Abstract
Recent proteomic analyses are revealing the dynamics of preribosome assembly. Following cleavage at processing site A2, which generates the 20S pre-rRNA (the immediate precursor to the 18S rRNA), early RRPs (ribosomal RNA processing factors) are released in bulk from the preribosomes, and the resulting pre-40S subunits are left associated with a limited set of proteins that we refer to as the SSU RRP complex. Dim2p, a core constituent of the SSU RRP complex and conserved KH-domain containing protein, is required for pre-rRNA processing and is associated with early nucleolar and late cytoplasmic pre-rRNA species. Consistently, Dim2p shuttles between the nucle(ol)us and the cytoplasm, a trafficking that is tightly regulated by growth. The association of Dim2p with the 18S rRNA dimethyltransferase Dim1p, as well as its requirement for pre-rRNA processing at cleavage sites A1 and A2 and for 18S rRNA dimethylation, suggest that Dim2p may recruit Dim1p to nucleolar pre-rRNAs through its KH domain.
Keywords: ribosome synthesis, pre-rRNA processing, KH-domain, nucleolus
INTRODUCTION
Ribosome biogenesis involves the synthesis, maturation, and assembly of four ribosomal RNAs (rRNAs) and ~80 ribosomal proteins (RPs). Mature rRNAs are generated from pre-rRNA precursors following a complex succession of endo- and exoribonucleolytic digestions (Fig. 1 ▶; for review, see Kressler et al. 1999; Venema and Tollervey 1999), specific residues are selected for ribose or base modification, and preribosomes are assembled from pre-rRNAs and RPs (for review, see Lafontaine and Tollervey 2001). The sites of RNA modification are largely selected through Watson-Crick base-pair interactions with members of conserved families of small nucleolar RNAs (snoRNAs; for review, see Kiss 2002). Preribosomes are released from the nucleolus, where most of these steps occur; diffuse through the nucleoplasm; and exit the nucleus through the nuclear pore complexes. Small and large ribosomal subunits transit to the cytoplasm independently.
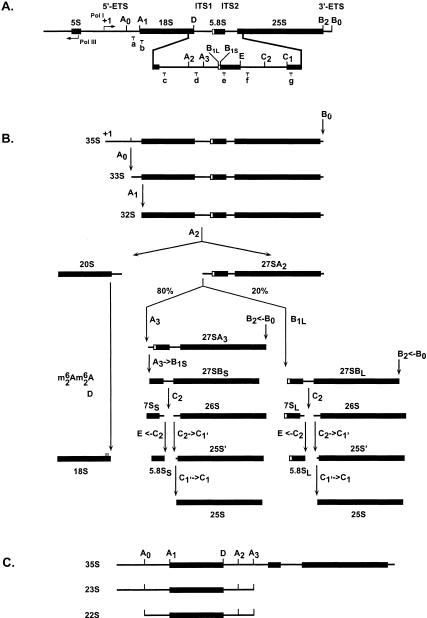
FIGURE 1.
Yeast rDNA locus and pre-rRNA processing pathway. (A) Structure of the rDNA. Three out of the four mature ribosomal rRNAs, the 18S, 5.8S, and 25S rRNAs, are encoded in a large Pol I transcript (35S) extending from the transcription start site (+1) to site B0. 18S and 5.8S–25S rRNAs are embedded into noncoding 5′- and 3′-external transcribed spacers (5′- and 3′-ETS) and internal transcribed spacers 1 and 2 (ITS1 and ITS2). 5S is produced as a 3′-extended precursor by RNA Pol III. Pre-rRNA processing sites are indicated as uppercase letters (A0–E); oligonucleotide probes used in Northern blot and primer extension are indicated in lowercase (a–g). (B) Pre-rRNA processing pathway. Following cotranscriptional cleavage in the 3′-ETS at site B0, the 35S pre-rRNA is processed in the 5′-ETS at sites A0 and A1, the 5′-mature end of 18S rRNA, and in the ITS1 at site A2, generating successively the 33S, 32S, 20S, and 27SA2 pre-rRNAs. The 20S pre-rRNA is exported to the cytoplasm, methylated by Dim1p, and processed at site D to generate the 18S rRNA. In a major pathway, accounting for up to 80% of the total processing, the 27SA2 is cleaved at site A3. This is followed by exoribonucleolytic digestion to site B1S, generating the major form of 5.8S (5.8SS). Processing in ITS2 is initiated by cleavage at site C2, which provides an entry site for exoribonucleolytic digestions to sites E and C1, the mature 3′-end of 5.8S, and the mature 5′-end of 25S, respectively. In an alternative minor pathway, the 27SA2 pre-rRNA is cleaved at site B1L; this results in the production of 5.8S rRNA molecules that are extended in 5′ by seven to eight nucleotides (5.8SL). Processing at site B2 is concomitant with the synthesis of 27SB. For further details, see Lafontaine (2003). (C) Aberrant pre-rRNA precursors detected in cells depleted for Dim2p. Inhibition of cleavage at sites A0–A2 results in the accumulation of the 23S RNA that extends from the transcription start site (+1) to site A3. The 22S RNA is accumulated on inhibition of cleavage at sites A1 and A2.
A basic scheme for ribosomal assembly, based on in vivo labeling and velocity gradients analysis, was established in the early 1970s (Udem and Warner 1973; Trapman et al. 1975). In this, a 90S particle, based on the RNA polymerase I (Pol I) primary transcript (35S in yeast), is converted into 43S and 66S preribosomes, precursors to the small and large ribosomal subunits, respectively. This vision remained unchallenged for more than three decades.
The recent development of new proteomic technologies has poised the field for rapid progress and led to the detailed characterization of several dozen species of nucle(ol)ar and cytoplasmic preribosomes (for review, see Fatica and Tollervey 2002; Lafontaine, Raue 2003; Tschochner and Hurt 2003). The large Pol I transcript is now known to be primarily bound by >30 RRPs, most of which almost exclusively involved in early pre-rRNA processing reactions and Small ribosomal SUbunit (SSU) synthesis. These 90S preribosomes largely overlap in composition with the recently described SSU processome, imaged as terminal balls on chromatin spreads and corresponding to the box C/D snoRNP U3 (Dragon et al. 2002; Grandi et al. 2002). Following cleavage at site A2, which generates the 20S and 27SA2 pre-rRNAs (Fig. 1 ▶), precursors to the small and large subunit rRNAs, the 90S preribosomes break up and the first set of >30 RRPs are released in bulk. The resulting pre-40S subunits are left associated with very few RRPs and acquire a limited number of novel factors. In fact, only eight major nonribosomal proteins are detected in nucle(ol)ar 40S preribosomes (Schafer et al. 2003), Rrp12p, Tsr1p (a putative GTPase related to Bms1p; Gelperin et al. 2001; Wegierski et al. 2001), Enp1p (Chen et al. 2003), Hrr25p, Nob1p, Rio2p (a putative protein kinase homologous to Rio1p; Vanrobays et al. 2001, 2003), Dim1p (a base methyltransferase; Lafontaine et al. 1995, 1998), and Dim2p/Yor145c (a KH-domain protein). Ltv1p, Nop14p/Noc5p, and Asc1p are also present but in substoichiometric amounts. We will hereafter refer to this protein set as the SSU RRP complex (Table 1 ▶). The kinetics of association of the various constituent of the SSU RRP complex with the pre-40S particles is distinctively different with Enp1p, Dim1p, Hrr25p and possibly Rrp12p binding to the pre-rRNAs at an early nucleolar stage, corresponding to the 90S preribosomes, and Nob1p, Rio2p, and Tsr1p associating later, after cleavages at sites A0–A2 (Schafer et al. 2003). Most constituents of the SSU RRP complex have known functions in 40S subunit synthesis (Lafontaine et al. 1995; Gelperin et al. 2001; Liu and Thiele 2001; Ihmels et al. 2002; Chen et al. 2003; Fatica et al. 2003; Schafer et al. 2003; Vanrobays et al. 2003).
TABLE 1.
Major components of the SSU RRP complex
| Names | Protein motifs, functions | Pre-rRNA processing defect | References |
| Dim1p/Ypl266c | SAM binding motif Base methyl-transferase | A1, A2 | Lafontaine et al. (1995) |
| Dim2p/Yor145c | KH motif, RNA binding, homology to Krr1p | A1, A2 | This work |
| Enp1p/Ybr247c | — | A0, A1, A2 | Chen et al. (2003) |
| Hrr25p/Ypl204w | protein kinase, a subunit of CK1 | Not determined | — |
| Nob1p/Yor056c | PIN motif, putative phosphodiesterase | A0, A1, A2, D | Fatica et al. (2003) |
| Rio2p/Ynl207w | RIO domain, putative protein kinase | D with marginal effects on A0, A1, A2 | Vanrobays et al. (2003) |
| Rrp12p/Ypl012w | — | Not determined | — |
| Tsr1p/Ydl060w | Homology to the GTPase Bms1p but without the P-loop | A0, A1, A2, D | Gelperin et al. (2001) |
The K homology (KH)-domain is a 45-amino-acid RNA-binding motif based on a nine-residues consensus (VIGxxGxxI). It is highly conserved and has been detected in all three domains of life (Gibson et al. 1993b; Siomi et al. 1993; Burd and Dreyfuss 1994; Adinolfi et al. 1999; Grishin 2001). A single amino acid substitution (I304N) in the KH domain of FMR1 led to fragile X syndrome, the most common cause of human hereditary mental retardation (De Boulle et al. 1993; Gibson et al. 1993a; Siomi et al. 1994).
Here, we report the functional characterization of Yor145c/Dim2p, a KH domain protein required for SSU synthesis and core component of the SSU RRP complex.
RESULTS
A late SSU RRP network
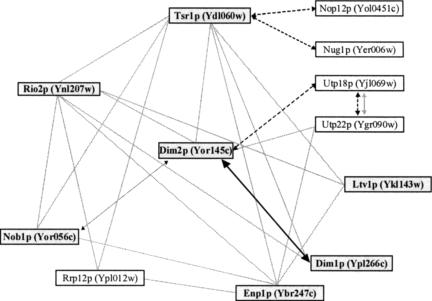
Recent proteomic analysis identified a network of physical interactions connecting about a dozen of proteins, most of which are known RRPs involved in small ribosomal subunit synthesis (Fig. 2 ▶; Table 1 ▶). Figure 2 ▶ was compiled from high-throughput copurification data sets (Gavin et al. 2002; Ho et al. 2002; Schafer et al. 2003; Vazquez et al. 2003). According to Samanta and Liang, Yor145c shares a statistically relevant number of common partners with Dim1p, defining a couple of “close functional partners” with potential related functions (cited in Vazquez et al. 2003). Yor145c was thus renamed Dim2p. Dim2p had not been tested for a function in ribosome synthesis when this work was initiated.
FIGURE 2.
The SSU RRP network. Physical and functional interactions between components of the SSU RRP complex. Following pre-rRNA processing at sites A0–A2, 20S-based preribosomes (or pre-40S subunits) are generated that are only associated with about a dozen of RRPs that we propose to refer to as the SSU RRP complex. Interactions between the various components of the SSU RRP complex are from as follows: in grey, Gavin et al. (2002); dashed lines, Ho et al. (2002); and dotted line, Tone and Toh (2002). Components in bold were identified in complexes recently described by Schafer et al. (2003). The interaction between Dim1p and Dim2p is from this work.
Dim2p is a conserved KH-domain protein homologous to Krr1p
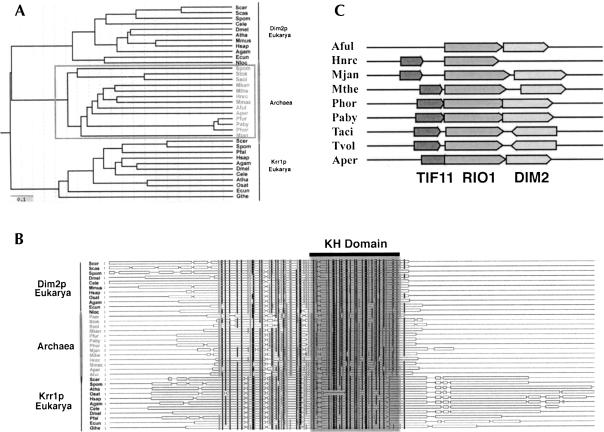
Blast analysis revealed that Dim2p is a conserved KH domain-containing protein with members throughout the archaeal and eukaryal domains (Fig. 3A ▶). The most conserved residues of the KH domain, involved in RNA binding (for review, see Adinolfi et al. 1999), are detected in Dim2p (Fig. 3B ▶). Striking homology with Krr1p, a eukaryotic-specific SSU RRP (Sasaki et al. 2000; Chan et al. 2001), indicates that these two proteins presumably derived from a common ancestor (Fig. 3A ▶). Krr1p and Dim2p are conserved in minimal eukaryotic genomes such as those of Giardia (12 Mb) and Encephalitozooan cuniculi (3 Mbp), attesting of the early divergence of these genes.
FIGURE 3.
Dim2p is a phylogenetically conserved KH-domain containing protein related to the eukaryotic-specific RRP Krr1p. (A, B) Phylogenetic analysis and multiple alignments were performed as described in the Material and Methods section. (A) 0.1 refers to the divergence index. (B) The KH-domain is highlighted. Residues in grey are 70%–90% similar; residues in black are either identical or show >90% similarity. (C) Structure of Archaeal genomes at the DIM2 locus. Species names are as follows: Scer, Saccharomyces cerevisiae; Scas, Saccharomyces casletellii; Spom, Schizosaccharomyces pombe; Dmel, Drosophila melanogaster; Osat, Oryza sativa; Mmus, Mus musculus; Hsap, Homo sapiens; Cele, Caenorhabditis elegans; Agam, Anopheles gambiae; Ecun, Encephalitozoon cuniculi; Nloc, Nosema locustae; Paer, Pyrobaculum aerophilum; Stok, Sulfolobus tokodaii; Mkan, Methanopyrus kandleri; Aper, Aeropyrum pernix; Pfur, Pyrococcus furiosus; Phor, Pyrococcus horikoshii; Paby, Pyrococcus abyssi; Mjan, Methanococcus jannaschii; Aful, Archaeoglobus fuldus; Ssol, Sulfolobus solfataricus; Hnrc, Halobacterium sp. NRC-1; Mmaz, Methanosarcina mazei; Atha, Arabidopsis thaliana; Pfal, Plasmodium falciparum; Mthe, Methanobacterium thermoautotrophicum; Taci, Thermoplasma acidophilum; Tvol, Thermoplasma volcanium; Hnrc, Halobacterium sp. NRC-1; and Gthe, Guillardia theta.
Within archaeal genomes, DIM2 is closely linked to RIO1, another SSU RRP (Vanrobays et al. 2001), suggesting a possible coregulation for expression and the existence of an operon (Fig. 3C ▶). In Archaea, DIM2 and RIO1 are either organized in a head-to-head or head-to-tail fashion, and in some species, these two open reading frames show striking overlaps. TIF11, involved in ribosome function, is also closely linked to these genes.
Dim2p shows a surprisingly dynamic subcellular distribution
The subcellular distribution of Dim2p was investigated in strains expressing a fluorescent construct and directly imaged in living cells (Fig. 4 ▶). A C-terminal GFP cassette was integrated on the chromosome at the DIM2 locus by PCR. The epitope-tagged construct was fully functional, as shown by its ability to rescue growth to wild-type levels (data not shown). A diploid strain, homozygous for the DIM2-GFP allele, was used in the localization studies for improved detection of the fluorescent signal.
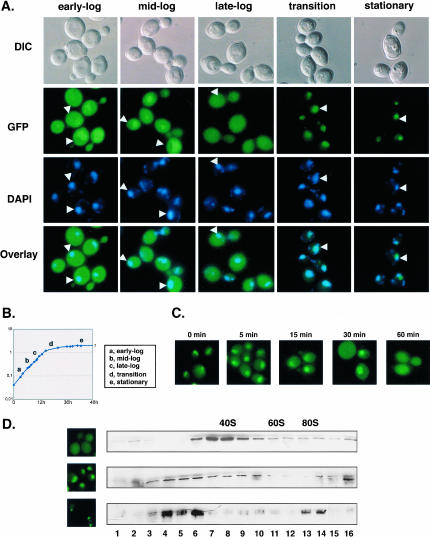
FIGURE 4.
Dim2p shows a dynamic subcellular distribution and a dynamic sedimentation profile throughout growth. (A–C) Subcellular distribution throughout growth. Cells expressing a functional Dim2p-GFP construct were inoculated in a complete, glucose-based medium (YPD) and examined for fluorescence under the microscope throughout growth. Cells were counter-stained with DAPI to visualize the bulk DNA. A phase (DIC) is provided. (B) Corresponding growth curve; cell growth was monitored by OD600. (C) Subcellular distribution dynamics. Cells expressing a Dim2p-GFP construct were grown to stationary phase, diluted to fresh medium, and inspected at regular intervals for 1 h. (D) Sedimentation profile throughout growth. Glass-beads lysates from cells expressing a functional Dim2p-GFP construct at different stages of growth were layered on 10%–30% glycerol gradients. Individual fractions were processed for Western-blot hybridization and decorated with an anti-GFP antibody to visualize the Dim2p-GFP fusion. Mid-log, transition to stationary, and stationary phase of growth (top, middle, and bottom, respectively) were analyzed; fluorescence panels are provided to the left as controls. Hybridization with antibodies specific to the small and large subunits, as well as the analysis of the contents in RNA of each fraction, allowed to position the peaks for 40S, 60S, and monosomes (data not shown).
A culture was inoculated in complete glucose-based medium (YPD), and cells were observed at regular intervals, starting at early-log to stationary phases of growth (Fig. 4A ▶); growth was monitored by OD600 (Fig. 4B ▶). Cells were counter-stained with DAPI to localize the DNA. In yeast, the nucleolus is typically detected as a crescent-shaped structure facing the chromosomal DNA.
Unexpectedly, Dim2p-GFP showed a particularly dynamic subcellular distribution. At early log-phase, Dim2p was detected both in the cytoplasm and nucleus, with a significant enrichment corresponding to the nucleolus. As cells progressed through the exponential phase of growth, Dim2p was progressively depleted from the cytoplasm and concentrated in the nucleus and nucleolus. When cells reached saturation, Dim2p was only found as a bright nucleolar focus (Fig. 4A ▶, stationary).
To investigate the dynamics of this subcellular redistribution, starved cells were diluted to fresh medium and allowed to grow for several generations (Fig. 4C ▶). The nuclear distribution was regained within 5 min; an equal distribution between the nucleus and the cytoplasm was restored after 1 h. This suggested that Dim2p shuttles between the nucleus and the cytoplasm, an assumption that recently turned out to be correct (Schafer et al. 2003).
Dim2p colocalizes with pre-40S subunits and 90S preribosomes
Total cellular extracts were prepared from cells expressing a Dim2p-GFP construct and fractionated on 10%–30% glycerol gradients (Fig. 4D ▶). Total RNA was extracted, separated on agarose/formaldehyde gels, and stained with ethidium bromide to position the peaks corresponding to 40S, 60S, and monosomes (data not shown). Proteins were extracted, resolved on PAGE, and transferred to nitrocellulose for Western-blot hybrization. Antibodies specific to ribosomal proteins from both subunits were used as controls. Membranes were hybridized with an antibody specific to the GFP moiety of the Dim2p construct.
In exponentially growing cells, Dim2p was distributed throughout the whole gradient, with a major peak around 40S that corresponded to pre-40S subunits and a fraction that colocalized with faster migrating particles, corresponding to 90S preribosomes (Fig. 4D ▶, upper panel). This analysis was repeated at later stages of growth, at the transition to stationary (middle panel) and at stationary (lower panel). As observed by fluorescence, Dim2-GFP also showed a dynamic behavior on glycerol gradient. Significantly, Dim2p-GFP was relocated to slower migrating particles that may correspond to proteinaceous intermediates in the preribosome assembly pathway. This distribution was even more striking at the stationary phase of growth. Despite repetitive attempts, the gradient profiles at high OD600 consistently showed an elevated level of proteolysis.
Dim2p is required for pre-rRNA processing
To test for a potential involvement of Dim2p in ribosome synthesis, a conditional strain was constructed. DIM2 was placed under the control of a GAL::-regulated promoter and expressed from a low copy plasmid in a dim2Δ background; Dim2p was further fused at its N-terminal end to the Protein A epitope (a twin Z domain was used), resulting in a GAL::ZZ-dim2 construct. Transcription driven from GAL:: promoters is strongly reduced in the presence of glucose in the growth medium.
Galactose-dependent growth was first tested on solid media. dim2Δ strains expressing either a plasmid-borne wild-type gene or the GAL::ZZ-dim2 fusion were grown in complete medium supplemented with either galactose or glucose (YPGal and YPD, respectively). Growth was only detected in permissive YPGal medium (data not shown); this was as expected for an essential gene (Grava et al. 2000). ZZ-Dim2p was functional as shown by its ability to rescue growth to wild-type levels in YPGal (data not shown).
The kinetics of Dim2p depletion was assessed in liquid medium by Western-blot. Following 10 h of transfer to the nonpermissive conditions, the growth rate of GAL::ZZ-dim2 strains was already substantially reduced with a doubling time (DT) >8 h (DT was ~2 h in permissive conditions), and the level of the ZZ-Dim2p fusion detectably affected (data not shown). Growth essentially ceased following 16 h of transfer. A low level of residual ZZ-Dim2p was detected at late depletion time points (data not shown).
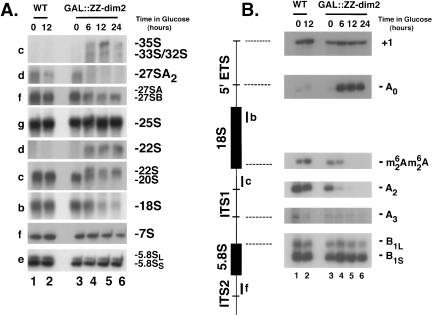
Pre-rRNA processing was assessed by Northern-blot hybridization, primer extension, and in vivo labeling (Figs. 5 ▶, 6 ▶).
FIGURE 5.
Pre-rRNA processing analysis in cells depleted for Dim2p. To achieve Dim2p depletion, a GAL::ZZ-dim2 strain was grown to mid-log phase in permissive conditions (YPGal); cells were thoroughly washed in water and transferred to prewarmed (30°C) YPD for up to 24 h. As a control, a wild-type isogenic strain was used. (A) Northern blot analysis. Total RNA was extracted at the time points of transfer indicated, separated on agarose/formaldehyde gels, and transferred to nylon membranes for Northern blot hybridizations. A collection of oligonucleotide probes (Fig. 1A ▶) specific to the pre-rRNAs and rRNAs were used. RNA species are indicated to the right; oligoprobes to the left. (B) Primer extension analysis. cDNA synthesis was initiated from oligonucleotides b, c, and f (Fig. 1A ▶). Signals corresponding to the transcription start site (+1), cleavage sites A0, A2, A3, B1S, and B1L, as well as for the base methylation carried out by Dim1p, are indicated.
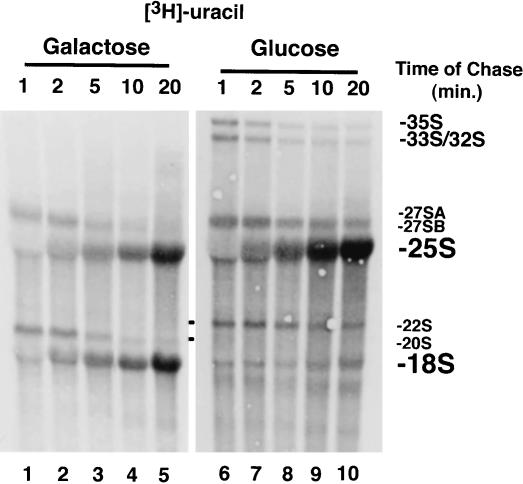
FIGURE 6.
Pulse-chase labeling analysis of pre-rRNA processing in cells depleted for Dim2p. Cells expressing a GAL-regulated epitope-tagged DIM2 construct (GAL::ZZ-dim2) were grown exponentially in complete medium, washed, and transferred to a glucose-based minimal medium lacking uracil for 19 h to achieve Dim2p depletion. Cells were pulse-labeled for 4 min with tritiated uracil and chased with an excess of cold uracil for up to 20 min. Total RNA was extracted and processed, separated on denaturing agarose gels, and transferred to high resolution membranes. Membranes were sprayed with tritium enhancer and exposed to film. For comparison, the experiment was performed in galactose in parallel.
GAL::ZZ-dim2 strains and a wild-type isogenic control were grown in YPGal and transferred to YPD to achieve Dim2p depletion (see above and Material and Methods). Total RNA was extracted at the time points indicated, separated on 1.2% agarose/formaldehyde gels, and transferred to nylon membranes for Northern-blot analysis. Membranes were hybridized with a collection of oligonucleotide probes specific to various subsets of precursors and mature rRNAs (Fig. 1A ▶).
In wild-type yeast cells, the primary Pol I transcript (35S) is rapidly cleaved in the 5′ external transcribed spacer (5′-ETS) at sites A0 and A1—the 5′ mature end of 18S rRNA—and in the internal transcribed spacer 1 (ITS1) at site A2 (Fig. 1B ▶). Cleavages at sites A0 and A1 generate the 33S and 32S pre-rRNAs, respectively. The 32S pre-rRNA is cleaved at site A2 to produce the 20S and 27SA2 pre-rRNAs, precursors to the small and large ribosomal subunit rRNAs, respectively.
The steady-state level of the 35S, 33S, and 32S pre-rRNAs was elevated in cells depleted for Dim2p by comparison to the wild-type isogenic control or to GAL::ZZ-dim2 strains grown in YPGal (Fig. 5A ▶, oligo c, cf. lanes 1–3 and 4–6), indicating a requirement for Dim2p in early pre-rRNA processing reactions.
The 20S and 27SA2 pre-rRNAs were readily detected in wild-type and GAL::ZZ-dim2 strains grown in permissive conditions; however, both species were strongly under accumulated in Dim2p-depleted cells (Fig. 5A ▶, oligos c and d, cf. lanes 1–3 and 4–6,). Cleavage at A2 therefore appeared strongly inhibited in the absence of Dim2p.
Hybridization with oligonucleotide d, specific to a sequence located between cleavage site A2 and A3, revealed the accumulation of the 22S RNA, an aberrant RNA species that extends between sites A0 and A3; 22S RNA accumulation is diagnostic of an inhibition of cleavage at sites A1 and A2 (Fig. 1C ▶). The 22S RNA is a dead-end intermediate that is not faithfully processed to 18S rRNA but rather degraded, presumably by the overlapping exoribonucleolytic activities of the Exosome complex (Allmang et al. 2000).
Hybridization with oligo c, which is specific to both the 20S pre-rRNA and the 22S RNA, showed that the accumulation of the 22S correlates with the depletion of 20S.
As a direct consequence of the inhibition of cleavage at sites A1 and A2, the steady-state level of 18S rRNA was substantially reduced in GAL::ZZ-dim2 strains grown in YPD (Fig. 5A ▶, oligo b, cf. lanes 1–3 and 4–6). PhosphorImager quantitation estimated the residual level of 18S rRNA to be of only ~5% of wild-type levels after a depletion prolonged for 24 h. By comparison, with the effects reported for other RRPs involved in SSU synthesis, including several core snoRNP components, this defect alone could easily accounts for the growth defect reported.
Processing downstream of cleavage site A2—which mostly concerns the synthesis of large ribosomal subunit rRNAs—was not greatly affected by the lack of Dim2p, and the steady-state level of the major intermediates involved in 5.8S–25S rRNA synthesis (notably, the 27SB and 7S pre-rRNAs), as well as the mature rRNAs, was essentially unaltered (Fig. 5A ▶, oligos e–g).
A low level of 23S RNA, which extends from the transcription start site (+1) to A3, is often detected in wild-type strains, indicating that a fraction of the pre-rRNA molecules is directly cleaved in the ITS1 at site A3; 23S was also detected in the absence of Dim2p but was not significantly increased (data not shown).
Concurring evidence for a defect in pre-rRNA processing at sites A1 and A2 was provided by a thorough primer extension analysis (Fig. 5B ▶). This allowed to examine the steady-state levels of low abundant and/or short-lived RNAs (e.g., 33S, 27SA3) and to detect species that are difficult to resolve on agarose gels (e.g., the short and long forms of several pre-rRNAs species; Fig. 1 ▶).
Oligonucleotide b, specific to the 5′ end of the 18S rRNA, was extended across the 5′-ETS; cDNA synthesis was specifically blocked at the transcription start site (+1), revealing the steady-state level of the primary transcript, and at site A0, corresponding to the 5′ end of the 33S pre-rRNA. Primer extension across ITS1 was performed from oligo f; this revealed cleavage at sites A2, A3, and B1. Alternative processing at site B1 generates short and long forms of pre-rRNAs that differ in length by seven to eight nucleotides (Fig. 1 ▶); these were well-resolved by this analysis.
Continuous cleavage at site A0 and inhibition of downstream processing sites were confirmed by a substantially increased signal at this site throughout the course of the depletion (Fig. 5B ▶, cf. lanes 1–3 and 4–7). The signal corresponding to RNA species ending at A2 was strongly codepleted with Dim2p, corroborating the inhibition of cleavage at this site. Cleavage at A3 and B1 appeared unaffected by the depletion of Dim2p, and the ratio of short versus long forms of the pre-rRNAs (~8:2) was unaltered. Position of cleavages all appeared conserved at the nucleotide level.
Finally, the kinetics of the pre-rRNA processing defect observed in strains lacking Dim2p was addressed by in vivo labeling (Fig. 6 ▶). Dim2p was depleted by growing GAL::ZZ-dim2 cells in a glucose-based minimal medium lacking uracil for 19 h (see Material and Methods). Cells were pulse-labeled for 4 min with tritiated uracil and chased with an excess of cold uracil for up to 20 min. Total RNA was extracted and processed as described above. For comparison, the experiment was performed in galactose in parallel.
In permissive conditions, the primary transcript was rapidly converted to 33S and 32S pre-rRNAs, and none of these RNA species were detected at the exposure presented in Figure 6 ▶. The 27S and 20S pre-rRNAs were readily detected and quantitatively chased into 25S and 18S rRNAs, respectively. In the absence of Dim2p, the 35S, 33S, and 32S pre-rRNAs were stably accumulated, due to their reduced processing rate. Processing of the 27SB precursors also significantly slowed down but with no apparent consequence for the overall accumulation of 25S rRNA. No 20S pre-rRNA was detected; rather, the 22S RNA was rapidly accumulated and appeared relatively stable over the time of the chase (20 min). The 22S RNA was not converted to 18S, leading to a substantial underaccumulation of this RNA.
None of the four snoRNAs involved in early pre-rRNA processing reactions in yeast, the box C/D snoRNAs U3 and U14 and the box H/ACA snoRNAs snR10 and snR30, were affected for their stability in cells depleted for Dim2p (data not shown).
In conclusion, Dim2p is required for the early pre-rRNA processing reactions at sites A1 and A2. In the absence of Dim2p, pre-rRNAs are directly cleaved in the ITS1 at site A3, generating the aberrant 22S RNA that is no longer processed. Consequently, cells deprived of Dim2p show a strong deficit in 18S rRNA, the small ribosomal subunit rRNA.
Dim2p is associated with Dim1p and is required for 18S rRNA dimethylation
Cells depleted for the 18S rRNA dimethyltransferase Dim1p virtually showed an identical pre-rRNA processing defect as those depleted for Dim2p, in particular, the accumulation of the 22S aberrant RNA diagnostic of pre-rRNA processing inhibitions at sites A1 and A2 (Lafontaine et al. 1995, 1998). This prompted us to test for physical and/or functional interactions between the two proteins, that is, to investigate whether Dim2p is associated with Dim1p in vivo and whether Dim2p is required for 18S rRNA demethylation.
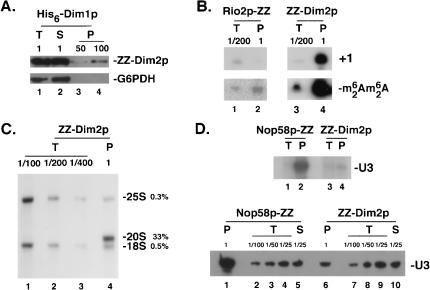
The association between Dim1p and Dim2p was tested by coprecipitation (Fig. 7A ▶). A total lysate was prepared in 150 mM salts from a ZZ-Dim2p strain transformed with a plasmid expressing a functional His6-Dim1p fusion under the control of its own promoter and incubated with metal affinity resin (see Material and Methods). Aliquots of total (T), supernatant (S), and pellet (P) fractions were loaded on PAGE, treated for Western blot, and decorated with an antibody specific for the protein A moiety of the ZZ-Dim2p fusion. Dim2p was efficiently recovered in the pellet fractions. As a control, the membrane was probed with an antibody specific to glucose-6-phosphate dehydrogenase (Zwf1p/G6PDH).
FIGURE 7.
Protein and RNA partners of Dim2p. Dim2p is stably associated with the 18S rRNA dimethyltransferase Dim1p as well as with the 35S primary transcripts, the 20S, and dimethylated 20S pre-rRNAs. A significant level of interaction with the box C/D snoRNA U3 was also noted. (A) Dim2p is stably associated with Dim1p. A crude cell lysate from cells expressing a ZZ-Dim2p fusion and a His6-Dim1p construct was incubated with metal affinity resin; the ZZ-Dim2p fusion was specifically recovered in the pellet fractions as detected by Western blotting with an antibody specific to the Protein A moiety of the fusion. As a control, an antibody specific to glucose-6-phosphate dehydrogenase (G6PDH) was used. (B, C) Dim2p is specifically associated with early and late pre-rRNAs. Crude cell lysates from strains expressing a functional ZZ-Dim2p fusion were used in a coprecipitation experiment with IgG-coated agarose beads (see Material and Methods). RNA from the total (T), supernatant (S), and pellet (P) fractions were either used in a primer extension experiment that selectively select for the 35S (+1) or the dimethylated 20S pre-rRNAs (oligonucleotides h and c respectively, B) or processed for Northern-blot hybridization with probes specific to the 20S pre-rRNA and the mature 18S and 25S rRNAs (C). Serial dilutions of the input were loaded as controls. (C) Phospor Imager quantitation of the material recovered revealed that over one third of the total amount of 20S pre-rRNA engaged in the pull-down assay was specifically coprecipitated with the ZZ-Dim2p fusion. For comparison, the low level of nonspecific coprecipitation observed for the mature rRNAs was of only ~0.5% and ~0.3% for the 18S and 25S rRNAs, respectively. (D) Dim2p interacts specifically with the box C/D snoRNA U3. Interaction between Dim2p and the snoRNAs was tested both by pCp-labeling (top) and Northern-blot (bottom). As a control, a strain expressing a core box C/D snoRNP protein (Nop58p-ZZ) was used.
The methylation status of pre-rRNAs accumulated in strains depleted for Dim2p was assessed by primer extension (Fig. 5B ▶). cDNA synthesis was initiated from primer c that is complementary to the 5′-end portion of ITS1; the reverse transcriptase used in this assay was previously shown to be specifically blocked by the modification (Henry et al. 1994; Lafontaine et al. 1995). A signal corresponding to the modification was readily detected in wild-type strains whether grown in YPGal or YPD (Fig. 5B ▶, lanes 1,2). For GAL::ZZ-dim2 strains, the modification was only detected in permissive conditions and at the early time points of transfer to glucose (Fig. 5B ▶, lanes 3,4).
We concluded that Dim1p and Dim2p are associated in vivo and that pre-rRNAs accumulated in the absence of Dim2p are not methylated.
Dim2p follows the preribosomes from early nucleolar to late cytoplasmic stages
RNA partners of Dim2p were also inspected by coprecipitation. Interactions with nucleolar RNA species (35S and U3) and a cytoplasmic pre-rRNA (methylated 20S) were tested by primer extension, Northern-blot, and/or pCp labeling (Fig. 7B–D ▶).
Glass-beads lysates from strains expressing a ZZ-Dim2p fusion were incubated with IgG-coated agarose beads in the presence of 150 mM salts. RNA from the total (T), supernatant (S), and pellet (P) fractions was extracted and processed for primer extension (Fig. 7B ▶), Northern-blot hybridization (Fig. 7C,D ▶, top), or pCp labeling (Fig. 7D ▶, bottom). Serial dilutions of the input were loaded to estimate the efficiency of recovery of the various RNAs coprecipitated. We concluded that Dim2p is tightly bound to the 35S, 20S, and methylated 20S pre-rRNAs. By comparison, Rio2p, another component of the SSU RRP complex, was also stably associated with methylated 20S pre-rRNAs but did not coprecipitate any 35S RNA. A significant level (~5%) of interaction with the box C/D snoRNA U3 was also noted; other snoRNAs were not efficiently coprecipitated (data not shown).
In conclusion, the association of Dim2p with the 35S and methylated 20S pre-rRNAs indicates that the protein joins the preribosomes at an early nucleolar stage and follows the particles to their site of final maturation in the cytoplasm.
DISCUSSION
KH-proteins and ribosome synthesis
The KH module is a well-characterized RNA binding domain, and unsurprisingly, KH-proteins have been involved in most steps of gene expression (see Kiledjian et al. 1995; Ostareck-Lederer et al. 1998; Irie et al. 2002; Kong et al. 2003; Oleynikov and Singer 2003); however, very few had been involved in ribosome synthesis (Sasaki et al. 2000).
Conditional inactivation of Dim2p led to a severe pre-rRNA processing defect. Cleavage of the pre-rRNAs was inhibited at the early processing sites A1 and A2, leading to a substantial deficit in 20S and 27SA2 pre-rRNAs and mature 18S rRNA with the concomitant accumulation of an aberrant 22S dead-end intermediate. This is similar to the phenotype described for cells depleted for Dim1p, a methyl-transferase that selects two adjacent adenosines for m62Am62A dimethylation at the 18S rRNA 3′ terminal stem–loop (Lafontaine et al. 1998 and references therein).
Dim2p is well conserved in eukaryotes, and orthologs are detected throughout the Archaeal domain of life. In addition, Dim2p shows striking homology with Krr1p, a eukaryotic-specific KH-protein involved in SSU synthesis (Sasaki et al. 2000). Other such couples of related nonredundant RRPs have been described, suggesting the possible handover of preribosomes from one partner to the next (e.g., the GTPases Bms1p and Tsr1p and the protein kinases Rio1p and Rio2p; Gelperin et al. 2001; Vanrobays et al. 2001, 2003; Wegierski et al. 2001). A similar mechanism has been proposed for several RRPs homologous to RPs or translation factors (e.g., Rlp7p/Rpl7p, Imp3p/Rps9p, Yho52p/Rpl1p, Rlp24p/Rpl24p, Efl1p/EF-2; for discussion, see Gadal et al. 2002).
Dim2p follows the preribosomes from the nucleolus to the cytoplasm
Subcellular distribution studies with a Dim2p-GFP construct demonstrated that the protein shows a highly dynamic behavior with respect to growth. The protein was detected both in the cytoplasm and the nucleus in exponentially growing cells and was concentrated to the nucleus and nucleolus as cells reached the stationary phase of growth. This suggested that Dim2p constantly shuttles between the nucle(ol)us and the cytoplasm and that this trafficking comes to a halt with the decreased need in ribosome synthesis that correlates with the stationary phase of growth. That Dim2p is indeed a nucleocytoplasmic shuttling protein was recently confirmed by its nuclear accumulation in strains defective for transport (Schafer et al. 2003). Furthermore, coprecipitation experiments demonstrated that Dim2p is associated with early and late pre-rRNAs, that is, nucleolar 35S primary transcripts and cytoplasmic methylated 20S pre-rRNAs. Collectively, these data indicate that Dim2p joins the preribosomes at an early nucleolar stage of assembly and follows the particles up to the cytoplasm.
Could Dim2p recruit Dim1p to the preribosomes?
The 18S rRNA dimethylation is a late modification that occurs on cytoplasmic 20S pre-rRNAs; however, the methyltransferase involved is additionally required for nucleolar pre-rRNA cleavages. We therefore postulated that nucleolar pre-rRNA molecules that have bound Dim1p and are thus programmed for cytoplasmic modification are positively discriminated, in a quality-control mechanism, for processing at sites A1 and A2 (Lafontaine et al. 1998 and references therein).
The association of Dim2p with early nucleolar pre-rRNAs and late cytoplasmic species, as well as its interaction with Dim1p and its requirement for 18S rRNA dimethylation, suggests that Dim2p could recruit Dim1p to the pre-rRNAs, a function that would be mediated by its KH-domain and account for its requirement in pre-rRNA processing.
A connection to proteasome function?
The relocation of Dim2p to the nucleolus, which occurs as cells reach the stationary phase of growth, coincides with the well-characterized “diauxic shift” (fermentation-based growth is limited by the depletion of glucose in the medium; see Herman 2002). In these conditions, the rate of ribosome synthesis is severely reduced, providing a possible explanation for the reduced nucleocytoplasmic shuttling of Dim2p and its nucleolar accumulation. It is quite remarkable in this respect that Nob1p (see below and Table 1 ▶), another component of the SSU RRP complex, is only detected in exponentially growing cells and is being degraded, notably by the action of the yeast proteasome, during the transition to the stationary phase (Tone et al. 2000).
A similar dynamic localization under different growth conditions was noted for Rix7p, a putative AAA-ATPase involved in LSU synthesis. Rix7p was mostly detected as a perinuclear rim in early-log phase, within the whole nucleus in mid-log and concentrated in the nucleolus in starved cells (Gadal et al. 2001). Importantly, this effect is not generic to the RRPs, as the conditions of growth did not affect the nucleolar localization of other SSU RRP such as Rio1p, Rio2p, Nop1p, Gar1p, and Nsr1p (E. Vanrobays and D.L.J. Lafontaine, unpubl.) and the LSU RRP (Noc1p; Milkereit et al. 2001).
Several lines of evidence suggest the existence of connections between small ribosomal subunit synthesis and proteasome function: (1) the Nin one binding protein (Nob1p), was originally isolated in a two-hybrid screen with Nin1p/Rpn12p, a subunit of the regulatory particle of the yeast proteasome (Tone and Toh 2002); (2) Dim2p is also known as Partner of Nob1p (Pno1p) and interacts physically both in two-hybrid and coprecipitation with Nob1p (Tone and Toh 2002); both Nob1p and Pno1p are required for proteasome biogenesis; (3) Krr1p was identified in a two-hybrid screen using Tom1p, a putative ubiquitine ligase, as a bait (Sasaki et al. 2000); and (4) the ribosomal proteins RPS0A/B, which are required for 18S rRNA synthesis, interact functionally with Tom1p (Tabb et al. 2001).
These interactions between SSU RRP components, proteasome subunits, and a putative ubiquitin ligase, suggest that the proteasome could be recruited to misassembled preribosomes, stalled in maturation, in order to trigger the rapid degradation of the associated proteins.
While this manuscript was in its final stage of preparation, Dim2p (Rrp20p) was isolated in a screen for mutants that die on loss of mitochondrial DNA (Senapin et al. 2003) and characterized in a large-scale proteomic analysis (Peng et al. 2003). Strikingly, a single amino acid substitution in the KH-domain (G235N) of Rrp20p led to a severe slow-growth phenotype at all temperatures tested, demonstrating the importance of this motif for protein function (Senapin et al. 2003).
MATERIAL AND METHODS
Sequence analysis
The multiple alignment presented in Figure 3 ▶ was performed with Multalign (http://prodes.toulouse.inra.fr/multalin/multalin). Phylogeny was addressed with PhTree (http://www.genebee.msu.su/services/phtree_reduced.html). Archaeal genomes were searched with COG at NCBI (http://www.ncbi.nlm.nih.gov/COG/). SMART (http://smart.embl-heidelberg.de/) was used to localize the KH-domain.
Yeast
Yeast media were standard yeast extract/peptone (YP) supplemented with either glucose (YPD) or galactose and sucrose (YPGal; 2% of each) and minimal yeast nitrogen base (YNB), supplemented with amino acids and bases as required. Fluoro-orotate–resistant clones were selected on YNB proline medium containing 0.6 g/L fluoro-orotic acid (5-FOA).
Yeast strains were as follows: YO590, a/α ura3-52/ura3-52 LEU2/LEU2 TRP1/TRP1 his3Δ200/HIS3 DIM2-GFP:KANMX6/DIM2-GFP:KANMX6;YO592, α ura3-52 leu2Δ1 TRP1 his3Δ200 dim2::KANMX4 (pYCG-DIM2 URA3); YO594, α ura3-52 leu2Δ1 TRP1 HIS3 (a generous gift from B. Winsor [Strasbourg, France]; Grava et al. 2000); and YO595, α ura3-52 leu2Δ1 TRP1 his3Δ200 dim2::KANMX4 (pGal::ZZ-DIM2 LEU2); YO596, α ura3-52 leu2Δ1 TRP1 his3Δ200 dim2::KANMX4 (pGal-ZZ-dim2 LEU2).
DNA constructs
Plasmid pGAL::ZZ-dim2 was constructed by PCR-directed homologous recombination as previously described (Oldenburg et al. 1997). Plasmid pGALPATA1L (CEN, ARS, LEU2) (a generous gift from K. Hellmuth and E. Hurt [Heidelberg, Germany]) was linearized with NcoI, and the DIM2 open reading frame was PCR-amplified (pfu polymerase Promega) from wild-type genomic DNA with the oligonucleotides 3′-YOR145c, ATAGATCTCTCGAG CTCGAATTCGGATCCCCGGGGCCTCCATGGCCATAAGCC CACAAATTATGTTTA and 5′-YOR145c, TTCGATATCCCAAC GACCGAAAACCTGTATTTTCAGGGCGCCCATATGGTTGCG CCTACTGCTTTGAA, flanked on the 5′ and 3′ ends by homologous sequences to plasmid pGALPATA1L. The linearized plasmid and PCR fragment were cotransformed into strain YO592; transformants were selected for leucine prototrophy and resistance to 5-FOA on glucose.
A functional His6-Dim1p fusion was generated by PCR and cloned into pFL39 (ARS/CEN-TRP1).
RNA methods
RNA isolation, Northern-blot hybridization, primer extension, and in vivo pulse-chase labeling experiments were essentially carried out as described previously (Tollervey 1987; Tollervey et al. 1991). The oligonucleotide probes used for Northern-blot hybridizations (Fig. 1A ▶) were as follows: a, CACCCATTCCCTCTTGC TAG; b, CATGGCTTAATCTTTGAGAC; c, TTAAGCGCAGGC CCGGCTGG; d, GATTGCTCGAATGCCCAAAG; e, TGCGTTC AAAGATTCGATG; f, GGCCAGCAATTTCAAGTTA; g, CTCAC GACGGTCTAAACCC; and h, ggcagatctgacgatcacc.
For the pulse-chase labeling, strain YO595 (GAL::ZZ-dim2) was pregrown at 30°C in YPGal medium and shifted to YPD for 12 h. Cells were washed and allowed for an addition 7 h of growth in minimal YNB medium (either glucose- or galactose-based) with uracil before being washed and shifted to identical media lacking uracil. Five milliliters of cultures was aliquoted and labeled for 4 min with 150 μCi {3H}-uracil; 0.6 mL cold uracil at 2 mg/mL was added, and 1 ml samples were collected at 1, 3, 5, 10, and 20 min and processed as described in Tollervey et al. (1991). 3′-end labeling was performed as described in Ganot et al. (1997).
Glycerol gradients analysis
Total yeast extracts and sedimentation profiles on 10% to 30% glycerol gradients were prepared as described in Vanrobays et al. (2003); 18 fractions were collected manually from the top of the gradients.
Western blot analysis
Proteins from total extracts, gradient fractions, or coprecipitation experiments were separated on 12% poly-acrylamide/SDS gels and transferred to Hybond-C membranes (AmershamBioSciences). GFP was detected by using a mouse monoclonal anti-GFP antibody (Roche) diluted at 1:1000; ribosomal proteins rpL3 and rpS8 were detected with a mouse monoclonal anti-rpL3 antiboby diluted at 1:5000 and a rabbit polyclonal anti-rpS8 antiboby diluted at 1:2000 (a kind gift from J. Warner, Albert Einstein, New York, and from G. Dieci, University of Parma, respectively). Protein A(ZZ)-epitope tagged proteins were detected with the rabbit HRP conjugated anti-Protein A antibody (PAP, Dako SA) diluted at 1:5000. Zwf1p/G6PDH was detected with a rabbit polyclonal antibody (Sigma) at the working dilution of 1:1000. The ECL kit (AmershamBioSciences) was used for detection.
Coprecipitation
Coprecipitation were performed exactly as described in Vanrobays et al. (2003). The Talon metal affinity resin (BD, Clontech) was used for the His6-Dim1p experiment.
Microscopy
A Leica (DMRB) microscope equipped with a Coolsnap-CCD camera was used to take the pictures.
Acknowledgments
Giovanna Jaramillo-Gutierrez (IBMM, ULB) is acknowledged for Northern-blot hybridizations and PhosphorImager quantitations; Sebastien Ferreira-Cerca, for support in the initial stages of this work. We thank Barbara Winsor (IBMC, CNRS, Strasbourg), Jonathan Warner (Albert Einstein, NY), and K. Hellmuth and E. Hurt (BZH, Heidelberg) for sharing valuable material; Yves Henry (LBME, CNRS, Toulouse), Stephan Visser, and Luis-Antonio Urrestarazu (IBMM, ULB), for helpful comments and critical reading of the manuscript. Research in MCF Laboratory is supported by the CNRS, the Université Paul Sabatier, and grants from the ‘Programme de Recherche Fondamentale en Microbiologie et Maladies Infectieuses et Parasitaires du Ministére de l’Education Nationale’ (MENRT) and ACI Microbiology. Research in DLJL Laboratory is supported by the Fonds National de la Recherche Scientifique, Université Libre de Bruxelles, and the following private charities: Alice and David van Buuren and Defay and Banque Nationale de Belgique. E.V. was supported by University Paul Sabatier and the EMBO (fellowship ALTF728-2002).
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.rnajournal.org/cgi/doi/10.1261/rna.5162204.
REFERENCES
- Adinolfi, S., Bagni, C., Castiglione Morelli, M.A., Fraternali, F., Musco, G., and Pastore, A. 1999. Novel RNA-binding motif: The KH module. Biopolymers 51: 153–164. [DOI] [PubMed] [Google Scholar]
- Allmang, C., Mitchell, P., Petfalski, E., and Tollervey, D. 2000. Degradation of ribosomal RNA precursors by the exosome. Nucleic Acids Res. 28: 1684–1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burd, C.G. and Dreyfuss, G. 1994. Conserved structures and diversity of functions of RNA-binding proteins. Science 265: 615–621. [DOI] [PubMed] [Google Scholar]
- Chan, H.Y., Brogna, S., and O’Kane, C.J. 2001. Dribble, the Drosophila KRR1p homologue, is involved in rRNA processing. Mol. Biol. Cell. 12: 1409–1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, W., Bucaria, J., Band, D.A., Sutton, A., and Sternglanz, R. 2003. Enp1, a yeast protein associated with U3 and U14 snoRNAs, is required for pre-rRNA processing and 40S subunit synthesis. Nucleic Acids Res. 31: 690–699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Boulle, K., Verkerk, A.J., Reyniers, E., Vits, L., Hendrickx, J., Van Roy, B., Van den Bos, F., de Graaff, E., Oostra, B.A., and Willems, P.J. 1993. A point mutation in the FMR-1 gene associated with fragile X mental retardation. Nat. Genet. 3: 31–35. [DOI] [PubMed] [Google Scholar]
- Dragon, F., Gallagher, J.E., Compagnone-Post, P.A., Mitchell, B.M., Porwancher, K.A., Wehner, K.A., Wormsley, S., Settlage, R.E., Shabanowitz, J., Osheim, Y., et al. 2002. A large nucleolar U3 ribonucleoprotein required for 18S ribosomal RNA biogenesis. Nature 417: 967–970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fatica, A. and Tollervey, D. 2002. Making ribosomes. Curr. Opin. Cell Biol. 14: 313–318. [DOI] [PubMed] [Google Scholar]
- Fatica, A., Oeffinger, M., Dlakic, M., and Tollervey, D. 2003. Nob1p is required for cleavage of the 3′ end of 18S rRNA. Mol. Cell. Biol. 23: 1798–1807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gadal, O., Strauss, D., Braspenning, J., Hoepfner, D., Petfalski, E., Philippsen, P., Tollervey, D., and Hurt, E. 2001. A nuclear AAA-type ATPase (Rix7p) is required for biogenesis and nuclear export of 60S ribosomal subunits. EMBO J. 20: 3695–3704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gadal, O., Strauss, D., Petfalski, E., Gleizes, P.E., Gas, N., Tollervey, D., and Hurt, E. 2002. Rlp7p is associated with 60S preribosomes, restricted to the granular component of the nucleolus, and required for pre-rRNA processing. J. Cell. Biol. 157: 941–951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganot, P., Caizergues-Ferrer, M., and Kiss, T. 1997. The family of box ACA small nucleolar RNAs is defined by an evolutionarily conserved secondary structure and ubiquitous sequence elements essential for RNA accumulation. Genes Dev. 11: 941–956. [DOI] [PubMed] [Google Scholar]
- Gavin, A.C., Bosche, M., Krause, R., Grandi, P., Marzioch, M., Bauer, A., Schultz, J., Rick, J.M., Michon, A.M., Cruciat, C.M., et al. 2002. Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature 415: 141–147. [DOI] [PubMed] [Google Scholar]
- Gelperin, D., Horton, L., Beckman, J., Hensold, J., and Lemmon, S.K. 2001. Bms1p, a novel GTP-binding protein, and the related Tsr1p are required for distinct steps of 40S ribosome biogenesis in yeast. RNA 7: 1268–1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson, T.J., Rice, P.M., Thompson, J.D., and Heringa, J. 1993a. KH domains within the FMR1 sequence suggest that fragile X syndrome stems from a defect in RNA metabolism. Trends Biochem. Sci. 18: 331–333. [DOI] [PubMed] [Google Scholar]
- Gibson, T.J., Thompson, J.D., and Heringa, J. 1993b. The KH domain occurs in a diverse set of RNA-binding proteins that include the antiterminator NusA and is probably involved in binding to nucleic acid. FEBS Lett. 324: 361–366. [DOI] [PubMed] [Google Scholar]
- Grandi, P., Rybin, V., Bassler, J., Petfalski, E., Strauss, D., Marzioch, M., Schafer, T., Kuster, B., Tschochner, H., Tollervey, D., et al. 2002. 90S pre-ribosomes include the 35S pre-rRNA, the U3 snoRNP, and 40S subunit processing factors but predominantly lack 60S synthesis factors. Mol. Cell 10: 105–115. [DOI] [PubMed] [Google Scholar]
- Grava, S., Dumoulin, P., Madania, A., Tarassov, I., and Winsor, B. 2000. Functional analysis of six genes from chromosomes XIV and XV of Saccharomyces cerevisiae reveals YOR145c as an essential gene and YNL059c/ARP5 as a strain-dependent essential gene encoding nuclear proteins. Yeast 16: 1025–1033. [DOI] [PubMed] [Google Scholar]
- Grishin, N.V. 2001. KH domain: One motif, two folds. Nucleic Acids Res. 29: 638–643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry, Y., Wood, H., Morrissey, J.P., Petfalski, E., Kearsey, S., and Tollervey, D. 1994. The 5′ end of yeast 5.8S rRNA is generated by exonucleases from an upstream cleavage site. EMBO J. 13: 2452– 2463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herman, P.K. 2002. Stationary phase in yeast. Curr. Opin. Microbiol. 5: 602–607. [DOI] [PubMed] [Google Scholar]
- Ho, Y., Gruhler, A., Heilbut, A., Bader, G.D., Moore, L., Adams, S.L., Millar, A., Taylor, P., Bennett, K., Boutilier, K., et al. 2002. Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature 415: 180–183. [DOI] [PubMed] [Google Scholar]
- Ihmels, J., Friedlander, G., Bergmann, S., Sarig, O., Ziv, Y., and Barkai, N. 2002. Revealing modular organization in the yeast transcriptional network. Nat. Genet. 31: 370–377. [DOI] [PubMed] [Google Scholar]
- Irie, K., Tadauchi, T., Takizawa, P.A., Vale, R.D., Matsumoto, K., and Herskowitz, I. 2002. The Khd1 protein, which has three KH RNA-binding motifs, is required for proper localization of ASH1 mRNA in yeast. EMBO J. 21: 1158–1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiledjian, M., Wang, X., and Liebhaber, S.A. 1995. Identification of two KH domain proteins in the α-globin mRNP stability complex. EMBO J. 14: 4357–4364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiss, T. 2002. Small nucleolar RNAs: An abundant group of noncoding RNAs with diverse cellular functions. Cell 109: 145–148. [DOI] [PubMed] [Google Scholar]
- Kong, J., Ji, X., and Liebhaber, S.A. 2003. The KH-domain protein α CP has a direct role in mRNA stabilization independent of its cognate binding site. Mol. Cell Biol. 23: 1125–1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kressler, D., Linder, P., and de la Cruz, J. 1999. Protein trans-acting factors involved in ribosome biogenesis in Saccharomyces cerevisiae. Mol. Cell. Biol. 19: 7897–7912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lafontaine, D.L.J. 2004. Eukaryotic ribosome synthesis. In Protein synthesis and ribosome structure (ed. K. Nierhaus). Wiley-InterScience (in press).
- Lafontaine, D.L.J. and Tollervey, D. 2001. The function and synthesis of ribosomes. Nat. Rev. Mol. Cell. Biol. 2: 514–520. [DOI] [PubMed] [Google Scholar]
- Lafontaine, D., Vandenhaute, J., and Tollervey, D. 1995. The 18S rRNA dimethylase Dim1p is required for pre-ribosomal RNA processing in yeast. Genes Dev. 9: 2470–2481. [DOI] [PubMed] [Google Scholar]
- Lafontaine, D.L.J., Preiss, T., and Tollervey, D. 1998. Yeast 18S rRNA dimethylase Dim1p: A quality control mechanism in ribosome synthesis? Mol. Cell. Biol. 18: 2360–2370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, P.C. and Thiele, D.J. 2001. Novel stress-responsive genes EMG1 and NOP14 encode conserved, interacting proteins required for 40S ribosome biogenesis. Mol. Biol. Cell. 12: 3644–3657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milkereit, P., Gadal, O., Podtelejnikov, A., Trumtel, S., Gas, N., Petfalski, E., Tollervey, D., Mann, M., Hurt, E., and Tschochner, H. 2001. Maturation and intranuclear transport of pre-ribosomes requires noc proteins. Cell 105: 499–509. [DOI] [PubMed] [Google Scholar]
- Oldenburg, K.R., Vo, K.T., Michaelis, S., and Paddon, C. 1997. Recombination-mediated PCR-directed plasmid construction in vivo in yeast. Nucleic Acids Res. 25: 451–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oleynikov, Y. and Singer, R.H. 2003. Real-time visualization of ZBP1 association with β-actin mRNA during transcription and localization. Curr. Biol. 13: 199–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostareck-Lederer, A., Ostareck, D.H., and Hentze, M.W. 1998. Cytoplasmic regulatory functions of the KH-domain proteins hnRNPs K and E1/E2. Trends Biochem. Sci. 23: 409–411. [DOI] [PubMed] [Google Scholar]
- Peng, W.T., Robinson, M.D., Mnaimneh, S., Krogan, N.J., Cagney, G., Morris, Q., Davierwala, A.P., Grigull, J., Yang, X., Zhang, W., et al. 2003. A panoramic view of yeast noncoding RNA processing. Cell 113: 919–933. [DOI] [PubMed] [Google Scholar]
- Raue, H.A. 2003. Pre-ribosomal RNA processing and assembly in Saccharomyces cerevisiae. In The nucleolus (ed. M.O. Olson). http://www.eurekah.com.
- Sasaki, T., Toh, E.A., and Kikuchi, Y. 2000. Yeast Krr1p physically and functionally interacts with a novel essential Kri1p, and both proteins are required for 40S ribosome biogenesis in the nucleolus. Mol. Cell. Biol. 20: 7971–7979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schafer, T., Strauss, D., Petfalski, E., Tollervey, D., and Hurt, E. 2003. The path from nucleolar 90S to cytoplasmic 40S pre-ribosomes. EMBO J. 22: 1370–1380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senapin, S., Clark-Walker, G.D., Chen, X.J., Seraphin, B., and Daugeron, M.C. 2003. RRP20, a component of the 90S preribosome, is required for pre-18S rRNA processing in Saccharomyces cerevisiae. Nucleic Acids Res. 31: 2524–2533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siomi, H., Matunis, M.J., Michael, W.M., and Dreyfuss, G. 1993. The pre-mRNA binding K protein contains a novel evolutionarily conserved motif. Nucleic Acids Res. 21: 1193–1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siomi, H., Choi, M., Siomi, M.C., Nussbaum, R.L., and Dreyfuss, G. 1994. Essential role for KH domains in RNA binding: Impaired RNA binding by a mutation in the KH domain of FMR1 that causes fragile X syndrome. Cell 77: 33–39. [DOI] [PubMed] [Google Scholar]
- Tabb, A.L., Utsugi, T., Wooten-Kee, C.R., Sasaki, T., Edling, S.A., Gump, W., Kikuchi, Y., and Ellis, S.R. 2001. Genes encoding ribosomal proteins Rps0A/B of Saccharomyces cerevisiae interact with TOM1 mutants defective in ribosome synthesis. Genetics 157: 1107–1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tollervey, D. 1987. A yeast small nuclear RNA is required for normal processing of pre-ribosomal RNA. EMBO J. 6: 4169–4175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tollervey, D., Lehtonen, H., Carmo-Fonseca, M., and Hurt, E.C. 1991. The small nucleolar RNP protein NOP1 (fibrillarin) is required for pre-rRNA processing in yeast. EMBO J. 10: 573–583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tone, Y. and Toh, E.A. 2002. Nob1p is required for biogenesis of the 26S proteasome and degraded upon its maturation in Saccharomyces cerevisiae. Genes Dev. 16: 3142–3157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tone, Y., Tanahashi, N., Tanaka, K., Fujimuro, M., Yokosawa, H., and Toh-e, A. 2000. Nob1p, a new essential protein, associates with the 26S proteasome of growing Saccharomyces cerevisiae cells. Gene 243: 37–45. [DOI] [PubMed] [Google Scholar]
- Trapman, J., Retel, J., and Planta, R.J. 1975. Ribosomal precursor particles from yeast. Exp. Cell. Res. 90: 95–104. [DOI] [PubMed] [Google Scholar]
- Tschochner, H. and Hurt, E. 2003. Pre-ribosomes on the road from the nucleolus to the cytoplasm. Trends Cell. Biol. 13: 255–263. [DOI] [PubMed] [Google Scholar]
- Udem, S.A. and Warner, J.R. 1973. The cytoplasmic maturation of a ribosomal precursor ribonucleic acid in yeast. J. Biol. Chem. 248: 1412–1416. [PubMed] [Google Scholar]
- Vanrobays, E., Gleizes, P.E., Bousquet-Antonelli, C., Noaillac-Depeyre, J., Caizergues-Ferrer, M., and Gelugne, J.P. 2001. Processing of 20S pre-rRNA to 18S ribosomal RNA in yeast requires Rrp10p, an essential non-ribosomal cytoplasmic protein. EMBO J. 20: 4204–4213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanrobays, E., Gelugne, J.P., Gleizes, P.E. and Caizergues-Ferrer, M. 2003. Late Cytoplasmic maturation of the small ribosomal subunit requires RIO proteins in Saccharomyces cerevisiae. Mol. Cell. Biol. 23: 2083–2095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vazquez, A., Flammini, A., Maritan, A., and Vespignani, A. 2003. Global protein function prediction from protein–protein interaction networks. Nat. Biotechnol. 21: 697–700. [DOI] [PubMed] [Google Scholar]
- Venema, J. and Tollervey, D. 1999. Ribosome synthesis in Saccharomyces cerevisiae. Ann. Rev. Genet. 33: 261–311. [DOI] [PubMed] [Google Scholar]
- Wegierski, T., Billy, E., Nasr, F., and Filipowicz, W. 2001. Bms1p, a G-domain-containing protein, associates with Rcl1p and is required for 18S rRNA biogenesis in yeast. RNA 7: 1254–1267. [DOI] [PMC free article] [PubMed] [Google Scholar]