with Fe(II)-BABE were dialyzed exhaustively against protein storage buffer (with no EDTA and no glycerol), and then washed several times with storage buffer (with no EDTA and no glycerol) on Microcon YM30 (Millipore); the samples derivatized with phenanthroline were purified on a G-25 Sephadex (1.5 mL) spin column, and then washed several times with storage buffer (with no EDTA) on Microcon YM30 (Millipore). Before use, the phenanthroline-derivatized proteins were incubated for 2 min on ice with a 20-fold molar excess of Cu(SO4)2.
The extent of protein modification was determined by comparing the reactivity of the free thiols of the protein before and after derivatization using the Ellman’s reagent 5,5′-dithio-bis-[2-nitrobenzoic acid] (DTNB). This reaction was carried out by adding 10 μL of DTNB (4 mg/mL in 100 mM Na2HPO4) to 90 μL of ~11.1 μM protein solution (fc ~10μM), and after 10 min incubation at 22°C, quantifying the colored chromophore formed upon reaction of DTNB with the sulfhydryl group by determining the A412 nm (taking the absorbance of the modification buffer as the baseline) before measuring the absorbance. The extinction coefficient ɛ412 of the chromophore 2-nitro-5-thiobenzoate anion (TNB2−) was taken to be = 13,600 M−1 cm−1 (Riddles et al. 1983).
Initiation complex formation
After preincubating the ribosomes (20 min at 42°C) in 10 mM Tris-HCl (pH 7.4) buffer containing MgAc215 mM and KCl 100 mM to activate them, the 30S or 70S initiation complexes were prepared by mixing, in 30 μL final volume, 50 pmole of 30S subunits (or 70S ribosomes), 50 pmole each of IF1 and IF3, 50 pmole fMet-tRNA (or 35S-labeled fMet-tRNA), 100 pmole of 022 mRNA and 50 pmole of IF2. The conditions under which the initiation complexes are formed are as follows: 50 mM Tris-HCl (pH 7.7), 80 mM NH4Cl; 7.5 mM MgAc2, and 1 mM GTP. After a 10-min incubation at 37°C, the initiation complexes were filtered on 0.45-μm membrane filters (Schleicher & Schuell NC45, 24 mm diameter), or alternatively placed on ice, where the subsequent cleavage reaction was carried out. For the filter-binding assay activity test, the filters were washed twice with a 10 mM Tris-HCl (pH 7.1) buffer containing MgAc2 7 mM; NH4Cl 100 mM, and β-mercaptoethanol 2 mM, and then counted with by liquid scintillation.
Fe(II)-BABE cleavage
A total of 1 μL of 250 mM ascorbic acid and 1 μL of 2.5% H2O2 were placed on the wall of a tube containing 30 μL of 30S or 70S initiation complexes, which was pulse centrifuged to start the Fenton reaction. After 15 min on ice, the reaction was stopped by addition of 100 μL of Stop solution (NaOAc 300 mM, SDS 2%, 30% glycerol), quickly followed by phenol extraction of the rRNAs.
Phenanthroline cleavage
A total of 1 μL of 140 mM mercaptopropionic acid (MPA) was added to the initiation complexes containing the IF2-phenanthroline derivatives. The cleavage reaction, carried out for 3 h on ice, was stopped by addition of 100 μL of stop solution before the phenol extraction of the rRNA.
Computer modeling
The docking of IF2 on 30S subunits and 70S ribosomes was carried out to a first approximation by positioning the amino acid residues on the crystal structure of IF2 (Roll-Mecak et al. 2000) that we derivatized with chemical nucleases close to those nucleotides that were cleaved in our assays on the crystal structures of the 30S subunit (Schluenzen et al. 2000; Wimberly et al. 2000) using Swiss PDB Viewer (Guex and Peitsch 1997). Although it is possible that interdomain movement can occur in IF2, we did not attempt conformational changes in IF2 as we docked it to the ribosome.
The model was refined and adapted to fit the 70S structure using Ribosome Builder software (WK, WEH, and JSL, to be described elsewhere) as follows. First, the high-resolution structures for the 30S (Schluenzen et al. 2000; Wimberly et al. 2000) and 50S (Ban et al. 2000) subunits were aligned by superimposing them on the published backbone structure of the 70S ribosome (Yusupov et al. 2001). Then, starting from the IF2:30S complex, the position of IF2 was adjusted to avoid steric conflicts with the 50S subunit, which brought the GI domain of IF2 in proximity to H89 and the sarcin-ricin loop of 23S.
Acknowledgments
We thank Martha J. Rice for technical support, and Jing Yuan, Scott Hennelly, Bill Bowen, and Jerneja Tomsic for fruitful discussions and suggestions. Financial support of the Italian MURST (PRIN 2000) to C.O.G. and NIH Grant # GM35717 to W.E.H. is gratefully acknowledged.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.rnajournal.org/cgi/doi/10.1261/rna.2116303.
REFERENCES
- Abel, K., Yoder, M.D., Hilgenfeld, R., and Jurnak, F. 1996. An α to β conformational switch in EF-Tu. Structure 4: 1153–1159. [DOI] [PubMed] [Google Scholar]
- Ævarsson, A., Brazhnikov, E., Garber, M., Zheltonosova, J., Chirgadze, Y., al-Karadaghi, S., Svensson, L.A., and Liljas, A. 1994. Three-dimensional structure of the ribosomal translocase: Elongation factor G from Thermus thermophilus. EMBO J. 13: 3669–3677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agrawal, R.K., Penczek, P., Grassucci, R.A., and Frank, J. 1998. Visualization of elongation factor G on the Escherichia coli 70S ribosome: The mechanism of translocation. Proc. Natl. Acad. Sci. 95: 6134–6138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agrawal, R.K., Heagle, A.B., Penczek, P., Grassucci, R.A., and Frank, J. 1999. EF-G-dependent GTP hydrolysis induces translocation accompanied by large conformational changes in the 70S ribosome. Nat. Struct. Biol. 6: 643–647. [DOI] [PubMed] [Google Scholar]
- al-Karadaghi, S., Aevarsson, A., Garber, M., Zheltonosova, J., and Liljas, A. 1996. The structure of elongation factor G in complex with GDP: Conformational flexibility and nucleotide exchange. Structure 4: 555–565. [DOI] [PubMed] [Google Scholar]
- Ban, N., Nissen, P., Hansen, J., Capel, M., Moore, P.B., and Steitz, T.A. 1999. Placement of protein and RNA structures into a 5 Å-resolution map of the 50S ribosomal subunit. Nature 400: 841–847. [DOI] [PubMed] [Google Scholar]
- Ban, N., Nissen, P., Hansen, J., Moore, P.B., and Steitz, T.A. 2000. The complete atomic structure of the large ribosomal subunit at 2.4 Å resolution. Science 289: 905–920. [DOI] [PubMed] [Google Scholar]
- Berchtold, H., Reshetnikova, L., Reiser, C.O., Schirmer, N.K., Sprinzl, M., and Hilgenfeld, R. 1993. Crystal structure of active elongation factor Tu reveals major domain rearrangements. Nature 365: 126–132. [DOI] [PubMed] [Google Scholar]
- Biou, V., Shu, F., and Ramakrishnan, V. 1995. X-ray crystallography shows that translational initiation factor IF3 consists of two compact α/β domains linked by an α-helix. EMBO J. 14: 4056–4064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boelens, R. and Gualerzi, C.O. 2002. Stucture and function of bacterial initiation factors. Curr. Prot. Pep. Sci. 3: 107–119. [DOI] [PubMed] [Google Scholar]
- Boileau, G., Butler, P., Hershey, J.W., and Traut, R.R. 1983. Direct cross-links between initiation factors 1, 2, and 3 and ribosomal proteins promoted by 2-iminothiolane. Biochemistry 22: 3162–3170. [DOI] [PubMed] [Google Scholar]
- Bollen, A., Heimark, R.L., Cozzone, A., Traut, R.R., and Hershey, J.W. 1975. Cross-linking of initiation factor IF-2 to Escherichia coli 30 S ribosomal proteins with dimethylsuberimidate. J. Biol. Chem. 250: 4310–4314. [PubMed] [Google Scholar]
- Bowen, W.S., Hill, W.E., and Lodmell, J.S. 2001. Comparison of rRNA cleavage by complementary 1,10-phenanthroline-Cu(II)- and EDTA-Fe(II)-derivatized oligonucleotides. Methods 25: 344–350. [DOI] [PubMed] [Google Scholar]
- Bradford, M.M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72: 248–254. [DOI] [PubMed] [Google Scholar]
- Brock, S., Szkaradkiewicz, K., and Sprinzl, M. 1998. Initiation factors of protein biosynthesis in bacteria and their structural relationship to elongation and termination factors. Mol. Microbiol. 29: 409–417. [DOI] [PubMed] [Google Scholar]
- Brombach, M., Gualerzi, C.O., Nakamura, Y., and Pon, C.L. 1986. Molecular cloning and sequence of the Bacillus stearothermophilus translational initiation factor IF2 gene. Mol. Gen. Genet. 205: 97–102. [DOI] [PubMed] [Google Scholar]
- Cameron, D.M., Thompson, J., March, P.E., and Dahlberg, A.E. 2002. Initiation factor IF2, thiostrepton and micrococcin prevent the binding of elongation factor G to the Escherichia coli ribosome. J. Mol. Biol. 319: 27–35. [DOI] [PubMed] [Google Scholar]
- Carter, A.P., Clemons, W.M., Brodersen, D.E., Morgan-Warren, R.J., Wimberly, B.T., and Ramakrishnan, V. 2000. Functional insights from the structure of the 30S ribosomal subunit and its interactions with antibiotics. Nature 407: 340–348. [DOI] [PubMed] [Google Scholar]
- Carter, A.P., Clemons Jr., W.M., Brodersen, D.E., Morgan-Warren, R.J., Hartsch, T., Wimberly, B.T., and Ramakrishnan, V. 2001. Crystal structure of an initiation factor bound to the 30S ribosomal subunit. Science 291: 498–501. [DOI] [PubMed] [Google Scholar]
- Cenatiempo, Y., Deville, F., Dondon, J., Grunberg-Manago, M., Sacerdot, C., Hershey, J.W., Hansen, H.F., Petersen, H.U., Clark, B.F., Kjeldgaard, M., et al. 1987. The protein synthesis initiation factor 2 G-domain. Study of a functionally active C-terminal 65-kilodalton fragment of IF2 from Escherichia coli. Biochemistry 26: 5070–5076. [DOI] [PubMed] [Google Scholar]
- Chen, C.B., Gorin, M.B., and Sigman, D.S. 1993. Sequence-specific scission of DNA by the chemical nuclease activity of 1,10-phenanthroline-copper(I) targeted by RNA. Proc. Natl. Acad. Sci. 90: 4206–4210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Culver, G.M., Heilek, G.M., and Noller, H.F. 1999. Probing the rRNA environment of ribosomal protein S5 across the subunit interface and inside the 30 S subunit using tethered Fe(II). J. Mol. Biol. 286: 355–364. [DOI] [PubMed] [Google Scholar]
- Czworkowski, J., Wang, J., Steitz, T.A., and Moore, P.B. 1994. The crystal structure of elongation factor G complexed with GDP, at 2.7 Å resolution. EMBO J. 13: 3661–3668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dallas, A. and Noller, H.F. 2001. Interaction of translation initiation factor 3 with the 30S ribosomal subunit. Mol. Cell 8: 855–864. [DOI] [PubMed] [Google Scholar]
- Garcia, C., Fortier, P.L., Blanquet, S., Lallemand, J.Y., and Dardel, F. 1995a. 1H and 15N resonance assignments and structure of the N-terminal domain of Escherichia coli initiation factor 3. Eur. J. Biochem. 228: 395–402. [PubMed] [Google Scholar]
- ———. 1995b. Solution structure of the ribosome-binding domain of E. coli translation initiation factor IF3. Homology with the U1A protein of the eukaryotic spliceosome. J. Mol. Biol. 254: 247–259. [DOI] [PubMed] [Google Scholar]
- Grill, S., Gualerzi, C.O., Londei, P., and Blasi, U. 2000. Selective stimulation of translation of leaderless mRNA by initiation factor 2: Evolutionary implications for translation. EMBO J. 19: 4101–4110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gualerzi, C.O., Severini, M., Spurio, R., La Teana, A., and Pon, C.L. 1991. Molecular dissection of translation initiation factor IF2. Evidence for two structural and functional domains. J. Biol. Chem. 266: 16356–16362. [PubMed] [Google Scholar]
- Gualerzi, C.O., Brandi, L., Caserta, E., La Teana, A., Spurio, R., Tomsic, J., and Pon, C.L. 2000. Translation initiation in bacteria. In The ribosome: Structure, function, antibiotics, and cellular interactions (eds. R.A. Garrett et al.), pp. 477–494. ASM Press, Washington, D.C.
- Gualerzi, C.O., Brandi, L., Caserta, E., Garofolo, C., Lammi, M., La Teana, A., Petrelli, D., Spurio, R., Tomsic, J., and Pon, C.L. 2001. Initiation factors in the early events of mRNA translation in bactera. Cold Spring Harbor Symp. Quant. Biol. 66: 363–376. [DOI] [PubMed] [Google Scholar]
- Guenneugues, M., Caserta, E., Brandi, L., Spurio, R., Meunier, S., Pon, C.L., Boelens, R., and Gualerzi, C.O. 2000. Mapping the fMet-tRNA(f)(Met) binding site of initiation factor IF2. EMBO J. 19: 5233–5240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guex, N and Peitsch, M.C. 1997. SWISS-MODEL and the SwissPdbViewer: An environment for comparative protein modeling. Electrophoresis 18: 2714–2723. [DOI] [PubMed] [Google Scholar]
- Hall, K.B. and Fox, R.O. 1999. Directed cleavage of RNA with protein-tethered EDTA-Fe. Methods 18: 78–84. [DOI] [PubMed] [Google Scholar]
- Heilek, G.M. and Noller, H.F. 1996. Site-directed hydroxyl radical probing of the rRNA neighborhood of ribosomal protein S5. Science 272: 1659–1662. [DOI] [PubMed] [Google Scholar]
- Heilek, G.M., Marusak, R., Meares, C.F., and Noller, H.F. 1995. Directed hydroxyl radical probing of 16S rRNA using Fe(II) tethered to ribosomal protein S4. Proc. Natl. Acad. Sci. 92: 1113–1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heimark, R.L., Kahan, L., Johnston, K., Hershey, J.W., and Traut, R.R. 1976. Cross-linking of initiation factor IF3 to proteins of the Escherichia coli 30 S ribosomal subunit. J. Mol. Biol. 105: 219–230. [DOI] [PubMed] [Google Scholar]
- Imlay, J.A. and Linn, S. 1988. DNA damage and oxygen radical toxicity. Science 240: 1302–1309. [DOI] [PubMed] [Google Scholar]
- Johnson, G.R.A. and Nazhat, N.B. 1987. Kinetics and mechanism of the reaction of the Bis(1,10-phenanthroline)copper(I) Ion with hydrogen peroxide in aqueous solution. J. Am. Chem. Soc. 109: 1990–1994. [Google Scholar]
- Kawashima, T., Berthet-Colominas, C., Wulff, M., Cusack, S., and Leberman, R. 1996. The structure of the Escherichia coli EF-Tu.EF-Ts complex at 2.5 Å resolution. Nature 379: 511–518. [DOI] [PubMed] [Google Scholar]
- Kjeldgaard, M. and Nyborg, J. 1992. Refined structure of elongation factor EF-Tu from Escherichia coli. J. Mol. Biol. 223: 721–742. [DOI] [PubMed] [Google Scholar]
- Kjeldgaard, M., Nissen, P., Thirup, S., and Nyborg, J. 1993. The crystal structure of elongation factor EF-Tu from Thermus aquaticus in the GTP conformation. Structure 1: 35–50. [DOI] [PubMed] [Google Scholar]
- Krafft, C., Diehl, A., Laettig, S., Behlke, J., Heinemann, U., Pon, C.L., Gualerzi, C.O., and Welfle, H. 2000. Interaction of fMet-tRNA(fMet) with the C-terminal domain of translational initiation factor IF2 from Bacillus stearothermophilus. FEBS Lett. 471: 128–132. [DOI] [PubMed] [Google Scholar]
- La Teana, A., Pon, C.L., and Gualerzi, C.O. 1993. Translation of mRNAs with degenerate initiation triplet AUU displays high initiation factor 2 dependence and is subject to initiation factor 3 repression. Proc. Natl. Acad. Sci. 90: 4161–4165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Teana, A., Gualerzi, C.O., and Dahlberg, A.E. 2001. Initiation factor IF 2 binds to the alpha-sarcin loop and helix 89 of Escherichia coli 23S ribosomal RNA. RNA 7: 1173–1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lancaster, L., Kiel, M.C., Kaji, A., and Noller, H.F. 2002. Orientation of ribosome recycling factor in the ribosome from directed hydroxyl radical probing. Cell 111: 129–140. [DOI] [PubMed] [Google Scholar]
- Laursen, B.S., Mortensen, K.K., Sperling-Petersen, H.U., and Hoffman, D.W. 2003. A conserved structural motif at the N-terminus of bacterial translation initiation factor IF2. J. Biol. Chem. 24: 24. [DOI] [PubMed] [Google Scholar]
- Lieberman, K.R., Firpo, M.A., Herr, A.J., Nguyenle, T., Atkins, J.F., Gesteland, R.F., and Noller, H.F. 2000. The 23 S rRNA environment of ribosomal protein L9 in the 50 S ribosomal subunit. J. Mol. Biol. 297: 1129–1143. [DOI] [PubMed] [Google Scholar]
- Mazumder, A., Chen, C.B., Gaynor, R., and Sigman, D.S. 1992. 1,10-Phenanthroline-copper, a footprinting reagent for single-stranded regions of RNAs. Biochem. Biophys. Res. Commun. 187: 1503–1509. [DOI] [PubMed] [Google Scholar]
- McCutcheon, J.P., Agrawal, R.K., Philips, S.M., Grassucci, R.A., Gerchman, S.E., Clemons Jr., W.M., Ramakrishnan, V., and Frank, J. 1999. Location of translational initiation factor IF3 on the small ribosomal subunit. Proc. Natl. Acad. Sci. 96: 4301–4306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meunier, S., Spurio, R., Czisch, M., Wechselberger, R., Guenneugues, M., Gualerzi, C.O., and Boelens, R. 2000. Structure of the fMet-tRNA(fMet)-binding domain of B. stearothermophilus initiation factor IF2. EMBO J. 19: 1918–1926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misselwitz, R., Welfe, K., Krafft, C., Gualerzi, C.O., and Welfle, H. 1997. Translational initiation factor IF2 from Bacillus stearothermophilus: A spectroscopic and microcalorimetric study of the C-domain. Biochemistry 36: 3170–3178. [DOI] [PubMed] [Google Scholar]
- Misselwitz, R., Welfle, K., Krafft, C., Welfle, H., Brandi, L., Caserta, E., and Gualerzi, C.O. 1999. The fMet-tRNA binding domain of translational initiation factor IF2: Role and environment of its two Cys residues. FEBS Lett. 459: 332–336. [DOI] [PubMed] [Google Scholar]
- Moazed, D., Robertson, J.M., and Noller, H.F. 1988. Interaction of elongation factors EF-G and EF-Tu with a conserved loop in 23S RNA. Nature 334: 362–364. [DOI] [PubMed] [Google Scholar]
- Moazed, D., Samaha, R.R., Gualerzi, C., and Noller, H.F. 1995. Specific protection of 16 S rRNA by translational initiation factors. J. Mol. Biol. 248: 207–210. [DOI] [PubMed] [Google Scholar]
- Moreno, J.M., Drskjotersen, L., Kristensen, J.E., Mortensen, K.K., and Sperling-Petersen, H.U. 1999. Characterization of the domains of E. coli initiation factor IF2 responsible for recognition of the ribosome. FEBS Lett. 455: 130–134. [DOI] [PubMed] [Google Scholar]
- Moreno, J.M., Sorensen, H.P., Mortensen, K.K., and Sperling-Petersen, H.U. 2000. Macromolecular mimicry in translation initiation: A model for the initiation factor IF2 on the ribosome. IUBMB Life 50: 347–354. [DOI] [PubMed] [Google Scholar]
- Moser, H.E. and Dervan, P.B. 1987. Sequence-specific cleavage of double helical DNA by triple helix formation. Science 238: 645–650. [DOI] [PubMed] [Google Scholar]
- Nissen, P., Kjeldgaard, M., Thirup, S., Polekhina, G., Reshetnikova, L., Clark, B.F., and Nyborg, J. 1995. Crystal structure of the ternary complex of Phe-tRNAPhe, EF-Tu, and a GTP analog. Science 270: 1464–1472. [DOI] [PubMed] [Google Scholar]
- Nissen, P., Thirup, S., Kjeldgaard, M., and Nyborg, J. 1999. The crystal structure of Cys-tRNACys-EF-Tu-GDPNP reveals general and specific features in the ternary complex and in tRNA. Structure Fold Des. 7: 143–156. [DOI] [PubMed] [Google Scholar]
- Perrin, D.M., Mazumder, A., Sadeghi, F., and Sigman, D.S. 1994. Hybridization of a complementary ribooligonucleotide to the transcription start site of the lacUV-5-Escherichia coli RNA polymerase open complex. Potential for gene-specific inactivation reagents. Biochemistry 33: 3848–3854. [DOI] [PubMed] [Google Scholar]
- Pioletti, M., Schlunzen, F., Harms, J., Zarivach, R., Gluhmann, M., Avila, H., Bashan, A., Bartels, H., Auerbach, T., Jacobi, C., et al. 2001. Crystal structures of complexes of the small ribosomal subunit with tetracycline, edeine and IF3. EMBO J. 20: 1829–1839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polekhina, G., Thirup, S., Kjeldgaard, M., Nissen, P., Lippmann, C., and Nyborg, J. 1996. Helix unwinding in the effector region of elongation factor EF-Tu-GDP. Structure 4: 1141–1151. [DOI] [PubMed] [Google Scholar]
- Pon, C.L., Paci, M., Pawlik, R.T., and Gualerz, C.O. 1985. Structure-function relationship in Escherichia coli initiation factors. Biochemical and biophysical characterization of the interaction between IF-2 and guanosine nucleotides. J. Biol. Chem. 260: 8918–8924. [PubMed] [Google Scholar]
- Pope, L.E. and Sigman, D.S. 1984. Secondary structure specificity of the nuclease activity of the 1,10-phenanthroline-copper complex. Proc. Natl. Acad. Sci. 81: 3–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rana, T.M. and Meares, C.F. 1991. Transfer of oxygen from an artificial protease to peptide carbon during proteolysis. Proc. Natl. Acad. Sci. 88: 10578–10582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riddles, P.W., Blakeley, R.L., and Zerner, B. 1983. Reassessment of Ellman’s reagent. Methods Enzymol. 91: 49–60. [DOI] [PubMed] [Google Scholar]
- Rodnina, M.V., Semenkov, Y.P., and Wintermeyer, W. 1994. Purification of fMet-tRNA(fMet) by fast protein liquid chromatography. Anal. Biochem. 219: 380–381. [DOI] [PubMed] [Google Scholar]
- Roll-Mecak, A., Cao, C., Dever, T.E., and Burley, S.K. 2000. X-Ray structures of the universal translation initiation factor IF2/eIF5B: Conformational changes on GDP and GTP binding. Cell 103: 781–792. [DOI] [PubMed] [Google Scholar]
- Roll-Mecak, A., Shin, B.S, Dever, T.E., and Burley, S.K. 2001. Engaging the ribosome: Universal IFs of translation. Trends Biochem. Sci. 26: 705–709. [DOI] [PubMed] [Google Scholar]
- Schluenzen, F., Tocilj, A., Zarivach, R., Harms, J., Gluehmann, M., Janell, D., Bashan, A., Bartels, H., Agmon, I., Franceschi, F., et al. 2000. Structure of functionally activated small ribosomal subunit at 3.3 Å resolution. Cell 102: 615–623. [DOI] [PubMed] [Google Scholar]
- Selmer, M., Al-Karadaghi, S., Hirokawa, G., Kaji, A., and Liljas, A. 1999. Crystal structure of Thermotoga maritima ribosome recycling factor:Aa tRNA mimic. Science 286: 2349–2352. [DOI] [PubMed] [Google Scholar]
- Sette, M., van Tilborg, P., Spurio, R., Kaptein, R., Paci, M., Gualerzi, C.O., and Boelens, R. 1997. The structure of the translational initiation factor IF1 from E. coli contains an oligomer-binding motif. EMBO J. 16: 1436–1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Severini, M., Choli, T., La Teana, A., and Gualerzi, C.O. 1992. Proteolysis of Bacillus stearothermophilus IF2 and specific protection by fMet-tRNA. FEBS Lett. 297: 226–228. [DOI] [PubMed] [Google Scholar]
- Spurio, R., Brandi, L., Caserta, E., Pon, C.L., Gualerzi, C.O., Misselwitz, R., Krafft, C., Welfle, K., and Welfle, H. 2000. The C-terminal subdomain (IF2 C-2) contains the entire fMet-tRNA binding site of initiation factor IF2. J. Biol. Chem. 275: 2447–2454. [DOI] [PubMed] [Google Scholar]
- Stark, H., Rodnina, M.V., Rinke-Appel, J., Brimacombe, R., Wintermeyer, W., and van Heel, M. 1997. Visualization of elongation factor Tu on the Escherichia coli ribosome. Nature 389: 403–406. [DOI] [PubMed] [Google Scholar]
- Stark, H., Rodnina, M.V., Wieden, H.J., van Heel, M., and Wintermeyer, W. 2000. Large-scale movement of elongation factor G and extensive conformational change of the ribosome during translocation. Cell 100: 301–309. [DOI] [PubMed] [Google Scholar]
- Tomsic, J. 2002. “Role of IF2 dependent GTPase during late events of procaryotic translation initiation.” Ph.D. thesis, University of Camerino, Camerino, Italy.
- Tomsic, J., Vitali, L.A., Daviter, T., Savelsbergh, A., Spurio, R., Striebeck, P., Wintermeyer, W., Rodnina, M.V., and Gualerzi, C.O. 2000. Late events of translation initiation in bacteria: A kinetic analysis. EMBO J. 19: 2127–2136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vestergaard, B., Van, L.B., Andersen, G.R., Nyborg, J., Buckingham, R.H., and Kjeldgaard, M. 2001. Bacterial polypeptide release factor RF2 is structurally distinct from eukaryotic eRF1. Mol. Cell 8: 1375–1382. [DOI] [PubMed] [Google Scholar]
- Wakao, H., Romby, P., Westhof, E., Laalami, S., Grunberg-Manago, M., Ebel, J.P., Ehresmann, C., and Ehresmann, B. 1989. The solution structure of the Escherichia coli initiator tRNA and its interactions with initiation factor 2 and the ribosomal 30 S subunit. J. Biol. Chem. 264: 20363–20371. [PubMed] [Google Scholar]
- Wakao, H., Romby, P., Laalami, S., Ebel, J.P., Ehresmann, C., and Ehresmann, B. 1990. Binding of initiation factor 2 and initiator tRNA to the Escherichia coli 30S ribosomal subunit induces allosteric transitions in 16S rRNA. Biochemistry 29: 8144–8151. [DOI] [PubMed] [Google Scholar]
- Wakao, H., Romby, P., Ebel, J.P., Grunberg-Manago, M., Ehresmann, C., and Ehresmann, B. 1991. Topography of the Escherichia coli ribosomal 30S subunit-initiation factor 2 complex. Biochimie 73: 991–1000. [DOI] [PubMed] [Google Scholar]
- Wilson, K.S. and Noller, H.F. 1998. Mapping the position of translational elongation factor EF-G in the ribosome by directed hydroxyl radical probing. Cell 92: 131–139. [DOI] [PubMed] [Google Scholar]
- Wimberly, B.T., Brodersen, D.E., Clemons Jr., W.M., Morgan-Warren, R.J., Carter, A.P., Vonrhein, C., Hartsch, T., and Ramakrishnan, V. 2000. Structure of the 30S ribosomal subunit. Nature 407: 327–339. [DOI] [PubMed] [Google Scholar]
- Yusupov, M.M., Yusupova, G.Z., Baucom, A., Lieberman, K., Earnest, T.N., Cate, J.H., and Noller, H.F. 2001. Crystal structure of the ribosome at 5.5 Å resolution. Science 292: 883–896. [DOI] [PubMed] [Google Scholar]
- Yusupova, G., Reinbolt, J., Wakao, H., Laalami, S., Grunberg-Manago, M., Romby, P., Ehresmann, B., and Ehresmann, C. 1996. Topography of the Escherichia coli initiation factor 2/fMet-tRNA(f)(Met) complex as studied by cross-linking. Biochemistry 35: 2978–2984. [DOI] [PubMed] [Google Scholar]