Abstract
Studies over the past years indicate that there is extensive coupling between nuclear export of mRNA and pre-mRNA processing. Here, we visualized the distribution of exon junction complex (EJC) proteins and RNA export factors relative to sites of abundant pre-mRNA synthesis in the nucleus. We analyzed both HeLa cells infected with adenovirus and murine erythroleukemia (MEL) cells stably transfected with the human β-globin gene. Using in situ hybridization and confocal microscopy, we observe accumulation of EJC proteins (REF/Aly, Y14, SRm160, UAP56, RNPS1, and Magoh) and core spliceosome components (U snRNPs) at sites of transcription. This suggests that EJC proteins bind stably to pre-mRNA cotranscriptionally. No concentration of the export factors NXF1/TAP, p15, and Dbp5 was detected on nascent transcripts, arguing that in mammalian cells these proteins bind the mRNA shortly before or after release from the sites of transcription. These results also suggest that binding of EJC proteins to the mRNA is not sufficient to recruit TAP-p15, consistent with recent findings showing that the EJC does not play a crucial role in mRNA export. Contrasting to the results obtained in MEL cells expressing normal human β-globin transcripts, mutant pre-mRNAs defective in splicing and 3′end processing do not colocalize with SRm160, REF, UAP56, or Sm proteins. This shows that the accumulation of EJC proteins at transcription sites requires efficient processing of the nascent pre-mRNAs, arguing that transcription per se is not sufficient for the stable assembly of the EJC.
Keywords: pre-mRNA splicing, mRNA export, exon junction complex, NXF1/TAP
INTRODUCTION
In eukaryotes, messenger RNAs are transcribed in the nucleus as precursor forms (pre-mRNAs). Immediately upon synthesis, nascent transcripts associate with proteins forming ribonucleoprotein (RNP) particles, the protein content of which evolves throughout the lifetime of a mRNA (for a recent review, see Dreyfuss et al. 2002). The proteins associated with messenger ribonucleoprotein particles (mRNPs) play key roles in all aspects of the RNA metabolism, from nucleocytoplasmic transport to cytoplasmic localization, translational efficiency, and decay. In particular, a specific subset of proteins including SRm160, RNPS1, Y14, Mago, DEK, REF/Aly, and UAP56 form the exon junction complex (EJC), which associates with mRNAs as a consequence of splicing (Blencowe et al. 1998; Mayeda et al. 1999; Kataoka et al. 2000; Le Hir et al. 2000a,b; McGarvey et al. 2000; Zhou et al. 2000; Luo et al. 2001). SRm160 and RNPS1 were originally characterized as activators of pre-mRNA splicing (Fleckner et al. 1997; Blencowe et al. 1998; Mayeda et al. 1999). More recently, SRm160 was shown to promote transcript 3′-end cleavage (McCracken et al. 2002), whereas RNPS1 couples splicing to nonsense-mediated decay (Lykke-Andersen et al. 2001). Y14 is involved in mRNA quality control via the nonsense-mediated mRNA decay (NMD) pathway (Lykke-Andersen et al. 2001), and together with Mago, is required for the proper cytoplasmic localization of oskar mRNA during Drosophila development (Hachet and Ephrussi 2001; Mohr et al. 2001). Y14 and Mago form a tight heterodimer in vivo (Lau et al. 2003), and the association between the two proteins is essential for function in NMD (Fribourg et al. 2003). DEK has been involved in multiple functions ranging from splicing (McGarvey et al. 2000) to chromatin structure (Alexiadis et al. 2000) and transcriptional regulation (Faulkner et al. 2001). REF/Aly has been identified as a chaperone that regulates the activity of bZIP transcription factors (Virbasius et al. 1999), and also as a factor that facilitates the nuclear export of mRNA by interacting with the export receptor NXF1/TAP (for review, see Reed and Hurt 2002). UAP56 is a putative RNA helicase also implicated as a splicing factor required for early spliceosome assembly (Kistler and Guthrie 2001; Libri et al. 2001; Zhang and Green 2001). Contrasting with NXF1/TAP and UAP56, the depletion of which causes nuclear accumulation of mRNA, REF/Aly, RNPS1, SRm160, and Y14 are dispensable for bulk mRNA export (Gatfield and Izaurralde 2002; Longman et al. 2003; MacMorris et al. 2003). Accordingly, SR proteins were identified recently as adaptors that, in addition to REF/Aly, can mediate the interaction between NXF1/TAP and cellular mRNAs (Huang et al. 2003).
Translocation of mRNAs through the nuclear pore complex requires binding of an heterodimer composed of NXF1/TAP and NXT1/p15 (Mex67 and Mtr2 in Saccharomyces cerevisiae). NXF1/TAP, which interacts both with RNA-binding adapter proteins and components of the nuclear pore complex, is believed to be the major receptor for the export of mRNAs to the cytoplasm (for recent reviews, see Izaurralde 2002; Lei and Silver 2002b; Reed and Hurt 2002; Cullen 2003; Stutz and Izaurralde 2003). Another protein that interacts simultaneously with mRNPs and nucleoporins is Dbp5/Rat8 (Snay-Hodge et al. 1998; Tseng et al. 1998). Although in yeast Dbp5 is essential for mRNA export to the cytoplasm (Snay-Hodge et al. 1998; Tseng et al. 1998), depletion of the Drosophila homolog of Dbp5 does not result in the bulk nuclear retention of mRNAs (Gatfield et al. 2001).
How export-competent mRNPs assemble in the living cell nucleus and travel to the nuclear pores remains poorly understood. Here, we visualize the localization of EJC proteins and mRNA export factors in the nucleus of mammalian cells producing abundant pre-mRNA transcripts. First, we analyzed HeLa cells infected with adenovirus, because this virus recruits the host transcription and processing machinery to the sites of viral pre-mRNA synthesis (Jimenez-Garcia and Spector 1993; Pombo et al. 1994). In addition, we made use of murine erythroleukemia (MEL) cells stably transfected with the human β-globin gene under the control of the locus control region (LCR). The human β-globin gene, which integrates in the host cell genome as a tandem array, expresses at physiological levels upon induction of MEL cells to undergo terminal erythroid differentiation (Collis et al. 1990). Using in situ hybridization and confocal microscopy, we observe colocalization of EJC proteins (REF/Aly, Y14, SRm160, UAP56, RNPS1, and Magoh) and core spliceosome components (U snRNPs) at sites in the nucleus containing nascent adenoviral and β-globin transcripts. In contrast, the export factors NXF1/TAP, p15, and Dbp5 are not detected at sites of transcription. The results further show that REF/Aly, SRm160, UAP56, and spliceosomal snRNPs fail to accumulate at the site of transcription of a processing-defective β-globin mutant. Taken together, these data suggest that EJC proteins bind cotranscriptionally to mRNPs, and that efficient pre-mRNA processing is required for assembly of EJC proteins onto nascent transcripts in vivo.
RESULTS
Components of the EJC, but not NXF1/TAP, p15, or Dbp5, are recruited to sites of adenoviral transcription in the nucleus of infected HeLa cells
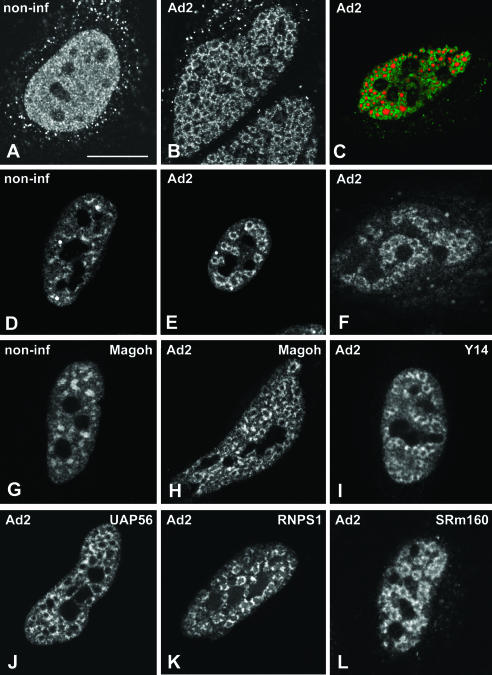
Adenovirus type 2 (Ad2) causes a productive infection of HeLa cells that proceeds through an infectious cycle of ~36 h. This cycle is conventionally divided into early and late stages, separated by the onset of viral DNA replication (for review, see Philipson et al. 1975). Adenoviruses enter the host cells by receptor-mediated endocytosis, penetrate the cytoplasm from endosomes, and deliver their DNA genome into the nucleus (Greber et al. 1993). Upon entry in the nucleus, transcription of the viral genome starts immediately, and the viral mRNAs transcribed at this early stage direct the synthesis of a small number of proteins that promote viral DNA replication. After the onset of viral DNA replication, which occurs at ~8 h post-infection, the remaining viral genomic information is expressed, yielding large quantities of the structural proteins that will eventually constitute new virus particles (Philipson et al. 1975; Flint 1986). During this period, the normal nuclear architecture undergoes a series of progressive changes (for review, see Pombo and Carmo-Fonseca 1995). The replication of Ad DNA produces single-stranded intermediates (ssDNA) that accumulate, forming viral inclusions in the nucleoplasm. These inclusions are readily identified using antibodies against the viral ssDNA-binding protein DBP. Following incorporation of a modified uridine-analog, it is possible to visualize by fluorescence microscopy the sites of RNA synthesis in the nucleus (Pombo et al. 1994). In noninfected cells, newly synthesized RNAs incorporating bromouridine are detected widespread throughout the nucleoplasm (Fig. 1A ▶). In infected cells, nascent transcripts form ring-like structures (Fig. 1B ▶) that surround the inclusions containing viral ssDNA (Fig. 1C ▶). Previous work has indicated that these ring-like structures represent nascent viral mRNA transcripts (Pombo et al. 1994).
FIGURE 1.
Localization of EJC proteins in adenovirus-infected cells. HeLa cells were either noninfected (A,D,G; non-inf) or infected with adenovirus for 14–16 h (B,C,E,F,H–L; Ad2). To detect nascent transcripts, cells were incubated with Br-U for 1 h (A–C). (C) A cell double-labeled for nascent transcripts (red) and viral ssDNA inclusions using anti-DBP antibody (green staining). To visualize spliceosome snRNPs, cells were immunolabeled with Y12 antibody directed against Sm proteins (D,E). Spliced viral mRNAs were detected by in situ hybridization using a probe (SJ1), which hybridizes with all mRNAs encoded by the highly expressed adenovirus major late transcription unit (Bridge et al. 1996) (F). The localization of EJC proteins was determined in cells expressing GFP–Magoh (G,H), zzY14 (I), GFP–UAP56 (J), GFP–RNPS1 (K). The distribution of SRm160 was visualized with specific rabbit polyclonal antibodies (L). Bar, 10 μm.
Consistent with previous evidence for cotranscriptional processing of pre-mRNA obtained in noninfected cells (Beyer and Osheim 1988; LeMaire and Thummel 1990; Wu et al. 1991; Xing et al. 1993; Bauren and Wieslander 1994; Zhang et al. 1994; Tennyson et al. 1995; Bauren et al. 1996; Huang and Spector 1996; Neugebauer and Roth 1997), in situ evidence suggests that splicing of Ad mRNAs occurs cotranscriptionally. Namely, spliceosome components including snRNPs and non-snRNP splicing proteins are recruited to the sites of viral mRNA synthesis (Pombo et al. 1994; Bridge et al. 1995; Gama-Carvalho et al. 1997, 2003). Furthermore, spliced major late-viral mRNAs are detected in the ring-like structures formed by the nascent transcripts (Bridge et al. 1996; Gama-Carvalho et al. 2003, Fig. 1D–F ▶).
Here we analyzed whether protein components of the EJC become concentrated at sites of Ad transcription. The results show that in noninfected cells, REF/Aly, Y14, SRm160, UAP56, RNPS1, and Magoh are localized throughout the nucleoplasm with higher concentration in nuclear speckles, and excluding nucleoli (Fig. 1G ▶; data not shown), as previously described (Blencowe et al. 1998; Loyer et al. 1998; Gatfield et al. 2001; Le Hir et al. 2001a; Rodrigues et al. 2001). At 14–16 h post-infection, each of these proteins is predominantly concentrated in ring-like structures (Fig. 1H–L ▶). Double-labeling experiments confirm that EJC proteins colocalize with spliced Ad mRNA and spliceosomal snRNPs at the ring-like structures formed by nascent viral transcripts (Fig. 2A–C ▶). In contrast to REF/Aly, Y14, SRm160, UAP56, RNPS1, and Magoh, the protein DEK appears more homogeneously distributed throughout the nucleoplasm with less pronounced accumulation in either nuclear speckles in noninfected cells or ring-like structures in Ad-infected cells (Fig. 2D–F ▶).
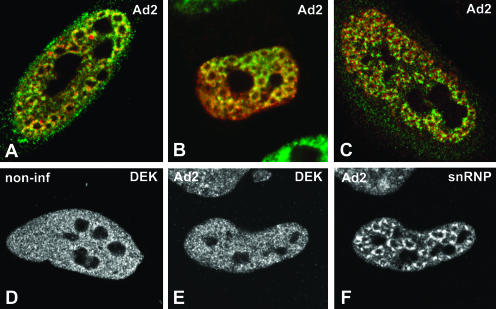
FIGURE 2.
Colocalization of EJC proteins and spliceosomal snRNPs in adenovirus-infected cells. (A–C) A superimposition of red and green images corresponding to double-labeling experiments performed in HeLa cells infected with adenovirus for 14–16 h. The distribution of EJC proteins is compared with that of spliceosomal snRNPs (A,B) and spliced viral major late mRNAs (C). (A) Cells were double labeled with antibodies directed against REF/Aly (green staining) and Sm proteins (red staining). (B) Cells expressing zzY14 (red staining) were immunolabeled with an antibody specific for the U2 snRNP B″ protein (green staining). (C) Cells were immunolabeled with anti-SRm160 antibodies (green staining) and hybridized with SJ1 probe (red staining). The distribution of DEK was analyzed by immunolabeling in noninfected (D) and infected (E) HeLa cells. The cell depicted in E was double labeled with antibody Y12, specific for Sm proteins (F).
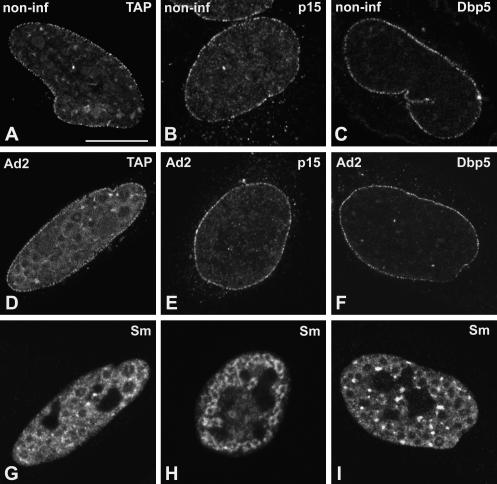
Unlike EJC components, the proteins NXF1/TAP, p15, and Dbp5 do not accumulate in nuclear speckles of noninfected cells, but rather, distribute homogeneously throughout the nucleoplasm with higher concentration at the nuclear rim (Schmitt et al. 1999; Bachi et al. 2000; Herold et al. 2000). Following treatment with TritonX-100 prior to fixation, the nucleoplasmic pool of NXF1/TAP, p15, and Dbp5 is largely solubilized and the proteins are predominantly detected at the nuclear rim (Fig. 3A–C ▶). This localization reflects interactions with the nuclear pore complexes, as previously described (Schmitt et al. 1999; Bachi et al. 2000; Herold et al. 2000). At 14–16 h after adenoviral infection, NXF1/TAP, p15, and Dbp5 remain predominantly localized at the nuclear rim (Fig. 3D–F ▶). Neither of these proteins is detected in the ring-like structures that concentrate spliceosomal snRNPs (Fig. 3G–I ▶).
FIGURE 3.
Localization of NXF1/TAP, p15, and Dbp5 in adenovirus-infected cells. HeLa cells were either noninfected (A–C; non-inf) or infected with adenovirus for 14–16 h (D–I; Ad2). Cells were transiently transfected with plasmids encoding GFP–TAP (A,D), zzp15 (B,E), and GFP–Dbp5 (C,F). The cells depicted in D, E, and F were double labeled with antibody Y12, specific for Sm proteins (G,H,I, respectively). Bar, 10 μm.
In summary, our results show that REF/Aly, Y14, SRm160, UAP56, RNPS1, and Magoh are all recruited to sites of adenoviral mRNA synthesis in the nucleus of infected HeLa cells. In contrast, NXF1/TAP, p15, and Dbp5 are not detected at sites of viral transcription.
Components of the EJC, but not NXF1/TAP or p15, are recruited to sites of β-globin transcription in the nucleus of MEL cells
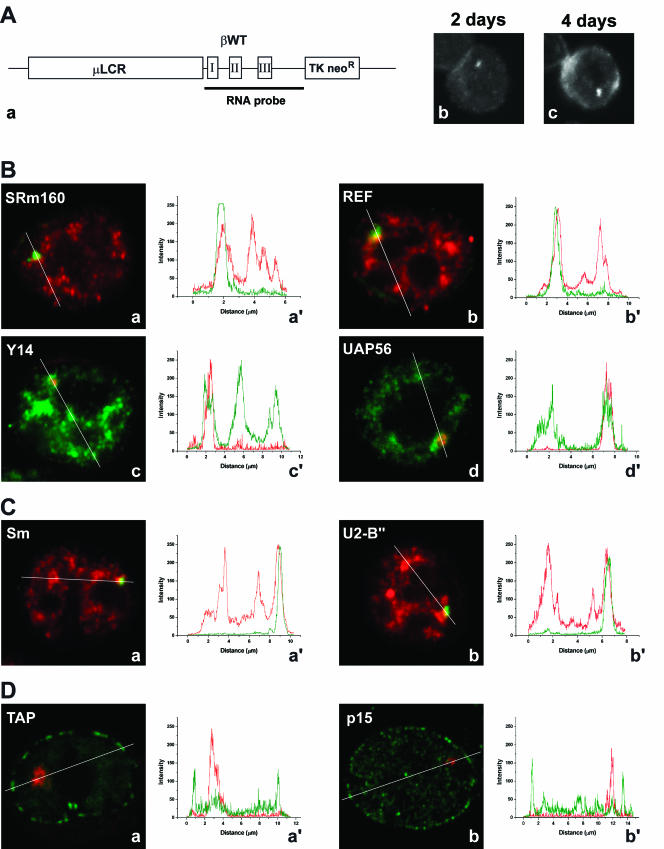
Having shown that EJC components accumulate at sites of adenoviral transcription, we next sought to visualize recruitment of these proteins to nascent cellular transcripts. As a model system, we used murine erythroleukemia (MEL) cells stably transfected with the human β-globin gene. The clone used in this study (MELβWT) harbors ~14 copies of the transgene as a tandem array (Custódio et al. 1999). Detection of the human β-globin transcripts by FISH was performed with a probe complementary to the transcribed sequence of the gene (RNA probe, Fig. 4A, a ▶). Expression of the human β-globin transgenes was induced by dimethylsulfoxide, which triggers terminal erythroid differentiation of the MEL cells (Antoniou 1991). In cells induced for 2 d, nascent β-globin transcripts are detected as a focus in the nucleus (Fig. 4A, b ▶). After induction for 4 d, the cells contain a focus in the nucleus corresponding to nascent β-globin transcripts and additional cytoplasmic staining (Fig. 4A, c ▶). To determine whether EJC proteins colocalize with nascent β-globin transcripts, MELβWT cells were induced to differentiate and sequentially hybridized with the RNA probe and immunolabeled with antibodies against SRm160 and REF (Fig. 4B, a and b ▶). Alternatively, cells were transfected with plasmids encoding tagged versions of Y14 and UAP56 (Fig. 4B, c and d ▶). The results indicate that each EJC protein colocalizes with the β-globin RNA focus in the nucleus. Quantification of the fluorescence intensity along a line that spans the nucleus across the RNA focus shows that the concentration of EJC proteins at the site of βglobin transcription is higher than in surrounding areas of the nucleoplasm (Fig 4B, a′–d′ ▶). Similar results were obtained for MEL cells labeled with antibodies against Sm proteins (Fig. 4C, a and a′ ▶) or U2snRNP B″ protein (Fig. 4C, b and b′ ▶).
FIGURE 4.
Localization of EJC proteins, NXF1/TAP, and p15 in the nucleus of MEL cells. (A, a) Schematic representation of the wild-type human β-globin construct. The wild-type human β-globin gene (βWT) is within the microlocus control region (μLCR) expression cassette (Collis et al. 1990). The TK neoR gene confers resistance to G418 in stable transfected MEL cells. The probe used to detect the human β-globin transcripts by FISH (RNA probe) corresponds to a DNA fragment complementary to the transcribed sequence of the gene. (b,c) Cells induced for 2 and 4 d, and hybridized with the RNA probe. (B) EJC proteins were detected either by indirect immunofluorescence using specific antibodies (a,b), or by transfection with zz or GFP-tagged constructs (c,d). (a,b) MELβWT were induced to differentiate for 3 d, hybridized with the RNA probe (green), and labeled with antibodies (red) against SRm160 (a) or REF (b). (c,d) Cells were first transfected with either zzY14 (c) or GFP–UAP56 (d), induced to differentiate for 2 d, and hybridized with the RNA probe directly labeled with Cy3 (red). The graphics in a′, b′, c′, and d′ correspond to a quantitative measurement of colocalization of the EJC proteins at the sites of human β-globin transcription. A line scan was made across the nucleus, including the site of β-globin transcription as indicated in the figures. The color of the lines in the graphics matches the color of the detection of RNA and protein in the cell. (C) MELβWT were induced to differentiate for 3 d, hybridized with the RNA probe (green), and labeled with antibodies (red) against either U2 snRNP B" with mAb 4G3 (b) or Sm with mAb Y12 (a). The graphics in a′, b′, correspond to a quantitative measurement of colocalization of spliceosomal proteins (red) at the sites of human β-globin transcription (green). (D) MELβWT were transfected with either GFP–TAP (a) or zzp15 (b), induced to differentiate for 2 d, and hybridized with the RNA probe directly labeled with Cy3 (red). The zz tag (green staining) was detected after in situ hybridization by indirect immunofluorescence with an antibody against protein A. The graphics in a′ and b′ correspond to a quantitative measurement of colocalization of NXF1/TAP or p15 (green) at the sites of human β-globin transcription (red).
Next, MELβWT cells expressing tagged NXF1/TAP or p15 were hybridized with the β-globin RNA probe (Fig. 4D, a and b ▶). NXF1/TAP and p15 are both detected homogeneously distributed throughout the nucleoplasm with higher accumulation at the nuclear rim, reflecting interactions with the nuclear pore complexes. Quantification of the fluorescence intensity along a line across the RNA focus clearly shows that NXF1/TAP and p15 do not concentrate at the site of β-globin transcription (Fig. 4D, a′ and b′ ▶).
EJC proteins are not recruited by splicing-defective mutant β-globin transcripts
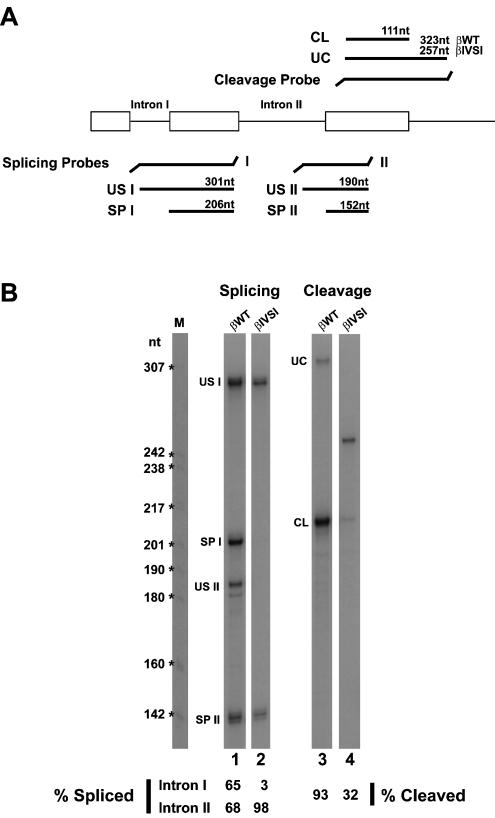
Current biochemical evidence indicates that EJC proteins are deposited onto mRNAs as a consequence of splicing (Le Hir et al. 2000a,b, 2001b; Kataoka et al. 2001; Kim et al. 2001). To study the role of splicing on recruitment of EJC proteins to sites of transcription in the nucleus, we have made use of MEL cells stably transfected with a human β-globin mutant gene that contains a normal first intron but is devoid of the second intron (termed βIVSI).
To analyze the processing of βIVSI pre-mRNAs in the nucleus of MEL cells, we performed RNase protection assays using nuclear RNA fractions from 4-day-induced cells and 32P-labeled antisense RNA probes (Fig. 5A ▶). The protection products were quantified by PhosphorImager, and the results show that for the wild type β-globin transcripts (βWT), the percentage of splicing of introns I and II is 65 and 68, respectively (Fig. 5B ▶, lane 1). Thus, splicing of both introns has occurred in the majority of wild type β-globin transcripts (βWT) present in the nucleus. Consistent with the genomic deletion of the second intron in the βIVSI mutant, no intron II sequences are detected in the corresponding transcripts (Fig. 5B ▶, lane 2). Unexpectedly, the first intron remains unspliced in all βIVSI transcripts (Fig. 5B ▶, lane 2). Furthermore, and according to data reported previously (Collis et al. 1990; Antoniou et al. 1998), the efficiency of 3′-end cleavage is reduced from 93% in βWT RNA to 32% in βIVSI transcripts (Fig. 5B ▶, lanes 3,4).
FIGURE 5.
RNase protection assays. (A) Schematic representation of the RNase protection probes used to analyze splicing and 3′-end cleavage of the human β-globin transcripts. The predicted RNase protection products are shown for each probe. (B) Nuclear RNA fractions (3 μg) of βwild-type (lanes 1,3) and βIVSI (lanes 2,4) cells induced to differentiate for 4 d were analyzed by RNase protection using either the splicing protection probes I and II simultaneously (lanes 1,2) or the cleavage probe (lanes 3,4). The identity of each RNA species is indicated. The uncleaved product is shorter for βIVSI, because this transgene terminates 45 bp past the poly(A)-addition site, whereas the βWT terminates at 1800 pb. This difference in the extent of 3′ sequences does not in itself compromise the efficiency of 3′ end formation (Antoniou et al. 1998). The amounts of unspliced (US), spliced (SP), uncleaved (UC), and cleaved (CL) RNAs were quantified by PhosphorImager and the values normalized for U content. The percentage of spliced intron I and intron II is indicated below lanes 1 and 2, and the percentage of cleaved RNA is indicated below lanes 3 and 4.
We have previously shown that βIVSI transcripts are not exported to the cytoplasm, being retained at the site of transcription (Custódio et al. 1999). We consider it unlikely that the reduced cleavage efficiency is the primary cause of nuclear retention, as another β-globin splicing mutant possessing a 5′-splice site mutation (GT→AC) in intron II, is correctly cleaved at normal rates, yet is also stalled at the site of transcription (Custódio et al. 1999). However, we cannot rule out the possibility that inefficient 3′-end processing contributes to retention of nascent β-globin RNA at the site of transcription.
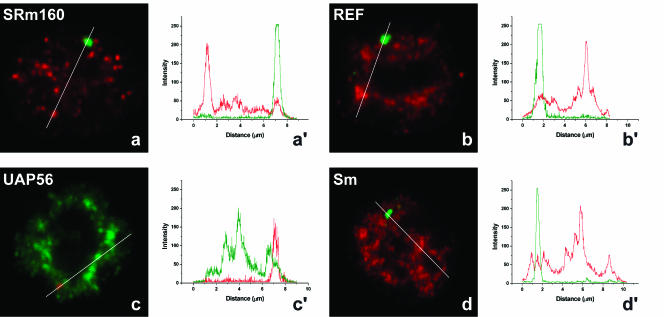
Having determined that there is no splicing of βIVSI transcripts in the nucleus of MEL cells, we next performed double-labeling experiments using a probe to detect β-globin RNA and antibodies or GFP to detect SRm160, REF, UAP56, and Sm. The results clearly show no significant colocalization of the RNA and protein signals (Fig. 6a–d ▶). Quantification of the fluorescence intensity along a line that spans the nucleus across the RNA focus confirms that there is no accumulation of EJC or Sm proteins at the site of transcription of the mutant β-globin gene (Fig. 6a′–d′ ▶). This suggests that, in the absence of efficient pre-mRNA processing, both spliceosomal components and EJC proteins fail to be properly assembled onto nascent β-globin transcripts in the nucleus.
FIGURE 6.
EJC proteins and spliceosomal snRNPs do not colocalize with mutant β-globin nascent transcripts. MELβIVSI were induced to differentiate for 3 d, hybridized with the RNA probe (green), and labeled with antibodies against SRm160 (a), REF (b), or Sm proteins (d; red). (c) Cells were first transfected with GFP–UAP56 (green), induced to differentiate for 2d, and hybridized with the RNA probe directly labeled with Cy3 (red). The graphics in a′, b′, c′, and d′ correspond to a quantitative measurement of colocalization of the EJC proteins at the sites of mutant β-globin transcription. A line scan was made across the nucleus as indicated in the figures, and the color of the lines in the graphics matches the color of the detection of RNA and protein in the cell.
DISCUSSION
In this work, we used a microscopy approach to study recruitment of the EJC to nascent transcripts in the nucleus of mammalian cells. We find that EJC proteins colocalize with protein components of the spliceosome at sites of transcription and splicing in the nucleoplasm, providing in vivo evidence that the EJC binds cotranscriptionally to mRNPs. We further report that mutant β-globin pre-mRNAs that are neither spliced nor released from the site of transcription fail to recruit spliceosome snRNPs and EJC proteins. This suggests that splicing of β-globin pre-mRNAs is required for efficient recruitment of the EJC and subsequent targeting of the resulting mRNP to the nuclear export pathway.
We show that the EJC proteins SRm160, RNPS1, Y14, Magoh, REF/Aly, and UAP56 accumulate in the nucleus at sites of abundant adenoviral and β-globin pre-mRNA synthesis. This is consistent with reports that the yeast homolog of REF/Aly, Yra1, associates with transcribed chromatin (Lei et al. 2001; Lei and Silver 2002a), and that both Yra1 and Sub2 (the yeast counterpart of UAP56) associate with the THO transcription complex (Strasser et al. 2002). A different result is observed for DEK, which appears more homogeneously distributed throughout the nucleoplasm, with no clear concentration at transcription sites. Remarkably, recent immunopurification analysis of nuclear mRNPs detected all EJC components except DEK (Lykke-Andersen et al. 2001; Lejeune et al. 2002). The failure to detect DEK using both microscopic and biochemical approaches suggests that either DEK associates loosely with the EJC or it is not part of this complex in vivo.
In contrast to EJC proteins, Dbp5, NXF1/TAP, and p15 fail to concentrate in the nucleus at sites of abundant pre-mRNA synthesis. Dbp5 was identified originally in yeast as a DEAD-box helicase implicated in mRNA export (for review, see Reed and Hurt 2002). Although early studies suggested that the function of metazoan Dbp5 is conserved, more recent analysis using RNA interference indicate that this protein is not essential for mRNA export in Drosophila (Gatfield et al. 2001). Like the yeast protein, human Dbp5 (hDbp5) shuttles between the nucleus and the cytoplasm, but at steady state, it is detected mainly in the cytoplasm (Schmitt et al. 1999). The Dbp5/hDbp5 protein appears enriched at the nuclear periphery, where it interacts with nucleoporins located at the cytoplasmic fibrils of nuclear pore complexes (Hodge et al. 1999; Schmitt et al. 1999; Strahm et al. 1999). Here, we observe that Dbp5 distribution is not altered upon infection of HeLa cells with adenovirus (Fig. 3C,F ▶). No Dbp5 labeling was detected at sites of nascent viral pre-mRNAs, suggesting that in mammalian cells, this protein is recruited to mRNPs shortly before or after release from the sites of transcription. However, a recent report indicates that a Dbp5 homolog in the dipteran Chironomus tentans (Ct-Dbp5) binds to the Balbiani ring pre-mRNP cotranscriptionally and accompanies the mRNP to and through the nuclear pores (Zhao et al. 2002). An involvement of yeast Dbp5 during the early steps of transcription was also suggested on the basis of genetic and physical interactions with the transcriptional factor IIH (Estruch and Cole 2003). Due to the sensitivity limits of our assay, we cannot exclude that some Dbp5 molecules may associate with nascent transcripts.
The heterodimer formed by vertebrate NXF1/TAP and NXT1/p15 represents to date the most important receptor involved in export of mRNA from the nucleus to the cytoplasm (Izaurralde 2002; Lei and Silver 2002b; Reed and Hurt 2002; Cullen 2003; Stutz and Izaurralde 2003). Several EJC proteins, including REF/Aly, Y14, and Magoh can bind to NXF1/TAP, suggesting that these factors may act as adaptors that recruit the export receptor to mRNAs (Dreyfuss et al. 2002; Reed and Hurt 2002). However, our observation that NXF1/TAP and NXT1/p15 do not accumulate at sites of transcription suggests that binding of the EJC proteins to the mRNA is not sufficient to recruit the TAP-p15 heterodimer. This is consistent with the recent finding that REF/Aly and other components of the EJC are dispensable for export of bulk mRNA in Drosophila (Gatfield and Izaurralde 2002) and Caenorhabditis elegans (Longman et al. 2003; MacMorris et al. 2003), indicating that additional adaptor proteins mediate the interaction between NXF1/TAP and mRNAs in metazoan. SR proteins were recently identified as adaptors that, in addition to REF/Aly, can mediate the interaction between NXF1/TAP and cellular mRNAs (Huang et al. 2003).
Although REF/Aly can interact directly with NXF1/TAP (Strasser and Hurt 2000; Stutz et al. 2000), and NXF1/TAP coimmunopurifies with EJC proteins in nuclear mRNP fractions (Lejeune et al. 2002), there is no evidence that the TAP-p15 heterodimer is recruited to mRNAs cotranscriptionally. In fact, NXF1/TAP was reported to bind very weakly to purified spliced mRNPs in vitro (Zhou et al. 2000). Our observation that NXF1/TAP and p15 do not concentrate at the sites of pre-mRNA transcription, therefore, favors the view that this heterodimer is recruited efficiently at a later stage in the export pathway.
At present, it is well established that EJC proteins are deposited onto mRNAs as a consequence of splicing (Le Hir et al. 2000a,b, 2001b; Kataoka et al. 2001; Kim and Dreyfus 2001). More recent studies described the timing of EJC assembly on spliced mRNA (Lejeune et al. 2002; Reichert et al. 2002). REF/Aly was shown to interact with pre-mRNA prior to spliceosome assembly, whereas Y14, Magoh, RNPS1, UAP56, and SRm160 are found in intermediate-containing spliceosomes (Reichert et al. 2002). Upon exon ligation, association of RNPS1, UAP56, and SRm160 are destabilized, whereas REF/Aly, Y14, and Magoh remain stably bound to the spliced exons (Reichert et al. 2002).
In a previous study, we have shown that transcripts encoded by a human β-globin mutant gene (βIVSI) that is devoid of the second intron fails to be released from the site of transcription (Custódio et al. 1999). Although pre-mRNAs encoded by this mutant contain a normal first intron, RNase protection assays show that splicing of this intron is almost completely inhibited in βIVSI transcripts (Fig. 5B ▶). Thus, the βIVSI mutant gene generates pre-mRNAs that are not spliced. Notably, βIVSI transcripts conserve the normal sequence spanning the second and third exons, but this exon–exon junction is no longer the product of a splicing reaction. In clear contrast with the results observed in MEL cells expressing normal human β-globin transcripts, βIVSI pre-mRNAs do not colocalize with SRm160, REF, UAP56, or Sm proteins. This argues that the accumulation of EJC proteins at transcription sites requires efficient pre-mRNA processing. However, there is previous evidence indicating that REF/Aly can associate with the region 20–24 nucleotides upstream of exon–exon junctions independent of splicing (Reichert et al. 2002), that UAP56 binds to nascent transcripts independent of the localization of introns (Kiesler et al. 2002), and that both REF/Aly and UAP56 are part of the TREX (transcription/export) complex, which is thought to be recruited during transcription (Strasser et al. 2002). Possibly, in our microscopic assay we fail to visualize proteins that are loosely associated with the RNA and detect only stable complexes. According to this view, our results would argue that the initial binding of REF/Aly and UAP56 to nascent transcripts is not sufficient for stable EJC assembly.
In addition to being splicing deficient, βIVSI transcripts are not properly 3′-end cleaved. This is consistent with recent findings showing that splicing stimulates mRNA biogenesis by enhancing mRNA 3′-end processing (Lu and Cullen 2003; Nott et al. 2003). Because this effect appears to be mediated by the EJC (Wiegand et al. 2003), failure of βIVSI transcripts to recruit EJC proteins may contribute to their inefficient cleavage.
In conclusion, we find that EJC proteins colocalize with protein components of the spliceosome at sites of transcription and splicing in the nucleoplasm, providing in vivo evidence that the EJC binds cotranscriptionally to mRNPs. We further report that mutant β-globin pre-mRNAs that are neither correctly processed nor released from the site of transcription fail to concentrate spliceosome snRNPs and EJC proteins. This suggests that processing of β-globin pre-mRNAs is required for efficient recruitment of the EJC and also for subsequent targeting of the resulting mRNP to the nuclear export pathway.
MATERIALS AND METHODS
HeLa cell culture, transfections, and Adenovirus infection
HeLa cells from ECACC were cultured as monolayers in minimum essential medium (MEM, GIBCO-BRL) supplemented with 10% fetal calf serum (FCS, GIBCO-BRL) and nonessential amino acids (Invitrogen Corporation). Experiments were performed with cells grown on 10 × 10 mm glass coverslips. Transfections were performed on subconfluent cells using FuGENE 6 reagent (Roche Biochemicals), and cells were fixed for analysis 24 h after transfection. Infections with Adenovirus Type 2 (Ad2, strain wt900) were performed as described previously (Pombo et al. 1994). Briefly, cells were incubated with Ad2 for 2 h in serum-free medium, and then supplemented with 10% FCS. For simultaneous Ad2 infection and transfection, the transfection mixture was added to the cells immediately after the addition of the FCS. For in situ detection of sites of transcription, the cells were incubated for 1 h in culture medium supplemented with 2 mM BrU (Sigma-Aldrich), and fixed for analysis.
MEL cell culture and transfections
The maintenance and induction of erythroid differentiation of the MEL cell lines were described previously (Antoniou 1991; Custódio et al. 1999). Transient transfections were performed with either FuGENE 6 reagent (Roche Biochemical) or Tfx-50 reagent (Promega). The erythroid differentiation was induced 5–6 h after transfection by the addition of 2% (v/v) DMSO to the culture medium, and allowed to proceed for 48 h.
DNA clones for transient transfections
The plasmids used for expression in HeLa and MEL cells were kindly provided by Elisa Izaurralde (EMBL-Heidelberg). NXF1/TAP, RNPS1, UAP56, Magoh, and Dbp5 full-length cDNAs were cloned in pEGFP (Clontech). Y14 and p15 cDNAs were cloned in the pRN3zz vector. The zz tag was detected by indirect immunofluorescence with an antibody against protein A.
In situ hybridization
HeLa cells were grown on glass coverslips, and MEL cells were washed in serum-free medium and allowed to adhere onto poly-L-lysine (Sigma) coated glass coverslips. Cells were fixed and permeabilized according to one of the following alternative protocols: (1) permeabilized with 0.5% Triton X-100 in CSK buffer (Fey et al. 1986) containing 0.1 mM PMSF for 1 min on ice, and fixed with 3.7% paraformaldehyde in CSK buffer for 10 min at room temperature; (2) fixed in 3.7% paraformaldehyde in PBS for 10 min, and permeabilized with 0.5% Triton X-100 in PBS for 10 min at room temperature.
Spliced major late adenoviral mRNAs were visualized using a 5′-biotinylated DNA oligonucleotide probe, as previously described (Bridge et al. 1996). Oligonucleotides are particularly suited to identify spliced mRNA molecules, as probes complementary to exon–exon splice junctions form unstable hybrids with the unspliced primary transcripts. The probe (termed SJ1) has the following sequence: CAACCGCGAGCCCAACAGCTG. All in situ hybridization procedures were based on the protocols previously described (Bridge et al. 1996), and the hybrids were detected with Cy3 avidin (1/200, Jackson ImmunoReseach Labs, Inc).
To detect the human β-globin transcripts, MEL cells were hybridized as previously described (Custódio et al. 1999). The probe used consists of a plasmid containing the genomic sequence of the human β-globin gene labeled with either digoxigenin-11-dUTP (Roche) or Cy3-AP3-dUTP (Amersham Pharmacia) by nick translation. Digoxigenin detection was either with Fluorescein-conjugated sheep anti-digoxigenin (1/100, Roche), followed by AlexaFluor488-conjugated goat anti-Fluorescein (1/200, Molecular Probes), or Cy3 conjugated mouse anti-digoxin (1/250, Jackson ImmunoReseach Labs, Inc.). After the detection steps, cells were fixed with 1% formaldehyde in PBS for 10 min, washed with PBS, and processed for indirect immunofluorescence.
Immunofluorescence
The following primary antibodies were used for immunofluorescence. Rabbit polyclonals directed against (1) SRm160 (1/250, Blencowe et al. 1998); (2) DEK (1/300, Fornerod et al. 1995); (3) REF (1/100, Rodrigues et al. 2001); (4) protein A (1/2000, Sigma); (5) adenoviral protein DBP (Linné et al. 1977). Mouse monoclonals directed against (1) Sm antigen of SnRNPs (mAb Y12; Lerner et al. 1981); (2) U2 snRNP specific protein B″ (mAb 4G3; Habets et al. 1989). Bromo-uridine incorporation was detected with a sheep polyclonal antibody directed against BrdU (1/200, Abcam).
The secondary antibodies used were AlexaFluor488-conjugated goat anti-rabbit IgG (1/200), TRITC-conjugated donkey anti-rabbit IgG (1/100, Jackson ImmunoResearch Labs, Inc.), AlexaFluo488-conjugated goat anti-mouse IgG (1/200, Jackson), Cy3-conjugated goat anti-mouse IgG (1/300, Jackson), FITC-conjugated goat anti-human IgG (1/100, Jackson), Cy5-conjugated rabbit anti-human IgG (1/100, Jackson), and FITC-conjugated donkey anti-sheep IgG (1/100, Jackson).
Microscopy
Images were acquired on a Zeiss LSM 510 confocal microscope using the PlanApochromat 63×/1.4 objective. FITC and AlexaFluor488 fluorescence was detected using the 488-nm line of the argon ion laser. The 543-nm line of the helium-neon laser was used to excite Cy3 and TRITC and the 633-nm line to excite Cy5.
RNase protection assay
Nuclear and cytoplasmic fractionation of MEL cells was as previously described (Antoniou et al. 1998). The nuclei were lysed with 4 M guanidinium thiocyanate, 25 mM tri-sodium citrate, 0.5% (w/v) N-lauryl sarcosine, 0.1 mM dithiothreitol (Huang and Carmichael 1996), and the homogenate sonicated (30 pulses using a small-diameter probe) to shear the DNA. The RNA was purified by phenol:chloroform (2.8:1) extraction, isopropanol and ethanol precipitations, and DNase digestion (RNase free, Roche). Human β-globin RNase protection probes (McCracken et al. 1997) were prepared by in vitro transcription with T7 RNA polymerase in the presence of [α-32P]UTP, and gel-purified prior to use. RNase protection assays were performed as previously described (McCracken et al. 2002). Briefly, 3 μg of nuclear RNA were incubated with the antisense RNA probe overnight at 50°C, and the hybridization products were digested with a mixture of RNase T1 (5 μg/mL) and RNase A (0.5 μg/mL) at 37°C for 1 h. The protected fragments were resolved on a 6% denaturing polyacrylamide gel, and the intensity of the bands quantified using a BioRad PhosphorImager. Following quantification of each gel band, background was subtracted and the values normalized for U content of the protected probe fragment.
Acknowledgments
We thank José Braga, José Rino, Helena Pina, and Maria do Carmo Silva for technical support and Susan McCracken (University of Toronto) for help with the RNase protection assay. We thank Elisa Izaurralde (EMBL-Heidelberg) for providing plasmids and antibodies. This study was supported by a grant (POCTI/MGI/36547/00) from Fundação para a Ciência e Tecnologia (FCT), Portugal, and by the European Commission (QLG2-CT-2001-01554).
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.rnajournal.org/cgi/doi/10.1261/rna.5258504.
REFERENCES
- Alexiadis, V., Waldmannm, T., Andersen, J., Mann, M., Knippers, R., and Gruss, C. 2000. The protein encoded by the proto-oncogene DEK changes the topology of chromatin and reduces the efficiency of DNA replication in a chromatin-specific manner. Genes & Dev. 14: 1308–1312. [PMC free article] [PubMed] [Google Scholar]
- Antoniou, A. 1991. Induction of erythroid-specific expression in murine erythroleukemia (MEL) cells. In Methods in molecular biology: Gene transfer and expression protocols. (ed. E.J. Murray), pp. 421–434. The Human Press, Inc., Clinfton, N.J. [DOI] [PubMed]
- Antoniou, M., Geraghty, F., Hurst, J., and Grosveld, F. 1998. Efficient 3′-end formation of human β-globin mRNA in vivo requires sequences within the last intron but occurs independently of the splicing reaction. Nucleic Acids Res. 26: 721–729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachi, A., Braun, I.C., Rodrigues, J.P., Pante, N., Ribbeck, K., von Kobbe, C., Kutay, U., Wilm, M., Gorlich, D., Carmo-Fonseca, M., et al. 2000. The C-terminal domain of TAP interacts with the nuclear pore complex and promotes export of specific CTE-bearing RNA substrates. RNA 6: 136–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauren, G. and Wieslander, L. 1994. Splicing of Balbiani ring 1 gene pre-mRNA occurs simultaneously with transcription. Cell 76: 183–192. [DOI] [PubMed] [Google Scholar]
- Bauren, G., Jiang, W.Q., Bernholm, K., Gu, F., and Wieslander, L. 1996. Demonstration of a dynamic, transcription-dependent organization of pre-mRNA splicing factors in polytene nuclei. J. Cell. Biol. 133: 929–941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beyer, A.L. and Osheim, Y.N. 1988. Splice site selection, rate of splicing, and alternative splicing on nascent transcripts. Genes & Dev. 2: 754–765. [DOI] [PubMed] [Google Scholar]
- Blencowe, B.J., Issner, R., Nickerson, J.A., and Sharp, P.A. 1998. A coactivator of pre-mRNA splicing. Genes & Dev. 12: 996–1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bridge, E., Xia, D.X., Carmo-Fonseca, M., Cardinali, B., Lamond, A.I., and Pettersson, U. 1995. Dynamic organization of splicing factors in adenovirus-infected cells. J. Virol. 69: 281–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bridge, E., Riedel, K.U., Johansson, B.M., and Pettersson, U. 1996. Spliced exons of adenovirus late RNAs colocalize with snRNP in a specific nuclear domain. J. Cell. Biol. 135: 303–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collis, P., Antoniou, M., and Grosveld, F. 1990. Definition of the minimal requirements within the human β-globin gene and the dominant control region for high level expression. EMBO J. 9:233–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullen, B.R. 2003. Nuclear RNA export. J. Cell. Sci. 116: 587–597. [DOI] [PubMed] [Google Scholar]
- Custódio, N., Carmo-Fonseca, M., Geraghty, F., Pereira, H.S., Grosveld, F., and Antoniou, M. 1999. Inefficient processing impairs release of RNA from the site of transcription. EMBO J. 18: 2855–2866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreyfuss, G., Kim, V.N., and Kataoka, N. 2002. Messenger-RNA-binding proteins and the messages they carry. Nat. Rev. Mol. Cell. Biol. 3: 195–205. [DOI] [PubMed] [Google Scholar]
- Estruch, F. and Cole, C.N. 2003. An early function during transcription for the yeast mRNA export factor Dbp5p/Rat8p suggested by its genetic and physical interactions with transcription factor IIH components. Mol. Biol. Cell 14: 1664–1676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faulkner, N.E., Hilfinger, J.M., and Markovitz, D.M. 2001. Protein phosphatase 2A activates the HIV-2 promoter through enhancer elements that include the pets site. J. Biol. Chem. 276: 25804– 25812. [DOI] [PubMed] [Google Scholar]
- Fey, E.G., Krochmalnic, G., and Penman, S. 1986. The nonchromatin substructures of the nucleus: The ribonucleoprotein (RNP)-containing and RNP-depleted matrices analyzed by sequential fractionation and resinless section electron microscopy. J. Cell. Biol. 102: 1654–1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleckner, J., Zhang, M., Valcarcel, J., and Green, M.R. 1997. U2AF65 recruits a novel human DEAD box protein required for the U2 snRNP-branchpoint interaction. Genes & Dev. 11: 1864–1872. [DOI] [PubMed] [Google Scholar]
- Flint, S.J. 1986. Regulation of adenovirus mRNA formation. Adv. Virus Res. 31: 169–228. [DOI] [PubMed] [Google Scholar]
- Fornerod, M., Boer, J., van Baal, S., Jaegle, M., von Lindern, M., Murti, K.G., Davis, D., Bonten, J., Buijs, A., and Grosveld, G. 1995. Relocation of the carboxy terminal part of CAN from the nuclear envelope to the nucleus as a result of leukemia-specific chromosome rearrangements. Oncogene 10: 1739–1748. [PubMed] [Google Scholar]
- Fribourg, S., Gatfield, D., Izaurralde, E., and Conti, E. 2003. A novel mode of RBD-protein recognition in the Y14-Mago complex. Nat. Struct. Biol. 10: 433–439. [DOI] [PubMed] [Google Scholar]
- Gama-Carvalho, M., Krauss, R.D., Chiang, L., Valcarcel, J., Green, M.R., and Carmo-Fonseca, M. 1997. Targeting of U2AF65 to sites of active splicing in the nucleus. J. Cell. Biol. 137: 975–987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gama-Carvalho, M., Condado, I., and Carmo-Fonseca, M. 2003. Regulation of adenovirus alternative RNA splicing correlates with a re-organization of splicing factors in the nucleus. Exp. Cell. Res. 289: 77–85. [DOI] [PubMed] [Google Scholar]
- Gatfield, D. and Izaurralde, E. 2002. REF1/Aly and the additional exon junction complex proteins are dispensable for nuclear mRNA export. J. Cell. Biol. 159: 579–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatfield, D., Le Hir, H., Schmitt, C., Braun, I.C., Kocher, T., Wilm, M., and Izaurralde, E. 2001. The DExH/D box protein HEL/UAP56 is essential for mRNA nuclear export in Drosophila. Curr. Biol. 11: 1716–1721. [DOI] [PubMed] [Google Scholar]
- Greber, U.F., Willetts, M., Webster, P., and Helenius, A. 1993. Stepwise dismantling of adenovirus 2 during entry into cells. Cell 75: 477–486. [DOI] [PubMed] [Google Scholar]
- Habets, W.J., Hoet, M.H., De Jong, B.A., Van der Kemp, A., and Van Venrooij, W.J. 1989. Mapping of B cell epitopes on small nuclear ribonucleoproteins that react with human autoantibodies as well as with experimentally-induced mouse monoclonal antibodies. J. Immunol. 143: 2560–2566. [PubMed] [Google Scholar]
- Hachet, O. and Ephrussi, A. 2001. Drosophila Y14 shuttles to the posterior of the oocyte and is required for oskar mRNA transport. Curr. Biol. 11: 1666–1674. [DOI] [PubMed] [Google Scholar]
- Herold, A., Suyama, M., Rodrigues, J.P., Braun, I.C., Kutay, U., Carmo-Fonseca, M., Bork, P., and Izaurralde, E. 2000. TAP (NXF1) belongs to a multigene family of putative RNA export factors with a conserved modular architecture. Mol. Cell. Biol. 20: 8996–9008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodge, C.A., Colot, H.V., Stafford, P., and Cole, C.N. 1999. Rat8p/Dbp5p is a shuttling transport factor that interacts with Rat7p/Nup159p and Gle1p and suppresses the mRNA export defect of xpo1–1 cells. EMBO J. 18: 5778–5788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, S. and Spector, D.L. 1996. Intron-dependent recruitment of pre-mRNA splicing factors to sites of transcription. J. Cell. Biol. 133: 719–732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, Y. and Carmichael, G.C. 1996. Role of polyadenylation in nucleocytoplasmic transport of mRNA. Mol. Cell. Biol. 16: 1534–1542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, Y., Gattoni, R., Stevenin, J., and Steitz, J.A. 2003. SR splicing factors serve as adapter proteins for TAP-dependent mRNA export. Mol. Cell 11: 837–843. [DOI] [PubMed] [Google Scholar]
- Izaurralde, E. 2002. A novel family of nuclear transport receptors mediates the export of messenger RNA to the cytoplasm. Eur. J. Cell. Biol. 81: 577–584. [DOI] [PubMed] [Google Scholar]
- Jimenez-Garcia, L.F. and Spector, D.L. 1993. In vivo evidence that transcription and splicing are coordinated by a recruiting mechanism. Cell 73: 47–59. [DOI] [PubMed] [Google Scholar]
- Kataoka, N., Yong, J., Kim, V.N., Velazquez, F., Perkinson, R.A., Wang, F., and Dreyfuss, G. 2000. Pre-mRNA splicing imprints mRNA in the nucleus with a novel RNA-binding protein that persists in the cytoplasm. Mol. Cell 6: 673–682. [DOI] [PubMed] [Google Scholar]
- Kataoka, N., Diem, M.D., Kim, V.N., Yong, J., and Dreyfuss, G. 2001. Magoh, a human homolog of Drosophila mago nashi protein, is a component of the splicing-dependent exon-exon junction complex. EMBO J. 20: 6424–6433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiesler, E., Miralles, F., and Visa, N. 2002. HEL/UAP56 binds cotranscriptionally to the Balbiani ring pre-mRNA in an intron-independent manner and accompanies the BR mRNP to the nuclear pore. Curr. Biol. 12: 859–862. [DOI] [PubMed] [Google Scholar]
- Kim, V.N. and Dreyfus, G. 2001. Nuclear mRNA binding proteins couple pre-mRNA splicing and post-splicing events. Mol. Cells 12: 1–10. [PubMed] [Google Scholar]
- Kim, V.N., Kataoka, N., and Dreyfuss, G. 2001. Role of the nonsense-mediated decay factor hUpf3 in the splicing- dependent exon-exon junction complex. Science 293: 1832–1836. [DOI] [PubMed] [Google Scholar]
- Kistler, A.L. and Guthrie, C. 2001. Deletion of MUD2, the yeast homolog of U2AF65, can bypass the requirement for sub2, an essential spliceosomal ATPase. Genes & Dev. 15: 42–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau, C.K., Diem, M.D., Dreyfuss, G., and Van Duyne, G.D. 2003. Structure of the y14-magoh core of the exon junction complex. Curr. Biol. 13: 933–941. [DOI] [PubMed] [Google Scholar]
- Le Hir, H., Izaurralde, E., Maquat, L.E., and Moore, M.J. 2000a. The spliceosome deposits multiple proteins 20–24 nucleotides upstream of mRNA exon-exon junctions. EMBO J. 19: 6860–6869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Hir, H., Moore, M.J., and Maquat, L.E. 2000b. Pre-mRNA splicing alters mRNP composition: Evidence for stable association of proteins at exon-exon junctions. Genes & Dev. 14: 1098–1108. [PMC free article] [PubMed] [Google Scholar]
- Le Hir, H., Gatfield, D., Braun, I.C., Forler, D., and Izaurralde, E. 2001a. The protein Mago provides a link between splicing and mRNA localization. EMBO Rep. 2: 1119–1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Hir, H., Gatfield, D., Izaurralde, E., and Moore, M.J. 2001b. The exon-exon junction complex provides a binding platform for factors involved in mRNA export and nonsense-mediated mRNA decay. EMBO J. 20: 4987–4997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei, E.P. and Silver, P.A. 2002a. Intron status and 3′-end formation control cotranscriptional export of mRNA. Genes & Dev. 16: 2761–2766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ———. 2002b. Protein and RNA export from the nucleus. Dev. Cell 2: 261–272. [DOI] [PubMed] [Google Scholar]
- Lei, E.P., Krebber, H., and Silver, P.A. 2001. Messenger RNAs are recruited for nuclear export during transcription. Genes & Dev. 15: 1771–1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lejeune, F., Ishigaki, Y., Li, X., and Maquat, L.E. 2002. The exon junction complex is detected on CBP80-bound but not eIF4E-bound mRNA in mammalian cells: Dynamics of mRNP remodeling. EMBO J. 21: 3536–3545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeMaire, M.F. and Thummel, C.S. 1990. Splicing precedes polyadenylation during Drosophila E74A transcription. Mol. Cell. Biol. 10: 6059–6063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerner, E.A., Lerner, M.R., Janeway Jr., C.A., and Steitz, J.A. 1981. Monoclonal antibodies to nucleic acid-containing cellular constituents: probes for molecular biology and autoimmune disease. Proc. Natl. Acad. Sci. 78: 2737–2741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Libri, D., Graziani, N., Saguez, C., and Boulay, J. 2001. Multiple roles for the yeast SUB2/yUAP56 gene in splicing. Genes & Dev. 15: 36–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linné, T., Jornvall, H., and Philipson, L. 1977. Purification and characterization of the phosphorylated DNA-binding protein from adenovirus-type-2-infected cells. Eur. J. Biochem. 76: 481–490. [DOI] [PubMed] [Google Scholar]
- Longman, D., Johnstone, I.L., and Caceres, J.F. 2003. The Ref/Aly proteins are dispensable for mRNA export and development in Caenorhabditis elegans. RNA 9: 881–891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loyer, P., Trembley, J.H., Lahti, J.M., and Kidd, V.J. 1998. The RNP protein, RNPS1, associates with specific isoforms of the p34cdc2-related PITSLRE protein kinase in vivo. J. Cell. Sci. 111: 1495–1506. [DOI] [PubMed] [Google Scholar]
- Lu, S. and Cullen, B.R. 2003. Analysis of the stimulatory effect of splicing on mRNA production and utilization in mammalian cells. RNA 9: 618–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo, M.L., Zhou, Z., Magni, K., Christoforides, C., Rappsilber, J., Mann, M., and Reed, R. 2001. Pre-mRNA splicing and mRNA export linked by direct interactions between UAP56 and Aly. Nature 413: 644–647. [DOI] [PubMed] [Google Scholar]
- Lykke-Andersen, J., Shu, M.D., and Steitz, J.A. 2001. Communication of the position of exon-exon junctions to the mRNA surveillance machinery by the protein RNPS1. Science 293: 1836–1839. [DOI] [PubMed] [Google Scholar]
- MacMorris, M., Brocker, C., and Blumenthal, T. 2003. UAP56 levels affect viability and mRNA export in Caenorhabditis elegans. RNA 9: 847–857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayeda, A., Badolato, J., Kobayashi, R., Zhang, M.Q., Gardiner, E.M., and Krainer, A.R. 1999. Purification and characterization of human RNPS1: A general activator of pre-mRNA splicing. EMBO J. 18: 4560–4570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCracken, S., Fong, N., Rosonina, E., Yankulov, K., Brothers, G., Siderovski, D., Hessel, A., Foster, S., Shuman, S., and Bentley, D.L. 1997. 5′-Capping enzymes are targeted to pre-mRNA by binding to the phosphorylated carboxy-terminal domain of RNA polymerase II. Genes & Dev. 11: 3306–3318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCracken, S., Lambermon, M., and Blencowe, B.J. 2002. SRm160 splicing coactivator promotes transcript 3′-end cleavage. Mol. Cell. Biol. 22: 148–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGarvey, T., Rosonina, E., McCracken, S., Li, Q., Arnaout, R., Mientjes, E., Nickerson, J.A., Awrey, D., Greenblatt, J., Grosveld, G., et al. 2000. The acute myeloid leukemia-associated protein, DEK, forms a splicing-dependent interaction with exon-product complexes. J. Cell. Biol. 150: 309–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohr, S.E., Dillon, S.T., and Boswell, R.E. 2001. The RNA-binding protein Tsunagi interacts with Mago Nashi to establish polarity and localize oskar mRNA during Drosophila oogenesis. Genes & Dev. 15: 2886–2899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neugebauer, K.M. and Roth, M.B. 1997. Transcription units as RNA processing units. Genes & Dev. 11: 3279–3285. [DOI] [PubMed] [Google Scholar]
- Nott, A., Meislin, S.H., and Moore, M.J. 2003. A quantitative analysis of intron effects on mammalian gene expression. RNA 9: 607–617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philipson, L., Pettersson, U., and Lindberg, U. 1975. Molecular biology of adenoviruses. Virol. Monogr. 14: 1–115. [DOI] [PubMed] [Google Scholar]
- Pombo, A. and Carmo-Fonseca, M. 1995. Interaction of adenovirus with the nucleus of the host cell. Rev. Med. Virol. 5: 213–218. [Google Scholar]
- Pombo, A., Ferreira, J., Bridge, E., and Carmo-Fonseca, M. 1994. Adenovirus replication and transcription sites are spatially separated in the nucleus of infected cells. EMBO J. 13: 5075–5085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed, R. and Hurt, E. 2002. A conserved mRNA export machinery coupled to pre-mRNA splicing. Cell 108: 523–531. [DOI] [PubMed] [Google Scholar]
- Reichert, V.L., Le Hir, H., Jurica, M.S., and Moore, M.J. 2002. 5′ exon interactions within the human spliceosome establish a framework for exon junction complex structure and assembly. Genes & Dev. 16: 2778–2791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues, J.P., Rode, M., Gatfield, D., Blencowe, B.J., Carmo-Fonseca, M., and Izaurralde, E. 2001. REF proteins mediate the export of spliced and unspliced mRNAs from the nucleus. Proc. Natl. Acad. Sci. 98: 1030–1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt, C., von Kobbe, C., Bachi, A., Pante, N., Rodrigues, J.P., Boscheron, C., Rigaut, G., Wilm, M., Seraphin, B., Carmo-Fonseca, M., et al. 1999. Dbp5, a DEAD-box protein required for mRNA export, is recruited to the cytoplasmic fibrils of nuclear pore complex via a conserved interaction with CAN/Nup159p. EMBO J. 18: 4332–4347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snay-Hodge, C.A., Colot, H.V., Goldstein, A.L., and Cole, C.N. 1998. Dbp5p/Rat8p is a yeast nuclear pore-associated DEAD-box protein essential for RNA export. EMBO J. 17: 2663–2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strahm, Y., Fahrenkrog, B., Zenklusen, D., Rychner, E., Kantor, J., Rosbach, M., and Stutz, F. 1999. The RNA export factor Gle1p is located on the cytoplasmic fibrils of the NPC and physically interacts with the FG-nucleoporin Rip1p, the DEAD-box protein Rat8p/Dbp5p and a new protein Ymr 255p. EMBO J. 18: 5761–5777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strasser, K. and Hurt, E. 2000. Yra1p, a conserved nuclear RNA-binding protein, interacts directly with Mex67p and is required for mRNA export. EMBO J. 19: 410–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strasser, K., Masuda, S., Mason, P., Pfannstiel, J., Oppizzi, M., Rodriguez-Navarro, S., Rondon, A.G., Aguilera, A., Struhl, K., Reed, R., et al. 2002. TREX is a conserved complex coupling transcription with messenger RNA export. Nature 417: 304–308. [DOI] [PubMed] [Google Scholar]
- Stutz, F. and Izaurralde, E. 2003. The interplay of nuclear mRNP assembly, mRNA surveillance and export. Trends Cell. Biol. 13: 319–327. [DOI] [PubMed] [Google Scholar]
- Stutz, F., Bachi, A., Doerks, T., Braun, I.C., Seraphin, B., Wilm, M., Bork, P., and Izaurralde, E. 2000. REF, an evolutionary conserved family of hnRNP-like proteins, interacts with TAP/Mex67p and participates in mRNA nuclear export. RNA 6: 638–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tennyson, C.N., Klamut, H.J., and Worton, R.G. 1995. The human dystrophin gene requires 16 hours to be transcribed and is cotranscriptionally spliced. Nat. Genet. 9: 184–190. [DOI] [PubMed] [Google Scholar]
- Tseng, S.S., Weaver, P.L., Liu, Y., Hitomi, M., Tartakoff, A.M., and Chang, T.H. 1998. Dbp5p, a cytosolic RNA helicase, is required for poly(A)+ RNA export. EMBO J. 17: 2651–2662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Virbasius, C.M., Wagner, S., and Green, M.R. 1999. A human nuclear-localized chaperone that regulates dimerization, DNA binding, and transcriptional activity of bZIP proteins. Mol. Cell 4: 219–228. [DOI] [PubMed] [Google Scholar]
- Wiegand, H.L., Lu, S., and Cullen, B.R. 2003. Exon junction complexes mediate the enhancing effect of splicing on mRNA expression. Proc. Natl. Acad. Sci. 100: 11327–11332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, Z.A., Murphy, C., Callan, H.G., and Gall, J.G. 1991. Small nuclear ribonucleoproteins and heterogeneous nuclear ribonucleoproteins in the amphibian germinal vesicle: Loops, spheres, and snurposomes. J. Cell. Biol. 113: 465–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing, Y., Johnson, C.V., Dobner, P.R., and Lawrence, J.B. 1993. Higher level organization of individual gene transcription and RNA splicing. Science 259: 1326–1330. [DOI] [PubMed] [Google Scholar]
- Zhang, G., Taneja, K.L., Singer, R.H., and Green, M.R. 1994. Localization of pre-mRNA splicing in mammalian nuclei. Nature 372: 809–812. [DOI] [PubMed] [Google Scholar]
- Zhang, M. and Green, M.R. 2001. Identification and characterization of yUAP/Sub2p, a yeast homolog of the essential human pre-mRNA splicing factor hUAP56. Genes & Dev. 15: 30–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, J., Jin, S.B., Bjorkroth, B., Wieslander, L., and Daneholt, B. 2002. The mRNA export factor Dbp5 is associated with Balbiani ring mRNP from gene to cytoplasm. EMBO J. 21: 1177–1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, Z., Luo, M.J., Straesser, K., Katahira, J., Hurt, E., and Reed, R. 2000. The protein Aly links pre-messenger-RNA splicing to nuclear export in metazoans. Nature 407: 401–405. [DOI] [PubMed] [Google Scholar]