Abstract
Several hundred microRNAs (miRNAs) have recently been cloned from a wide range of organisms across phylogeny. Despite the high degree of conservation of miRNAs, their functions in general, and in mammals particularly, are just beginning to be defined. Here we show that an oligonucleotide DNA array can be successfully used for the simultaneous analysis of miRNA expression profiles from tissues or cells. From a subset of miRNAs expressed in the brain we designed an oligonucleotide array spotted with probes specific for 44 mature miRNAs. These arrays demonstrated precise regulation of miRNA expression at mammalian brain developmental epochs. About 20% of the probed miRNAs changed significantly in their expression during normal brain development, and two of them, miR-9 and miR-131, were dysregulated in presenilin-1 null mice exhibiting severe brain developmental defects. Transcripts with regulated expression patterns on the arrays were validated by Northern blots. Additionally, a bioinformatic analysis of developmentally regulated miRNAs suggested potential mRNA targets. The arrays also revealed miRNAs distributed to translating polyribosomes in primary neurons where they are likely to modulate translation. Therefore, oligonucleotide arrays provide a new tool for studying miRNA expression in a variety of biological and pathobiological settings. Creating clusters of coexpressed miRNAs will contribute to understanding their regulation, functions, and discovery of mRNA targets.
Keywords: MicroRNA, development, oligonucleotide array, mRNA targets
INTRODUCTION
miRNAs are 19–23-nucleotide transcripts cleaved from a ~70 nt hairpin precursor with signature 3′ hydroxy and 5′ phosphate termini of an RNase III cleavage event that leaves a two-nucleotide overhang. The enzyme responsible for this cleavage is Dicer (Hutvagner et al. 2001; Ketting et al. 2001). Many miRNAs, often with highly conserved sequences, are present in the genomes of Caenorhabditis elegans, Drosophila, mice, and humans (Lagos-Quintana et al. 2001, 2002; Lau et al. 2001; Lee and Ambros 2001; Mourelatos et al. 2002; Dostie et al. 2003). Some miRNAs are organized in the genome as clusters sometimes separated by intervals as short as a few nucleotides (Lagos-Quintana et al. 2001; Lau et al. 2001). The roles proposed for this novel class of tiny RNA molecules are diverse: they are likely to be involved in regulation of mRNA stability and translation, heterochromatin formation, genome rearrangement, and DNA excision (Baulcombe 2002). Two C. elegans miRNAs, lin-4 and let-7, control developmental timing by forming imperfect base pairing with elements within the 3′ UTR of target mRNAs and attenuating their translation (Lee et al. 1993; Wightman et al. 1993; Olsen and Ambros 1999; Reinhart et al. 2000; Slack et al. 2000). In plants, miRNAs have greater complementarity to their mRNA targets, and are likely to trigger the degradation of their targets similarly to the action of small interfering RNAs (siRNAs; Llave et al. 2002; Rhoades et al. 2002). In the animal kingdom, no genomic sequences have been identified with a perfect complementarity to known miRNAs.
miRNAs appear to have crucial roles in development. Inactivation of C. elegans Dicer (dcr-1) causes heterochronic phenotypes similar to let-7 and lin-4 mutations (Grishok et al. 2001). In Drosophila, bantam miRNA stimulates cell proliferation and tissue growth (Brennecke et al. 2003). In Arabidopsis, suppression of RNA silencing, which shares a common pathway with miRNA processing, results in developmental defects (Kasschau et al. 2003). However, no function has been demonstrated for miRNA in mammals yet. Many miRNAs have been isolated from mammalian embryonic neurons and mature brain, and some of them are neuronal-specific (Lagos-Quintana et al. 2002; Dostie et al. 2003). miRNA expression is regulated as a function of time course in neuronal cultures and several miRNAs have been observed in association with polyribosomes in primary neurons (J. Kim, A.M. Krichevsky, Y. Grad, G.D. Hayes, K.S. Kosik, G.M. Church, and G. Ruvkun, in prep.). The involvement of translational regulation in neuronal development, synaptic plasticity and possibly long-term memory storage (Okabe et al. 2001; Miller et al. 2002) could potentially require a role for posttranscriptional regulation by miRNAs. A particularly interesting connection of miRNAs to neuronal function is via the RNA binding protein Fragile X mental retardation protein (FMRP; O’Donnell and Warren 2002). Drosophila homolog of this protein associates with Dicer and with miRNAs and colocalizes with translating ribosomes (Caudy et al. 2002). Dysfunction related to miRNAs also occurs in spinal muscular atrophy, which is caused by deletions or loss-of-function mutations in the Survival of Motor Neuron (SMN) protein. This protein is part of a large ribonucleoprotein complex that contains numerous miRNAs (Mourelatos et al. 2002). To study the involvement of miRNAs in brain development, we have designed an oligonucleotide array capable of determining expression patterns of many neuronal miRNAs. We show here that specific miRNA expressions are precisely regulated during brain development and dysregulated in mice that lack presenilin.
RESULTS AND DISCUSSION
miRNA expression during corticogenesis
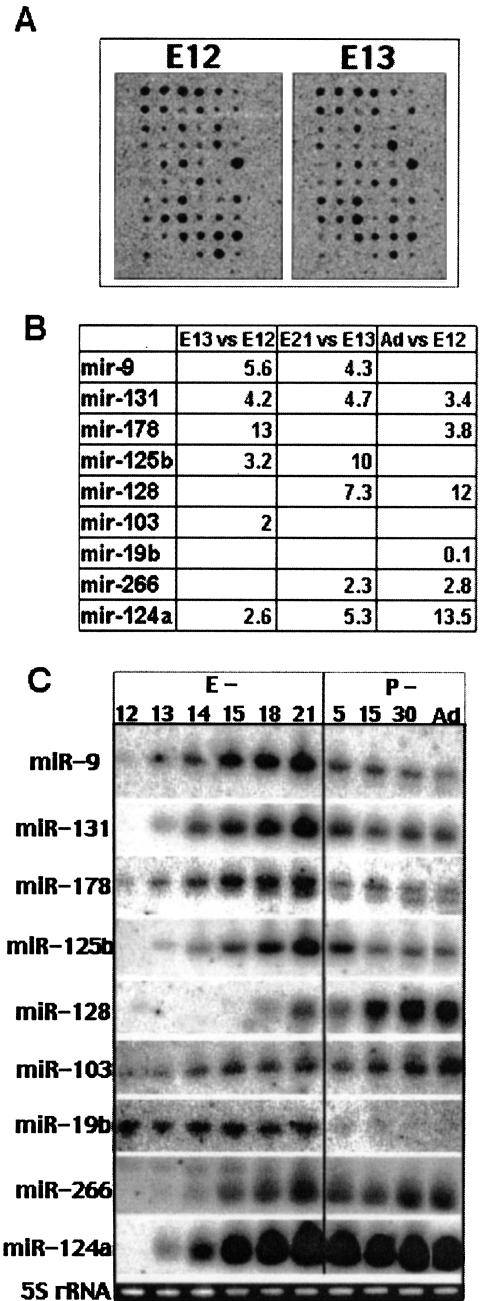
Probes for 44 selected miRNAs that were cloned from mouse brain (Lagos-Quintana et al. 2002) and from cultured rat primary neurons (J. Kim, A.M. Krichevsky, Y. Grad, G.D. Hayes, K.S. Kosik, G.M. Church, and G. Ruvkun, in prep.) were spotted onto a nylon membrane (Table 1 ▶). The following criteria were used for miRNA inclusion: conservation between species (many are conserved in the fly, worm, rodent, and humans), expression in mammalian brain tissue or cultured neurons, and detection by Northern blots (Lagos-Quintana et al. 2002). Specific probes for the loop region of a few highly abundant pre-miRNAs were also included to distinguish between a specific pre-miRNA and mature miRNA. First, miRNA expression profiling was performed at stages of cortical neurogenesis when cell proliferation, migration, regional specification, and the establishment of circuitry occur. For the purpose of detecting the expression of mature miRNA and for minimizing background, we used a labeled RNA sample highly enriched in low molecular weight (LMW) RNA molecules below 70 nt (Fig. 1 ▶). LMW RNA from the forebrain of prenatal (E12, E13, E21), juvenile (P5), and adult rats was labeled, hybridized to arrays, and analyzed. Nine candidate miRNAs appeared differentially expressed during brain development (Fig. 2A,B ▶) with more than a twofold change (p < 0.05). For example, the average increase in the miR-131 signal from E12 to E13 was 4.2-fold, and 4.7 from E13 to E21. After E21, the miR-131 signal decreased, and therefore in adults it appeared just 3.4-fold higher than at E12. MiR-124a signal increased more than 13-fold from E12 to E21, after which it remained stable. MiR-128 signal enhanced later in corticogenesis with an overall 12-fold increase between E12 and adult. MiR-19b signal was strong in E12 and E13, but nearly undetectable in the adult.
TABLE 1.
List of miRNAs included on the array
| miRNA | Sequence (5′ to 3′) |
| mir-9 | TCTTTGGTTATCTAGCTGTATGA |
| mir-16 | TAGCAGCACGTAAATATTGGCG |
| mir-19b | TGTGCAAATCCATGCAAAACTGA |
| mir-30b | TGTAAACATCCTACACTCAGC |
| mir-93 | AAAGTGCTGTTCGTGCAGGTAG |
| mir-98 | TGAGGTAGTAAGTTGTATTGTT |
| mir-99a | ACCCGTAGATCCGATCTTGT |
| mir-103 | AGCAGCATTGTACAGGGCTATGA |
| mir-124a | TAAGGCACGCGGTGAATGCCA |
| mir-125b | TCCCTGAGACCCTAACTTGTGA |
| mir-127 | TCGGATCCGTCTGAGCTTGGCT |
| mir-128 | TCACAGTGAACCGGTCTCTTT |
| mir-130 | CAGTGCAATGTTAAAAGGGCAT |
| mir-131 | TAAAGCTAGATAACCGAAAGT |
| mir-132 | TAACAGTCTACAGCCATGGTCGT |
| mir-138 | AGCTGGTGTTGTGAATC |
| mir-178 | AACATTCAACGCTGTCGGTGAG |
| mir-191 | CAACGGAATCCCAAAAGCAGCT |
| mir-223* | AAACATGAAGCGCTGCAACA |
| mir-224* | GCACATTACACGGTCGACCTCT |
| mir-226* | ACTGCCCCAGGTGCTGCTGG |
| mir-227* | CGCATCCCCTAGGGCATTGGTGT |
| mir-229* | CCTCTGGGCCCTTCCTCCAGT |
| mir-231* | CTATACGACCTGCTGCCTTTCTA |
| mir-232* | CTGGCCCTCTCTGCCCTTCCGT |
| mir-233* | AACACACCCAGCTAACCTTTTT |
| mir-234* | CAAAGCACAGGGCCTGCAGT |
| mir-239* | TACCACAGGGTAGAACCACGGACA |
| mir-240* | TCAAGAGCAATAACGAAAAATGT |
| mir-244* | TCCAGCATCAGTGATTTTGTTGA |
| mir-245* | TCCCTGTCCTCCAGGAGCTCATT |
| mir-248* | TCTCACACAGAAATCGCACCCGTC |
| mir-250* | TGATCTAGCCAAAGCCTGACCGT |
| mir-251* | TGCTGACCCCTAGTCCAGTGC |
| mir-253* | TTGTGCTTGATCTAACCATGTG |
| mir-254* | ACCATCGACCGTTGATTGTACC |
| mir-255* | TGTCCCTCTGGGTCGCCA |
| mir-257* | CAGCCCTGCTGTCTTAACCTCT |
| mir-258* | AAGCCCTTACCCCAAAAAGCAT |
| mir-260* | AGCTACATTGTCTGCTGGGTTTC |
| mir-263* | TCCCTGAGAGCCCTTTGAGCCTGT |
| mir-263*-like | TCCCTGAGGAGCCCTTTGAGCA |
| mir-266* | CAGTGCAATGATGAAAGGGCAT |
| mir-271* | TCCACCTAGCCGGGAGAACCA |
miRNAs recently cloned from rat neurons by J. Kim, A.M. Krichevsky, Y. Grad, G.D. Hayes, K.S. Kosik, G.M. Church, and G. Ruvkin (in prep.) without official names assigned yet are marked with asterisks.
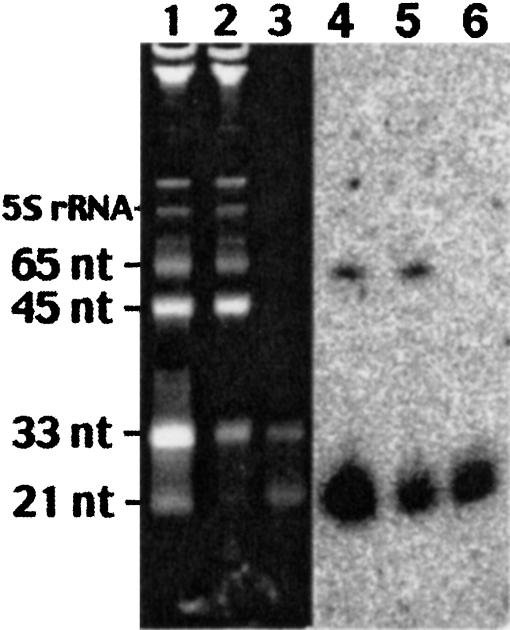
FIGURE 1.
RNA samples enriched in LMW molecules were used as a probe to hybridize to oligo arrays. Total RNA was filtrated through Microcon YM-100 to obtain a fraction enriched in LMW RNA. Total RNA input (lanes 1,4), retaining fraction (lanes 2,5), and LMW RNA (lanes 3,6) were resolved into 15% TBE-Urea RNA gels and stained with Ethidium Bromide (lanes 1–3) or analyzed by Northern blotting with a probe specific for miR-124a (lanes 4–6). In addition to miR-124a, its precursor is detected in lanes 4 and 5, but not in a lane 6. Synthetic RNA markers of indicated size were added to an input total RNA sample. tRNAs here comigrate with a synthetic marker of 65 nt.
FIGURE 2.
Oligonucleotide arrays reveal differential expression of miRNAs during corticogenesis. (A) Representative oligonucleotide arrays on which 33P-labeled LMW RNA samples from E12 and E13 rat forebrains were hybridized. The arrays were analyzed to predict differentially regulated miRNAs. (B) Expression ratios of differentially expressed miRNA as calculated from arrays. RNA at developmental stages E12, E13, E21, and adult were hybridized to arrays and analyzed. Expression ratios are shown only where the ratio exceeds twofold and the corresponding p-value is equal or below 0.05. (C) Northern blot hybridizations of total RNA confirm regulated expression patterns of nine miRNAs during brain prenatal (E) and postnatal (P) development. 5S rRNA was detected by ethidium bromide staining of the gels prior to transfer to verify equal loading of total RNA.
Northern blots validated the differential expression of all nine miRNAs (Fig. 2C ▶). While multiple samples were used for oligoarrays and usually single sample for Northerns, the fold differences detected by Northern blots were in most cases within the range predicted by the oligoarrays. The expression patterns fell into four categories: (1) postnatal expression, for example, miR-128; (2) prenatal expression, for example, miR-19b; (3) peak expression at E21, for example, miR-9, miR-125b, miR-131, miR-178; (4) increasing during the embryonic period followed by stable expression in the postnatal period, for example, miR-124a and miR-266; (5) gradually increasing expression, for example, miR-103. Because several factors, including transcription, maturation and degradation contribute to expression, these patterns suggest finely tuned regulatory mechanisms resulting in specific up- or down-regulation at precise developmental time points. Interestingly, miR-9, miR-125b, miR-131, and miR-178 showed a similar pattern of up-regulation from E12 to E21 followed by down-regulation after birth and a steady-state level after P5. The similar expression pattern of these miRNAs suggests that they share common regulatory elements. Two of these miRNAs, miR-9 and miR-131, are brain-enriched transcripts (Fig. 3A ▶) processed from both arms of the stem of a ~65 nt common precursor (Fig. 3B ▶). Usually just one mature miRNA can be detected as a stable product cleaved from a hairpin-shaped pre-miRNA. Therefore, similar expression patterns of miR-9 and miR-131 may reflect their cotranscription and comaturation from the same genetic unit. However, a slightly greater reduction in miR-9 expression compared to miR-131 occurred between E-21 and the adult (three- to sevenfold for miR-9; two- to fivefold for miR-131) possibly indicating a difference in their stability or processing. Interestingly, miR-9 is also down-regulated during development of the fly, the organism from which it was initially cloned (Lagos-Quintana et al. 2001).
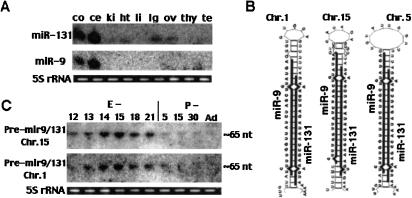
FIGURE 3.
Characterization of developmentally regulated miR-9 and miR-131 transcripts. (A) Northern blots of total RNA isolated from cerebral cortex (co), cerebellum (ce), kidney (ki), heart (ht), liver (li), lungs (lg), ovary (ov), thymus (thy), and testis (te), probed with miR-9 and mir-131. (B) Predicted secondary structures for putative precursors of multicopy human and rodent miR-9 and miR-131 cotranscribed pair. RNA secondary structure prediction was performed using mfold software. MiR-9 and miR-131 sequences are shown in bold. (C) Northern blots of total RNA isolated from pre- and postnatal developing brain with probes specific for the loop region of the predicted mir-9/miR-131 precursors. Oligonucleotides used as probes were 5′-TTATGAAGACTCCA CACCAC-3′ for chromosome 1 precursor, 5′-TTTATGAAGACCAATACAC-3′ for chromosome 5 precursor, and 5′-TTATGACGGCTCTGTGGCAC-3′ for chromosome 15 precursor. The putative precursor encoded on chromosome 5 was undetectable.
The miR-9/miR-131 pair is encoded on three different chromosomal loci that are identical in rodent and human (Homo sapiens chromosomes 1, 5, and 15; −Mus musculus chromosomes 3, 13, and 7). At all three loci the sequences of miR-9 and miR-131 are perfectly conserved, whereas their joining sequences are just moderately conserved at two loci (chromosomes 1 and 5), and differ strongly at a chromosome 15 locus (Fig. 3B ▶). Two different precursors of ~65 nt, pre-mir9/131-ch1 and pre-mir9/131-ch15, but not pre-mir9/131-ch5, are detected with probes specific for the loop regions (Fig. 3C ▶). Therefore, these precursors expression can be independently regulated during brain development, and it is possible that only one member of the pair is expressed from each locus. Although miR-9 and miR-131 expression patterns demonstrate their maximal level at E21, both precursors are more abundant at E14–E15, suggesting developmental regulation of their maturation. At all three loci the pre-miR-9/miR-131 stem strongly resembles the “classical” siRNA structure with two 3′ overhangs. Therefore, the precursor may be cleaved to a double-stranded siRNA-like duplex and miR-9/miR-131 may function as a double-stranded RNA. Alternatively, the processing may result in two single-stranded miRNAs (miR-9 and miR-131) with independent functions, or functionally interplaying with each other.
miRNA expression in Presenilin-1 knockout mice
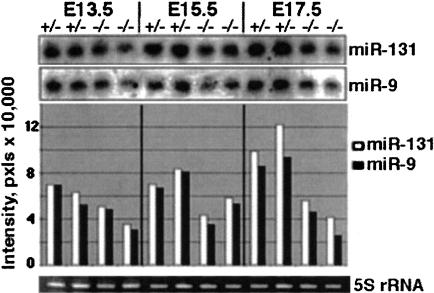
Because miRNAs demonstrate precisely regulated patterns of expression during brain development, we sought evidence for their dysregulation in a brain developmental disorder. Presenilin-1 (PS1) is part of a multiprotein complex that mediates intramembranous proteolysis and release of receptor endodomane for cell signaling. Because the Notch receptors are key substrates of the complex, PS1-deficient mice show lethal developmental defects in the CNS similar to those observed in mice deficient in Notch 1 (Shen et al. 1997). Although PS1(−/−) mice die shortly after birth and exhibit severe CNS defects and axial skeleton malformations, a 50% reduction of PS1 activity in PS1(+/−) mice does not lead to any detectable developmental abnormalities. Premature neuronal differentiation in the PS1(−/−) brain is associated with aberrant neuronal migration and disorganization of the laminar architecture of the developing cerebral hemisphere (Handler et al. 2000). Because at least 41 out of 44 miRNAs on the array were perfectly conserved between mouse and rat, we applied the array to the study of miRNA expression in PS1 knockout mice. These experiments demonstrated that expression of the developmentally regulated pair, miR-9 and miR-131, was reduced in the forebrains of E17.5 PS1-deficient mice compared to their PS1(+/−) littermate forebrains (data not shown). Northern blot analysis confirmed the more than twofold reduction in miR-9 and miR-131 expression in PS1(−/−) E17.5 brains (Fig. 4 ▶). The difference in miR-9 and miR-131 expression between PS1(+/−) and PS1(−/−) brains was less evident at E13.5 and enhanced over development from E13.5 to E17.5. The failure of miR-9 and miR-131 to up-regulate in PS1(−/−) embryonic brain development may be due to impaired Notch signaling. No differences in the expression of other miRNAs in PS1-deficient mice were detected.
FIGURE 4.
MiR-9 and miR-131 are disregulated in PS1 null mice. Northern blots of total brain RNA isolated from PS1(+/−) and PS1(−/−) embryos of indicated ages (two embryos per each group), probed for miR-9 and miR-131. PS1(+/−) heterozygous are phenotypically undistinguishable from wild-type animals. Bands were quantified and represented as bars in the lower panel.
The functions of mammalian miRNAs have been extrapolated from other species. Heterochronic RNAs in C. elegans control cell cycle progression and a particular sequence of cell divisions that is necessary for the generation of a specific cell type (Ambros 2000). Stem cell lineages appear to be a fundamental principle of development, and miRNAs are likely to play a role in modifying these lineages. miRNA arrays could reveal specific miRNAs that modify lineages as stem cells undergo staged differentiation in many organs including the brain. The dysregulation of the miR-9/miR-131 pair in the PS(−/−) mouse supports this idea. Here, the lack of PS1 leads to premature differentiation of neural progenitor cells, while neural proliferation and apoptotic cell death during neurogenesis are unaltered (Handler et al. 2000). This alteration in the outcome of lineage determination during development resembles the heterochronic phenotypes seen in C. elegans.
miRNAs are differentially associated with polyribosomes
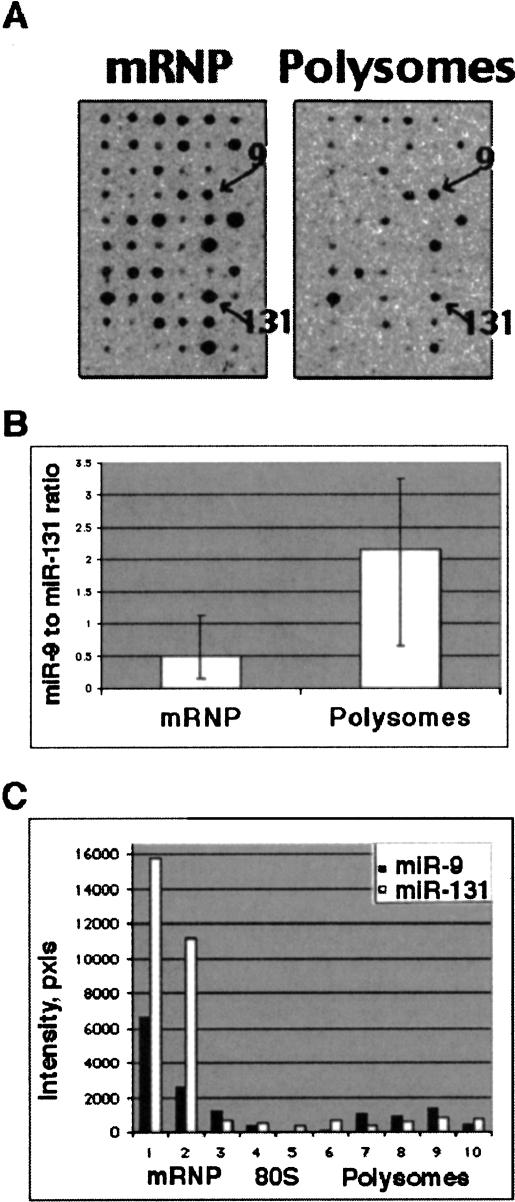
One of the proposed roles for miRNAs is the regulation of translation by forming imperfectly base-paired duplexes with mRNAs. In C. elegans both lin-4 miRNA and its target lin-14 mRNA associate with polyribosomes and lin-4 inhibits the translation of lin-14 mRNA after initiation of its translation (Ambros 2000). Therefore, we studied the association of miRNAs with polyribosomes. Cytosolic lysates from primary cerebrocortical cells (7 d in culture) were fractionated in 15%–45% sucrose gradient, and RNA was isolated separately from fractions of mRNP, ribosomal subunits and free (untranslating) ribosomes, polyribosomes, and RNA granules as described in (Krichevsky and Kosik 2001). LMW RNA samples from each of the four principal fractions were hybridized to the oligonucleotide arrays. Strong hybridization signals were detected with probes corresponding to mRNP- and polyribosomes-associated RNA (Fig. 5A ▶). Overall, a stronger signal was observed in the mRNPs; however, the proportion of specific miRNAs in polysomes versus mRNPs differed widely. Some miRNAs were overrepresented in polysomes, whereas others are underrepresented. The cosedimentation of miRNAs with polysomes strongly suggests their role in the regulation of translation.
FIGURE 5.
miRNAs are differentially associated with translating polyribosomes. (A) Representative oligo arrays showing hybridization with 33P-labeled LMW RNA that was isolated from mRNP and polysomal fractions of cultured primary neurons. Fifty percent of the RNA from the sucrose gradient fractions was used. Signals corresponding to miR-9 and miR-131 hybridizations are indicated. (B) Oligonucleotide arrays predict differential association of miR-9 and miR-131 with polyribosomes. mRNP and polysomal RNAs were isolated from three independent fractionation experiments, and each LMW RNA sample was hybridized in triplicate to the array membranes as shown in A. MiR-9 and miR-131 spots were quantified and the ratio of miR-9 to miR-131 signals was calculated for filters hybridized with polysomal RNA versus mRNP’s RNA. The bars represent an average of three completely independent experiments. (C) Northern blot hybridization of RNA isolated from sucrose gradient fractions with miR-9 and miR-131 specific probes. Fifty percent of RNA from the fraction was loaded per lane. Bands were quantified by densitometry and plotted, confirming that higher proportion of miR-9 than of miR-131 is associated with actively translating polysomes.
Interestingly, miR-9 and miR-131, while coexpressed in neurons demonstrated quantitative differences in their association with polysomes. Relatively, the miR-9 signal was stronger in polysomes than the miR-131 signal (Fig. 5A,B ▶). To validate this predicted difference in miR-9 and miR-131 distributions between nontranslating mRNPs and translating polyribosomes we analyzed these miRNAs with specific probes by Northern blots (Fig. 5C ▶). Indeed, a higher proportion of miR-9 was associated with polyribosomes relative to miR-131 (20%–25% of miR-9 versus 8%–9% of miR-131). These data suggest a complex hierarchical regulation of miR-9/miR-131. In addition to the regulation of miR-9/miR-131 transcription from multiple loci and maturation (most likely as single-stranded molecules), their functioning might be regulated at two additional levels. First, miR-9 may compete with miR-131 for binding to its target, and vise versa. Second, and more general, the dynamic association of miRNAs with different RNP complexes also may affect their targeting of different mRNA molecules.
Putative mRNA targets for developmentally regulated miRNAs
A major challenge in understanding miRNA function is target identification. The expression pattern of a miRNA may narrow the search for and contribute to the validation of targets. The miRNA and its putative target must be expressed in the same tissue. Putative targets may undergo posttranscriptional regulation that is coordinated with the miRNA expression. Combining this candidate strategy with a search of the NCBI databases revealed several messages with a high degree of complementarity to developmentally regulated neuronal miRNAs (Fig. 6 ▶). Some of those mRNAs are implicated in important neuronal functions. Calcineurin Aβ, an isoform of the calcineurin catalytic subunit, fits these target criteria for miR-131. Calcineurin is a major phosphatase of the central nervous system involved in a variety of neuronal signaling cascades, which plays a critical role in long-term depression (Zhuo et al. 1999). In humans and rodents, miR-131 demonstrates 19 nt and 18 nt, respectively, out of 21 nt complementarity to a sequence within the 3′ UTR of calcineurin Aβ mRNA (Fig. 6 ▶). Calcineurin A, and particularly its β isoform expression in developing brain is regulated posttranscriptionally, most likely at the translational level (Polli et al. 1991). Another putative target for miR-131 is a cofactor required for Sp1 transcriptional activation (20 nt out of 21 nt complementarity for both mouse and human mRNAs). mRNA for Id2 (Inhibitor of DNA binding), the protein antagonizing neuronal differentiation (Gleichmann et al. 2002), demonstrates 20 nt out of 23 nt complementarity to miR-9. A sequence within DNA helicase SMBP2 transcript is nearly complementary to miR-103. Mutation of this protein causes mouse neuromuscular degeneration (Cox et al. 1998), a disease similar to human spinal muscular atrophy and linked to miRNA-containing complex (see above). Otx1, murine homolog of the Drosophila transcription factor orthodenticle, is necessary for normal corticogenesis (Simeone et al. 2002), and may represent another putative target for miR-103. Similar to the majority of the predicted plant targets (Rhoades et al. 2002), the mentioned mRNAs encode either transcription factors or, like calcineurin, may affect transcription indirectly.
FIGURE 6.
Putative mRNA targets. Sequence alignments for neuronal developmentally regulated miRNAs and their putative human and mouse mRNA targets. Black boxes indicate sequences conserved between human and mice. DataBank accession numbers for mRNAs are indicated.
Oligonucleotide array analyses have proven an informative tool in assigning biological significance to an mRNA expression profile. MicroRNAs, which are likely to share related regulatory control elements with other genes, display expression profiles that uniquely correspond to developmental epochs of brain development. Creating clusters of coexpressed miRNAs, such as miR-9/131, miR-178 and miR-125b, will contribute to understanding their functions, finding mRNA targets and bioinformatic analysis of upstream regulatory motifs. The estimated number of the total miRNAs in vertebrates is ~250 molecules (Lim et al. 2003). This prediction is based on the observation that miRNAs derive from evolutionary conserved hairpin precursor RNAs. However, mammalian brain may develop unique regulatory molecules, and express many more miRNAs. In addition, many tiny noncoding RNA molecules that are not processed from a miRNA-like hairpin precursor and are not phylogenetically conserved have been recently cloned from C. elegans and likely to express in vertebrates as well (Ambros et al. 2003). In any case, annotating the genes for small noncoding RNAs will involve establishing of their expression profiles, and the use of oligonucleotide arrays is ideally suited to this task. We are currently applying a new version of arrays to profile 180 miRNAs that have been cloned from vertebrates.
MATERIALS AND METHODS
RNA preparation from primary cortical neurons and dissected brain
Meningeal coverings were removed, and prosencephalic tissue was dissected from embryonic rats and from embryonic PS1−/− and PS1+/− mice, and forebrains were removed from postnatal rats. Primary cerebrocortical neurons were cultured and fractionated on 15%–45% sucrose gradients as previously described (Krichevsky and Kosik 2001). Total RNA was isolated from brain tissues and fractionated cells (Krichevsky and Kosik 2001).
Oligonucleotide array for miRNA
Tri-mer oligonucleotides (antisense to microRNAs) of 54–72 nt at final concentration of 7 μM were spotted on the GeneScreen Plus (NEN) membranes with a 1536 pin plate replicator (V&P Scientific). To confirm the specificity of hybridization we also included a series of oligonucleotides with three mismatches (G → C or C → A) on the array. These mismatches resulted in a significant drop in signal from these spots compared to their cognates. Oligonucleotides were immobilized in 100 mM NaOH, the membranes were briefly neutralized in 5% SDS/0.5 M Tris (pH 7.5), and washed with 2 × SSC 0.5% SDS at RT and with 0.2% SDS at 72°C. Arrays were stored in 0.2% SDS at −4°C.
Five to ten micrograms of total RNA from brain tissue (or 50% of RNA isolated from each principal sucrose gradient fraction) were preheated at 80°C for 3 min, cooled on ice, and filtered through Microcon YM-100 concentrators to obtain a low molecular weight fraction of RNA enriched in molecules under 60 nt (Fig. 1 ▶). Nevertheless, we were able to detect several highly expressed pre-miRNA, as well as two tRNAs, which were spotted as controls. Also, a synthetic 21-nt RNA with a sequence that does not correspond to any miRNA, but is an exact complement to a random spotted sequence, was added to the RNA sample at a known concentration as a reference for normalization. The LMW RNA was end-labeled with 30 μCi of γ33P dATP (3000 Ci/mmole) by T4 polynucleotide kinase and purified using the QIAgen Nucleotide Removal Kit. γ33P dATP incorporation was determined using BioScan. Samples with abnormally low incorporation were excluded from the analysis.
For hybridization, membranes were first prehybridized in MicroHyb Hybridization Buffer (ResGen) at 37°C for at least 30 min followed by an overnight hybridization in the same solution containing RNA probe. Following hybridization membranes were washed twice with 2 × SSC/0.5% SDS at 37°C. The second wash was performed in 1 × SSC/0.5% SDS at 37°C. Membranes were exposed to a phosphor storage screen, scanned using Phosphor Imager, and hybridization signals were quantified using Image Quant software (Molecular Dynamics). After exposure the membranes were stripped with 0.2% SDS at 72°C, tested again by exposure to phosphoimager screen, and rehybridized three to five times.
Typically, an experiment included three independent RNA samples for each developmental time point. Each rat brain E12 and E13 sample was collected from three to four embryos, whereas other samples were prepared from separate embryos or animals. The standard deviation between triplicate samples was distributed as follows: <0.6 for all probes, <0.3 for 80% of probes, and <0.2 for 50% of probes. To ensure accuracy of the hybridizations, each RNA sample was hybridized with two or three membranes. Hybridization signals for each spot of the array and background values at 15 empty spots were measured. Raw data was further automatically processed in Microsoft Excel. Hybridization signals that failed to exceed the average background value by more than three standard deviations were excluded from analysis. Upon normalization, the remaining data was averaged among duplicate arrays, and the resulting three data sets each corresponding to an RNA sample were considered independent measurements for the purposes of the two-tailed, two-sample t-test when comparing different developmental stages. Also, the three data sets were averaged among three RNA samples to produce the set of “expression levels at a given developmental stage.” The expression ratio for each miRNA between given developmental stages was calculated as the ratio between the corresponding expression levels. An example of expression ratios is presented at Figure 2B ▶.
Northern blot analysis
Fifteen to twenty micrograms of total RNA or 50% of RNA isolated from each principal sucrose gradient fraction per lane was separated on a denaturing 15% polyacrilamide gel and transferred electrophoretically to a GeneScreen Plus membrane. Membranes were crosslinked and baked according to the manufacturer’s manual. Probes were prepared by T4 polynucleotide kinase labeling of antisense oligonucleotides with γ32P dATP (6000Ci/mmole). Prehybridizations and hybridizations were carried out using UltraHyb Hybridization Buffer (Ambion) at 37°C. Blots were washed, with the final wash in the range of 0.2–0.5 × SSC/0.1% SDS at 37°C and analyzed by phosphorimaging. Control probes corresponding to specific miRNAs but with three mismatches demonstrated absence of signal, indicating that there was little if any cross-hybridization between family members.
Identification of mRNAs with complementarity sites to regulated neuronal miRNAs
Fasta searches against the EMBL database for complementary targets were designed with ambiguous codes to allow for G–T base-pairing and limited gaps, following the example of known targets for lin-4 and let-7. Primary blast searches were also conducted with low penalty for gaps and increased match rewards. Secondary blast searches of the hits indicated if any corresponded to previously characterized mRNA.
Acknowledgments
We thank members of Kenneth Kosik laboratory for helpful discussions and comments on the manuscript, and members of Jie Shen laboratory for help with PS1 KO mice. We are grateful to James Eberwine for advice on miRNA arrays design. This work was supported in part by NIH and NSF Grants to K.S.K. and by NIH Grants R01AG18388, R01AG19787, and R01ES11343 to K.K.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.rnajournal.org/cgi/doi/10.1261/rna.5980303.
REFERENCES
- Ambros, V. 2000. Control of developmental timing in Caenorhabditis elegans. Curr. Opin. Genet. Dev. 10: 428–433. [DOI] [PubMed] [Google Scholar]
- Ambros, V., Lee, R.C., Lavanway, A., Williams, P.T., and Jewell, D. 2003. MicroRNAs and other tiny endogenous RNAs in C. elegans. Curr. Biol. 13: 807–818. [DOI] [PubMed] [Google Scholar]
- Baulcombe, D. 2002. DNA events. An RNA microcosm. Science 297: 2002–2003. [DOI] [PubMed] [Google Scholar]
- Brennecke, J., Hipfner, D.R., Stark, A., Russell, R.B., and Cohen, S.M. 2003. Bantam encodes a developmentally regulated microRNA that controls cell proliferation and regulates the proapoptotic gene hid in Drosophila. Cell 113: 25–36. [DOI] [PubMed] [Google Scholar]
- Caudy, A.A., Myers, M., Hannon, G.J., and Hammond, S.M. 2002. Fragile X-related protein and VIG associate with the RNA interference machinery. Genes & Dev. 16: 2491–2496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox, G.A., Mahaffey, C.L., and Frankel, W.N. 1998. Identification of the mouse neuromuscular degeneration gene and mapping of a second site suppressor allele. Neuron 21: 1327–1337. [DOI] [PubMed] [Google Scholar]
- Dostie, J., Mourelatos, Z., Yang, M., Sharma, A., and Dreyfuss, G. 2003. Numerous microRNPs in neuronal cells containing novel microRNAs. RNA 9: 180–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gleichmann, M., Buchheim, G., El-Bizri, H., Yokota, Y., Klockgether, T., Kugler, S., Bahr, M., Weller, M., and Schulz, J.B. 2002. Identification of inhibitor-of-differentiation 2 (Id2) as a modulator of neuronal apoptosis. J. Neurochem. 80: 755–762. [DOI] [PubMed] [Google Scholar]
- Grishok, A., Pasquinelli, A.E., Conte, D., Li, N., Parrish, S., Ha, I., Baillie, D.L., Fire, A., Ruvkun, G., and Mello, C.C. 2001. Genes and mechanisms related to RNA interference regulate expression of the small temporal RNAs that control C. elegans developmental timing. Cell 106: 23–34. [DOI] [PubMed] [Google Scholar]
- Handler, M., Yang, X., and Shen, J. 2000. Presenilin-1 regulates neuronal differentiation during neurogenesis. Development 127: 2593–2606. [DOI] [PubMed] [Google Scholar]
- Hutvagner, G., McLachlan, J., Pasquinelli, A.E., Balint, E., Tuschl, T., and Zamore, P.D. 2001. A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7 small temporal RNA. Science 293: 834–838. [DOI] [PubMed] [Google Scholar]
- Kasschau, K.D., Xie, Z., Allen, E., Llave, C., Chapman, E.J., Krizan, K.A., and Carrington, J.C. 2003. P1/HC-Pro, a viral suppressor of RNA silencing, interferes with Arabidopsis development and miRNA function. Dev. Cell 4: 205–217. [DOI] [PubMed] [Google Scholar]
- Ketting, R.F., Fischer, S.E., Bernstein, E., Sijen, T., Hannon, G.J., and Plasterk, R.H. 2001. Dicer functions in RNA interference and in synthesis of small RNA involved in developmental timing in C. elegans. Genes & Dev. 15: 2654–2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krichevsky, A.M., and Kosik, K.S. 2001. Neuronal RNA granules: A link between RNA localization and stimulation-dependent translation. Neuron 32: 683–696. [DOI] [PubMed] [Google Scholar]
- Lagos-Quintana, M., Rauhut, R., Lendeckel, W., and Tuschl, T. 2001. Identification of novel genes coding for small expressed RNAs. Science 294: 853–858. [DOI] [PubMed] [Google Scholar]
- Lagos-Quintana, M, Rauhut, R., Yalcin, A., Meyer, J., Lendeckel, W., and Tuschl, T. 2002. Identification of tissue-specific MicroRNAs from mouse. Curr. Biol. 12: 735–739. [DOI] [PubMed] [Google Scholar]
- Lau, N.C., Lim, L.P., Weinstein, E.G., and Bartel, D.P. 2001. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science 294: 858–862. [DOI] [PubMed] [Google Scholar]
- Lee, R.C. and Ambros, V. 2001. An extensive class of small RNAs in Caenorhabditis elegans. Science 294: 862–864. [DOI] [PubMed] [Google Scholar]
- Lee, R.C., Feinbaum, R.L., and Ambros, V. 1993. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 75: 843–854. [DOI] [PubMed] [Google Scholar]
- Lim, L.P., Glasner, M.E., Yekta, S., Burge, C.B., and Bartel, D.P. 2003. Vertebrate microRNA genes. Science 299: 1540. [DOI] [PubMed] [Google Scholar]
- Llave, C., Xie, Z., Kasschau, K.D., and Carrington, J.C. 2002. Cleavage of Scarecrow-like mRNA targets directed by a class of Arabidopsis miRNA. Science 297: 2053–2056. [DOI] [PubMed] [Google Scholar]
- Miller, S., Yasuda, M., Coats, J.K., Jones, Y., Martone, M.E., and Mayford, M. 2002. Disruption of dendritic translation of CaMKIIα impairs stabilization of synaptic plasticity and memory consolidation. Neuron 36: 507–519. [DOI] [PubMed] [Google Scholar]
- Mourelatos, Z., Dostie, J., Paushkin, S., Sharma, A., Charroux, B., Abel, L., Rappsilber, J., Mann, M., and Dreyfuss, G. 2002. miRNPs: A novel class of ribonucleoproteins containing numerous micro RNAs. Genes & Dev. 16: 720–728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Donnell, W.T. and Warren, S.T. 2002. A decade of molecular studies of fragile X syndrome. Annu. Rev. Neurosci. 25: 315–338. [DOI] [PubMed] [Google Scholar]
- Okabe, M., Imai, T., Kurusu, M., Hiromi, Y., and Okano, H. 2001. Translational repression determines a neuronal potential in Drosophila asymmetric cell division. Nature 411: 94–98. [DOI] [PubMed] [Google Scholar]
- Olsen, P.H. and Ambros, V. 1999. The lin-4 regulatory RNA controls developmental timing in Caenorhabditis elegans by blocking LIN-14 protein synthesis after the initiation of translation. Dev. Biol. 216: 671–680. [DOI] [PubMed] [Google Scholar]
- Polli, J.W., Billingsley, M.L., and Kincaid, R.L. 1991. Expression of the calmodulin-dependent phosphodiesterase, calcineurin, in rat brain: Developmental patterns and the role of nigrostriatal innervation. Dev. Brain Res. 61: 105–119. [DOI] [PubMed] [Google Scholar]
- Reinhart, B.J., Slack, F.J., Basson, M., Pasquinelli, A.E., Bettinger, J.C., Rougvie, A.E., Horvitz, H.R., and Ruvkun, G. 2000. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature 403: 901–906. [DOI] [PubMed] [Google Scholar]
- Rhoades, M., Reinhart, B., Lim, L., Burge, C., Bartel, B., and Bartel, D. 2002. Prediction of plant microRNA targets. Cell 110: 513. [DOI] [PubMed] [Google Scholar]
- Shen, J., Bronson, T., Chjen, D.F., Xia, W., Selkoe, D.J., and Tonegawa, S. 1997. Skeletal and CNS defects in presenilin-1-deficient mice. Cell 89: 629–639. [DOI] [PubMed] [Google Scholar]
- Simeone, A., Puelles, E., and Acampora, D. 2002. The Otx family. Curr. Opin. Genet. Dev. 12: 409–415. [DOI] [PubMed] [Google Scholar]
- Slack, F.J., Basson, M., Liu, Z., Ambros, V., Horvitz, H.R., and Ruvkun, G. 2000. The lin-41 RBCC gene acts in the C. elegans heterochronic pathway between the let-7 regulatory RNA and the LIN-29 transcription factor. Mol. Cell 5: 659–669. [DOI] [PubMed] [Google Scholar]
- Wightman, B., Ha, I., and Ruvkun, G. 1993. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell 75: 855–862. [DOI] [PubMed] [Google Scholar]
- Zhuo, M., Zhang, W., Son, H., Mansuy, I., Sobel, R.A., Seidman, J., and Kandel, E.R. 1999. A selective role of calcineurin aα in synaptic depotentiation in hippocampus. Proc. Natl. Acad. Sci. 96: 4650–4655. [DOI] [PMC free article] [PubMed] [Google Scholar]