Abstract
Cic1p/Nsa3p was previously reported to be associated with the 26S proteasome and required for the degradation of specific substrates, but was also shown to be associated with early pre-60S particles and to be localized to the nucleolus. Here we report that Cic1p/Nsa3p is required for the synthesis of 60S ribosome subunits. A temperature-sensitive lethal cic1–2 point mutation inhibits synthesis of the mature 5.8S and 25S rRNAs. Release of the pre-60S particles from the nucleolus to the nucleoplasm was also inhibited as judged by the nuclear accumulation of an Rpl11b-GFP reporter construct. We suggest that Cic1p/Nsa3p associates early with nascent preribosomal particles and is required for correct processing and nuclear release of large ribosomal subunit precursors.
Keywords: pre-rRNA, ribosome synthesis
INTRODUCTION
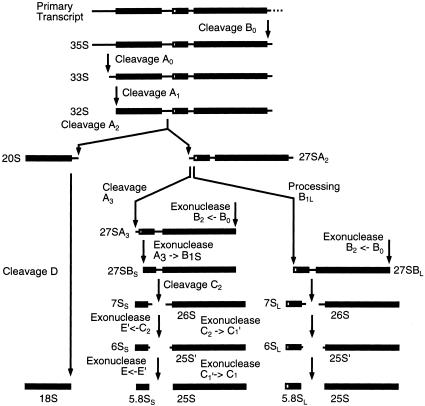
Making ribosome in eukaryotes is a complex process that commences in the nucleolus with the transcription of a large primary transcript (35S pre-rRNA in yeast) by RNA polymerase I (Pol I). The 35S pre-rRNA is processed at multiple sites (Fig. 1 ▶) to generate the mature 18S, 5.8S, and 25S rRNAs with concomitant modification of the mature rRNAs (for review, see Venema and Tollervey 1999). During these processing reactions, the rRNA intermediates associate with the 5S rRNA, about 80 ribosomal proteins, and a large number of nonribosomal proteins, to form preribosomal particles. The preribosomal particles are dynamically remodeled and matured as they transit from the nucleolus to the nucleoplasm and then are exported to the cytoplasm, where synthesis of the 60S and 40S ribosomal subunits is completed (Nissan et al. 2002; Saveanu et al. 2003; Schafer et al. 2003).
FIGURE 1.
Pre-rRNA processing pathway. In wild-type cells, the 35S pre-rRNA is cleaved at site A0 producing the 33S pre-rRNA. This molecule is rapidly cleaved at site A1 to produce the 32S, which is cleaved at site A2, releasing the 20S and 27SA2 pre-rRNAs. The 20S pre-rRNA is exported to the cytoplasm, where it is dimethylated by Dim1p and then cleaved at site D, by an unidentified enzyme, to generate the mature 18S rRNA. 27SA2 is processed via two alternative pathways. It can be cut at site A3 to generate 27SA3, which is then trimmed to site B1S, producing 27SBS. Alternatively, it can be processed to 27SBL by an as yet unknown mechanism. 27SBS and 27SBL are matured to the 5.8S and 25S following identical pathways. Cleavage at site C2 generates the 7S and 26S pre-rRNAs. The 7S pre-rRNA is digested 3′ to 5′ to 6S pre-rRNA and then to the mature 5.8S rRNA. The 26S pre-rRNA is digested 5′ to 3′ to the 25S` pre-rRNA and then to the mature 25S rRNA. For a review on pre-rRNA processing and the known processing enzymes, see Venema and Tollervey (1999).
The pathway of ribosome maturation in yeast has been characterized by extensive proteomic analyses of preribosomal particles (for review, see Fatica and Tollervey 2002; Milkereit et al. 2003). These analyses reveal that more than 160 different proteins are dynamically exchanged during ribosome maturation. A role in pre-rRNA processing had previously been demonstrated for many of these proteins, but a substantial number were either previously uncharacterized or had been identified as factors required for functions apparently unrelated to ribosome synthesis.
One such is Cic1p/Nsa3p, an essential protein previously reported to function as an adaptor for the 26S proteasome (Glickman and Ciechanover 2001) that is specifically required for degradation of SCF protein components (Jager et al. 2001). Despite this involvement in regulating the ubiquitin–proteasome system, Cic1p was localized to the nucleolus (Jager et al. 2001; Nissan et al. 2002), and the identification of Cic1p as a putative component of early pre-60S ribosomes (Baβler et al. 2001; Harnpicharnchai et al. 2001; Nissan et al. 2002; Saveanu et al. 2003) strongly suggested an additional or alternative role for Cic1p in ribosome synthesis.
Here we report that Cic1p is indeed required for maturation of large subunit rRNAs and is apparently also required for the release of the pre-60S ribosomal particles from the nucleus.
RESULTS AND DISCUSSION
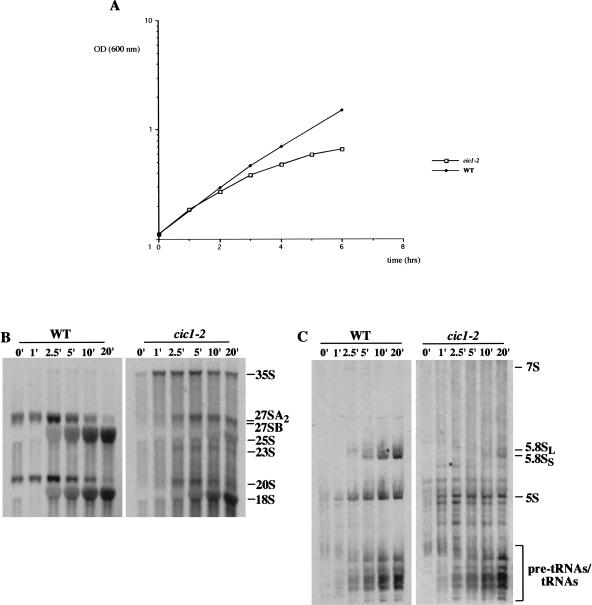
To study the requirement for Cic1p in rRNA synthesis, we utilized the previously described cic1–2 temperature-sensitive mutant strain (Jager et al. 2001). Growth of the cic1–2 strain was very similar to the otherwise isogenic wild-type strain at the permissive temperature of 25°C (data not shown), but almost ceased 6 h after transfer at the restrictive temperature of 37°C (Fig. 2A ▶).
FIGURE 2.
Inactivation of Cic1p/Nsa3p inhibits growth and rRNA synthesis. (A) Growth rate of cic1–2 (open squares) and wild-type (closed diamonds) strains following a shift from permissive to restrictive temperature. (B, C) Strains cic1–2 and wild type were growth at 25°C in YPD then shifted to 37°C for 4 h. Cell were pulse labeled with [5,6-3H] uracil for 1 min and then chased with an excess of cold uracil. Total RNA was extracted from cell samples harvested at the indicated time points and resolved on a 1.2% agarose/formaldehyde (B) and 6% acrylamide/urea gels (C). The position of mature rRNAs, pre-rRNAs, and tRNA species are indicated. Incorporation of [5,6-3H] uracil in the mutant strain was lower than in the wild type, and a twofold longer exposure of these lanes is shown.
Pre-rRNA processing was initially assessed by pulse-chase labeling in vivo with [5,6-3H]uracil, performed 4 h after transfer to 37°C. Analysis of high molecular weight RNA (Fig. 2B ▶) showed that in the wild-type strain, the 35S pre-rRNA was rapidly converted into the 27SA and 20S pre-rRNAs. The 27SA pre-rRNA is then converted to 27SB and the 25S rRNA, whereas 20S pre-rRNA is converted to 18S rRNA (see Fig. 1 ▶). By contrast, in the cic1–2 strain, processing of the 35S pre-rRNA was substantially slowed and the aberrant 23S RNA was readily detected. The 23S RNA originates from direct cleavage of the 35S pre-rRNA at site A3 when the cleavages at sites A0, A1, and A2 are delayed (Venema and Tollervey 1999). The 27SA pre-rRNA was synthesized with some delay but its conversion to 25S rRNA was strongly reduced. The 20S pre-rRNA and mature 18S rRNA appeared with delayed kinetics and reduced yield, but this reduction was much less marked than that seen for 27S and 25S.
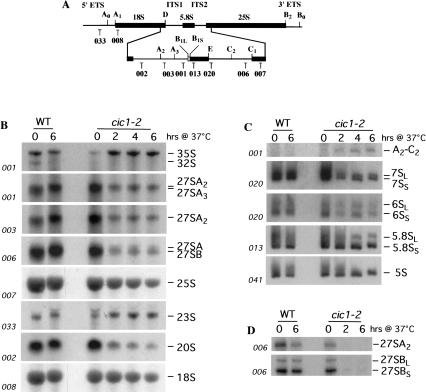
Analysis of low molecular weight RNA (Fig. 2C ▶) showed the processing of the 7S pre-rRNA to mature 5.8S rRNA in the wild-type strain. In the cic1–2 strain, the 7S pre-rRNAs was not detected and synthesis of 5.8S rRNA was strongly depleted. Synthesis of 5S rRNA was mildly reduced, presumably as a consequence of the growth inhibition. These data show that Cic1p is required for synthesis of the large subunit rRNAs. As seen for almost all other mutations defective in synthesis of the 60S subunit rRNAs, a delay in the early pre-rRNA processing steps was also observed. To further characterize the function of Cic1p in pre-rRNA processing, steady-state levels of mature rRNAs and precursors were assessed by Northern hybridization (Fig. 3 ▶). After transfer of the cic1–2 strain to 37°C for 2 h, the 35S pre-rRNA and 23S RNA were accumulated (Fig. 3B ▶), consistent with the pulse-chase analysis, whereas the 27SA2 and 20S pre-rRNAs were reduced. The 27SB and 27SA3 pre-rRNA were depleted as well. In good agreement with the reduced levels of 27SA2 and 27SB, primer extension analysis showed that in the cic1–2 strain, there is a strong reduction of the stops at site A2, B1L and B1S (Fig. 3D ▶). This suggests that the processing of all 27S pre-rRNAs is affected by Cic1p inactivation. Steady-state levels of mature 25S and, to a lesser extent, of 18S rRNAs were decreased. The defect in 18S synthesis is a common feature of strains with defects in 60S synthesis and may be secondary consequence of the reduced growth rate (Venema and Tollervey 1999). In the cic1–2 strain at 37°C there was a strong reduction of the 7S and 6S pre-rRNAs (Fig. 3C ▶), precursors of the 5.8S rRNA. In addition, a fragment that extends from A2 to C2 was accumulated (Fig. 3C ▶), indicating that inactivation of Cic1p allows an aberrant cleavage of 27SA2 at site C2 that reduce the conversion of 27SA2 into 27SB (Fig. 3B,D ▶) and 7S pre-rRNAs (Fig. 3C ▶). The A2-C2 molecule has been previously described for other components of early pre-60S ribosomal particles (Fatica et al. 2002) and indicates that Cic1p is required to maintain the normal order of processing in the 27SA2 molecule.
FIGURE 3.
Cic1p/Nsa3p inactivation impairs pre-rRNA processing. (A) Structure and processing sites of the 35S pre-rRNA. This precursor contains the sequences for the mature 18S, 5.8S, and 25S, which are separated by the two internal transcribed spacers ITS1 and ITS2 and flanked by the two external transcribed spacers 5′ETS and 3′ETS. The positions of the oligonucleotides probes utilized in Northern hybridizations and primer extension analysis are indicated. (B, C) Northern analysis of pre-rRNA processing. Strains cic1–2 and wild type were growth at 25°C in YPD then shifted to 37°C. Cells were harvested at the indicated times and total RNAs were extracted. Equal amounts of RNA (5 μg) were resolved on 1.2% agarose/formaldehyde gel (B) and 6% acrylamide/urea gel (C) and transferred to a nylon membrane. The membranes were consecutively hybridized with the probes indicated in panel A. The position of the mature rRNAs and pre-rRNAs are indicated. (D) Primer extension analysis of the level of the 27SA2 and 27SB pre-rRNAs, which were detected using primer 006.
We conclude that Cic1p/Nsa3p is required for synthesis of 25S rRNA and 7S pre-rRNA, suggesting that the pre-rRNAs are destabilized as a result of Cic1p/Nsa3p deficiency. We do not believe that this is a secondary effect due to proteasome inactivation because specific pre-rRNA processing defects were not observed in strains carrying mutations in the proteasome components Rpt2p, Rpt3p, Rpt4p, Rpt5p, Rpt6p, Cim5p, and Rpn11p (A. Fatica, unpubl.). Cic1p is a nucleolar protein that was found to be associated with many nonribosomal proteins required for early pre-rRNA processing events and with the 27SA2, 27SB, and 7S pre-rRNAs (Nissan et al. 2002), strongly supporting a direct role in ribosome synthesis.
Many factors required for ribosome assembly are necessary for the export of the preribosomal particles (Baβler et al. 2001; Gadal et al. 2001; Milkereit et al. 2001; Schafer et al. 2003).
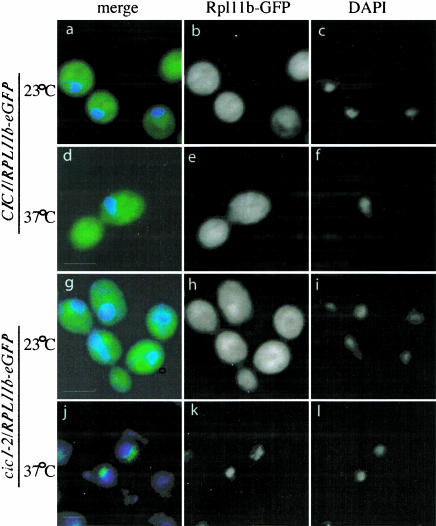
To assess the involvement of Cic1p in subunit transport, the wild-type and cic1–2 strains were transformed with a plasmid expressing an eGFP-tagged form of the 60S subunit ribosomal protein Rpl11b, and analyzed as previously described (Stage-Zimmermann et al. 2000). In the wild-type strain and in the cic1–2 strain at permissive temperature, Rpl11b-eGFP was predominately cytoplasmic (Fig. 4b,e,h ▶). Following transfer to 37°C for 4 h, the fusion protein showed nuclear localization (Fig. 4k ▶). The GFP-decorated region did not fully coincide with the nucleoplasm, identified by DAPI staining (Fig. 4j ▶), and generally showed the classic crescent shape of the yeast nucleolus.
FIGURE 4.
Cic1–2 accumulates Rpl11p-eGFP in the nucleolus. Cic1–2 and isogenic wild-type cells were transformed with Rpl11p-eGFP grown at 23°C, before a shift for 4 h to 37°C and inspection in the fluorescence microscope. (c, f, i, l) DAPI staining. (b, e, h, k) Localization of Rpl11p-eGFP. (a, d, g, j) Superimposition of the signals from Rpl11p-eGF and DAPI.
We conclude that in the cic1–2 strain, pre-60S particles accumulate in the nucleolus, indicating that Cic1p is required for the release of pre-60S from the nucleolus into the nucleoplasm. This transport defect is consistent with the fact that Cic1p/Nsa3p is present in early pre-60S particles that assemble in the nucleolus (Baβler et al. 2001; Harnpicharnchai et al. 2001; Nissan et al. 2002; Saveanu et al. 2003).
CONCLUSIONS
Cic1p (Nsa3p) was found associated with many nonribosomal proteins required for early pre-rRNA processing events in the pathway of 60S synthesis (Nissan et al. 2002) and was therefore predicted to be present in early pre-60S particles in the yeast nucleolus (Baβler et al. 2001; Harnpicharnchai et al. 2001; Nissan et al. 2002). Consistent with this model, we report that the cic1–2 mutant allele (Jager et al. 2001) inhibits the synthesis of the large subunit rRNAs. Pulse-chase, Northern, and primer extension analyses showed that Cic1p is required for normal maturation of the 27SA2 pre-rRNA to the 25S and 5.8S rRNAs. In addition, cic1–2 strains accumulated a GFP-tagged 60S reporter protein within the nucleolus, indicating a defect in release of pre-60S particles from the nucleolus into the nucleoplasm. A similar defect was previously seen for strains carrying a mutation in Noc1p (Milkereit et al. 2001), and Noc1p was coprecipitated with Cic1p (Nsa3p-TAP; Nissan et al. 2002) indicating their presence in the same early pre-60S particle.
Cic1p is one of a surprisingly large number of yeast ribosome synthesis factors that apparently function in additional cellular processes (Buscemi et al. 2000; Valasek et al. 2001; Zimmerman and Kellogg 2001; Du and Stillman 2002; Tone and Toh-E 2002; Zhang et al. 2002). Cic1p was initially identified as a protein required for degradation of specific substrates by the 26S proteasome (Jager et al. 2001). Interestingly, this is not the first preribosome component to show functional interactions with the ubiquitin–proteasome system. Nob1p was shown to be required for biogenesis of both the 40S ribosomal subunit and the 26S proteasome (Tone and Toh-E 2002; Fatica et al. 2003; Schafer et al. 2003). The nucleolar protein Krr1p, which is also required for 40S synthesis, was identified in a two-hybrid screen using Tom1p, a member of the E3 ubiquitin-ligase family (Sasaki et al. 2000). Moreover, three ribosomal proteins are synthesized as fusions with ubiquitin, whereas others undergo posttranslational ubiquitination that is regulated during the cell cycle (Finley et al. 1989; Spence et al. 2000). In these latter cases, ubiquitination does not function to target the acceptor protein for degradation but facilitates incorporation of the ribosomal proteins into functional subunits (Finley et al. 1989; Spence et al. 2000).
Many interactions between ribosome synthesis and the ubiquitin–proteasome system can be envisioned. Stoichiometry must be maintained between synthesis of the rRNAs and proteins and, at least under some conditions, this is largely ensured by the rapid degradation of excess ribosomal proteins (Ju and Warner 1994; Warner 1999), presumably via the proteasome. Moreover, preribosomes that are misassembled or in which the RNA is not correctly processed are generally very rapidly degraded, probably including the ribosomal protein components. Finally, ribosome biogenesis plays an important role in the regulation of cell growth and proliferation (Jorgensen et al. 2002; Ruggero and Pandolfi 2003). Communication between ribosome assembly factors and proteasome activity could be an important mechanism to synchronize ribosome synthesis with cell growth and division.
MATERIALS AND METHODS
RNA extraction, Northern hybridization, and primer extension
Standard techniques were employed for growth and handling of yeast. RNA was extracted as described previously (Kufel et al. 2000). Northern hybridization and primer extension were as described (Kufel et al. 2000). Standard 1.2% agarose/formaldehyde and 6% acrylamide/urea gels were used to analyze the high and low molecular weight RNA species, respectively.
Oligonucleotides
For RNA hybridizations and prime extensions, the following oligonucleotides were used:
001: 5′-CCAGTTACGAAAATTCTTG; 002: 5′-GCTCTTTGCTCT TGCC
003: 5′-TGTTACCTCTGGGCCC; 006: 5′-GGCCAGCAATTTCA AGTTA;
007: 5′-CTCCGCTTATTGATATGC; 008: 5′-CATGGCTTAATCT TTGAGAC;
013: 5′-GCGTTGTTCATCGATGC; 020: 5′-TGAGAAGGAAATG ACGCT;
033: 5′-CGCTGCTCACCAATGG; 041: 5′-CTACTCGGTCAGG CTC.
Pulse-chase labeling
Metabolic labeling of RNA was performed as described previously (Fatica et al. 2002). The strains cic1–2 and W303 (Jager et al. 2001) were transformed with a plasmid containing the URA3 gene and grown in glucose minimal medium lacking uracil. Cells were pregrown at permissive temperature (23°C) and transferred to restrictive temperature (37°C) for 4 h. Cells at 0.3 O.D.600 were labeled with [5,6-3H] uracil for 1 min followed by a chase with excess unlabeled uracil for 0, 1, 2.5, 5, 10, and 20 min. Standard 1.2% agarose/formaldehyde and 6% poly-acrylamide/urea gels were used to analyze the high and low molecular weight RNA species, respectively.
Rpl11b-eGFP localization assay
The assay was carried out as previously described (Stage-Zimmermann et al. 2000) with the following modifications. Cells were fixed in 3.7% formaldehyde for 30 min at room temperature before being washed in 1× PBS and were mounted using Vectashield-containing DAPI to stain the nuclei. Rpl11b-GFP was vizualized by fluorescence microscopy in the fluorescein channel using a Nikon microscope. Pictures were obtained using a Coolsnap HQ camera.
Acknowledgments
We thank Dieter H. Wolf for strains W303 and cic1–2, T. Rimaldi and M.H. Glickman for TS alleles of proteasome components, and Pamela Silver for plasmid-expressing Rpl11b-eGFP. D.T. and M.O. were supported by the Wellcome Trust. A.F. and I.B. were supported by grants from MURST (FIRB—p.n. RBNE015MPB and RBNE01KXC9), PRIN, and “Centro di eccellenza BEMM.”
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.rnajournal.org/cgi/doi/10.1261/rna.5130503.
REFERENCES
- Baβler, J., Granti, P., Gadal, O., Leβmann, T., Petfalski, E., Tollervey, D., and Lechner, J. 2001. Identification of a pre-ribosomal particle that is closely linked to nuclear export. Mol. Cell 8: 517–529. [DOI] [PubMed] [Google Scholar]
- Buscemi, G., Saracino, F., Masnada, D., and Carbone, M.L. 2000. The Saccharomyces cerevisiae SDA1 gene is required for actin cytoskeleton organization and cell cycle progression. J. Cell Sci. 113: 1199–1211. [DOI] [PubMed] [Google Scholar]
- Du, Y.C. and Stillman, B. 2002. Yph1p, an ORC-interacting protein: Potential links between cell proliferation control, DNA replication, and ribosome biogenesis. Cell 109: 835–848. [DOI] [PubMed] [Google Scholar]
- Fatica, A. and Tollervey, D. 2002. Making ribosome. Curr. Opin. Cell Biol. 14: 313–318. [DOI] [PubMed] [Google Scholar]
- Fatica, A., Cronshaw, A.D., Dlakic, M., and Tollervey, D. 2002. Ssf1p prevents premature processing of an early pre-60S ribosomal particle. Mol. Cell 9: 341–351. [DOI] [PubMed] [Google Scholar]
- Fatica, A., Oeffinger, M., Dlakic, M., and Tollervey, D. 2003. Nob1p is required for cleavage of the 3′ end of 18S rRNA. Mol. Cell. Biol. 23: 1798–1807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finley, D., Bartel, B., and Varshavsky, A. 1989. The tails of ubiquitin precursors are ribosomal proteins whose fusion to ubiquitin facilitates ribosome biogenesis. Nature 338: 394–401. [DOI] [PubMed] [Google Scholar]
- Gadal, O., Strauss, D., Braspenning, J., Hoepfner, D., Petfalski, E., Philippsen, P., Tollervey, D., and Hurt, E. 2001. A nuclear AAA-type ATPase (Rix7p) is required for biogenesis and nuclear export of 60S ribosomal subunits. EMBO J. 20: 3695–3704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glickman, M.H. and Ciechanover, A. 2001. The ubiquitin–proteasome proteolytic pathway: Destruction for the sake of construction. Physiol. Rev. 82: 373–428. [DOI] [PubMed] [Google Scholar]
- Harnpicharnchai, P., Jakovljevic, J., Horsey, E., Miles, T., Roman, J., Rout, M., Meagher, D., Imai, B., Guo, Y., Brame, C.J., et al. 2001. Composition and functional characterization of yeast 66s ribosome assembly intermediates. Mol. Cell 8: 663–670. [DOI] [PubMed] [Google Scholar]
- Jager, S., Strayle, J., Heinemeyer, W., and Wolf, D.H. 2001. Cic1, an adaptor protein specifically linking the 26S proteasome to its substrate, the SCF component Cdc4. EMBO J. 20: 4423–4431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgensen, P., Nishikawa, J.L., Breitkreutz, B.J., and Tyers, M. 2002. Systematic identification of pathways that couple cell growth and division in yeast. Science 297: 395–400. [DOI] [PubMed] [Google Scholar]
- Ju, Q. and Warner, J.R. 1994. Ribosome synthesis during the growth cycle of Saccharomyces cerevisiae. Yeast 10: 151–157. [DOI] [PubMed] [Google Scholar]
- Kufel, J., Allmang, C., Chanfreau, G., Petfalski, E., Lafontaine, D.L.J., and Tollervey, D. 2000. Precursors to the U3 snoRNA lack snoRNP proteins but are stabilized by La binding. Mol. Cell. Biol. 20: 5415–5424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milkereit, P., Gadal, O., Podtelejnikov, A., Trumtel, S., Gas, N., Petfalski, E., Tollervey, D., Mann, M., Hurt, E., and Tschochner, H. 2001. Maturation and intranuclear transport of pre-ribosomes requires Noc proteins. Cell 18: 499–509. [DOI] [PubMed] [Google Scholar]
- Milkereit, P., Kuhn, H., Gas, N., and Tschochner, H. 2003. The pre-ribosomal network. Nucleic Acids Res. 31: 799–804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nissan, T.A., Bassler, J., Petfalski, E., Tollervey, D., and Hurt, E. 2002. 60S pre-ribosome formation viewed from assembly in the nucleolus until export to the cytoplasm. EMBO J. 21: 5539–5547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruggero, D. and Pandolfi, P.P. 2003. Does the ribosome translate cancer? Nat. Rev. Cancer 3: 179–192. [DOI] [PubMed] [Google Scholar]
- Sasaki, T., Toh-E, A., and Kikuchi, Y. 2000. Yeast Krr1p physically and functionally interacts with a novel essential Kri1p, and both proteins are required for 40S ribosome biogenesis in the nucleolus. Mol. Cell. Biol. 20: 7971–7979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saveanu, C., Namane, A., Gleizes, P.E., Lebreton, A., Rousselle, J.C., Noaillac-Depeyre, J., Gas, N., Jacquier, A., and Fromont-Racine, M. 2003. Sequential protein association with nascent 60S ribosomal particles. Mol. Cell. Biol. 23: 4449–4460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schafer, T., Strauss, D., Petfalski, E., Tollervey, D., and Hurt, E. 2003. The path from nucleolar 90S to cytoplasmic 40S pre-ribosomes. EMBO J. 22: 1370–1380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spence, J., Gali, R.R., Dittmar, G., Sherman, F., Karin, M., and Finley, D. 2000. Cell cycle-regulated modification of the ribosome by a variant multiubiquitin chain. Cell 102: 67–76. [DOI] [PubMed] [Google Scholar]
- Stage-Zimmermann, T., Schmidt, U., and Silver, P.A. 2000. Factors affecting nuclear export of the 60S ribosomal subunit in vivo. Mol. Biol. Cell 11: 3777–3789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tone, Y. and Toh-E, A. 2002. Nob1p is required for biogenesis of the 26S proteasome and degraded upon its maturation in Saccharomyces cerevisiae. Genes & Dev. 16: 3142–3157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valasek, L., Hasek, J., Nielsen, K.H., and Hinnebusch, A.G. 2001. Dual function of eIF3j/Hcr1p in processing 20 S pre-rRNA and translation initiation. J. Biol. Chem. 276: 43351–43360. [DOI] [PubMed] [Google Scholar]
- Venema, J. and Tollervey, D. 1999. Ribosome synthesis in Saccharomyces cerevisiae. Annu. Rev. Gen. 33: 261–311. [DOI] [PubMed] [Google Scholar]
- Warner, J.R. 1999. The economics of ribosome biosynthesis in yeast. Trends Biochem. Sci. 24: 437–440. [DOI] [PubMed] [Google Scholar]
- Zhang, Y., Yu, Z., Fu, X., and Liang, C. 2002. Noc3p, a bHLH protein, plays an integral role in the initiation of DNA replication in budding yeast. Cell 109: 849–860. [DOI] [PubMed] [Google Scholar]
- Zimmerman, Z.A. and Kellogg, D.R. 2001. The Sda1 protein is required for passage through start. Mol. Biol. Cell 12: 201–219. [DOI] [PMC free article] [PubMed] [Google Scholar]