Abstract
A simple strategy is reported for 5′-adenylation of nearly any RNA sequence of indefinite length. The 5′-adenylated product (5′-AppRNA) is an activated RNA that is structurally similar to 5′-triphosphorylated RNA, which is usually prepared by in vitro transcription using T7 RNA polymerase. In the new 5′-adenylation strategy, the RNA substrate is first 5′-monophosphorylated either by T4 polynucleotide kinase, by in vitro transcription in the presence of excess GMP, or by appropriate derivatization during solid-phase synthesis. The RNA is then 5′-adenylated using ATP and T4 RNA ligase, in an interrupted version of the natural adenylation–ligation mechanism by which T4 RNA ligase joins two RNA substrates. Here, the final ligation step of the mechanism is inhibited with complementary DNA blocking oligonucleotide(s) that permit adenylation to occur with good yield. The 5′-AppRNA products of this approach should be valuable as activated RNAs for in vitro selection experiments as an alternative to 5′-triphosphorylated RNAs, among other likely applications. The 5′-terminal nucleotide of an RNA substrate to be adenylated using the new method is not restricted to guanosine, in contrast to 5′-triphosphorylated RNA prepared by in vitro transcription. Therefore, using the new approach, essentially any RNA obtained from solid-phase synthesis or other means can be activated by 5′-adenylation in a practical manner.
Keywords: adenylation, RNA ligation, T4 RNA ligase, triphosphate, blocking oligo
INTRODUCTION
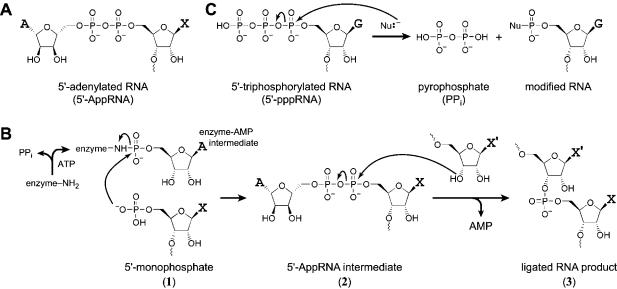
RNA with a 5′,5′-adenyl pyrophosphoryl cap structure—here termed 5′-adenylated1 RNA or 5′-AppRNA (Fig. 1A ▶)—is a key intermediate in the enzymatic joining of RNA substrates by T4 RNA ligase (Ohtsuka et al. 1976; Uhlenbeck and Gumport 1982). In the natural reaction mechanism, a lysine residue of T4 RNA ligase first reacts with ATP to form a covalent enzyme–AMP intermediate. This activated intermediate transfers its adenyl moiety to a 5′-monophosphorylated RNA terminus, yielding 5′-AppRNA (1 → 2 in Fig. 1B ▶). To complete the ligation reaction, the 5′-AppRNA then reacts with a 3′ hydroxyl of a second RNA substrate to form a 3′–5′ native phosphodiester bond (2 → 3). The same mechanistic pathway 1 → 2 → 3 is followed by many enzymes that ligate DNA or RNA (Weiss et al. 1968; Harvey et al. 1971; Lehman 1974; Higgins and Cozzarelli 1979). Because 5′-AppRNA plays a central role in this natural mechanism of nucleic acid ligation, a simple synthesis of 5′-AppRNA would facilitate experimental studies of such reactions.
FIGURE 1.
5′-Adenylated RNA. (A) The structure of 5′-AppRNA. X is the 5′-terminal nucleotide of the RNA substrate before adenylation. (B) The T4 RNA ligase mechanism, showing the 5′-AppRNA intermediate 2. X and X′ may be any nucleotides. (C) Nucleophilic displacement reaction on 5′-triphosphorylated RNA (5′-pppRNA). Nu, nucleophile. The 5′-terminal nucleotide of the RNA is shown as guanosine G because 5′-triphosphorylated RNAs are most typically prepared by in vitro transcription, which introduces G at this position. The nucleophilic substitution reaction on 5′-AppRNA is analogous, except with displacement of AMP instead of PPi (cf. 2 → 3 in B).
If the reactive 5′- and 3′-termini are in the same RNA molecule (intramolecular ligation), then the ligation reaction is a circularization. In vitro, circularization is often the major product formed by T4 RNA ligase, particularly for short oligonucleotides. Indeed circularization was the first T4 RNA ligase-catalyzed reaction to be studied (Silber et al. 1972; Kaufmann et al. 1974). The potential utility of T4 RNA ligase was immediately recognized for intermolecular ligation of mononucleotides or oligonucleotides to other oligonucleotides (Kaufmann and Littauer 1974; Walker and Uhlenbeck 1975; Uhlenbeck and Cameron 1977; England and Uhlenbeck 1978a; Hinton et al. 1978; Hinton and Gumport 1979; Moseman McCoy and Gumport 1980; Brennan et al. 1983; Romaniuk and Uhlenbeck 1983). Often this was in the context of labeling an oligonucleotide substrate with 32P or a fluorescent probe (Barrio et al. 1978; Bruce and Uhlenbeck 1978; England and Uhlenbeck 1978b). T4 RNA ligase is still used on occasion for preparative RNA ligation (Pan et al. 1991; Bain and Switzer 1992; Sherlin et al. 2001). A synthetic route to 5′-AppRNA will further enable these practical applications.
5′-Triphosphorylated RNA (5′-pppRNA) is an important RNA variant because it is synthesized in vitro using nucleotide triphosphate (NTP) monomers by T7 RNA polymerase (Milligan et al. 1987; Milligan and Uhlenbeck 1989) or by other phage polymerases (Krieg and Melton 1987; Stump and Hall 1993; Pokrovskaya and Gurevich 1994). Many in vitro selected ribozymes and deoxyribozymes catalyze reactions of RNA substrates that bear 5′triphosphates (Bartel and Szostak 1993; Ekland et al. 1995; Jaeger et al. 1999; Rogers and Joyce 1999; Tuschl et al. 2001; Reader and Joyce 2002). Natural ribozymes can sometimes use triphosphate substrates as well (Mörl et al. 1992). The use of NTP monomers has been mimicked by artificial RNA polymerase ribozymes (Ekland and Bartel 1996; Glasner et al. 2000; Johnston et al. 2001). In all of these cases, when a nucleophile attacks the 5′-triphosphate, the leaving group is pyrophosphate (PPi; Fig. 1C ▶). 5′-AppRNA is a structural analog of 5′-pppRNA that reacts similarly with a nucleophile, except that the leaving group is AMP rather than PPi (e.g., 2 → 3). Because both AMP and PPi are good leaving groups, a general synthesis of 5′-AppRNA will facilitate the preparation of activated nucleic acid substrates for use by in vitro selected ribozymes and deoxyribozymes (Hager and Szostak 1997). In addition, the 5′-terminal nucleotide of a T7 RNA polymerase transcript is typically 5′-pppG. A general 5′-adenylation method that does not specifically require a 5′-G but instead tolerates any 5′-terminal nucleotide would therefore expand even further the range of RNA sequences that may be explored as substrates for artificial nucleic acid enzymes.
5′-Activated RNA oligonucleotides with site-specific internal modifications will be useful for studying RNA structure, folding, and catalysis, in conjunction with developing RNA ligation methodology (Wang and Silverman 2003a,b; R.L. Coppins and S.K. Silverman, in prep.). However, in vitro transcription cannot insert a modified nucleoside site specifically at an arbitrary position within RNA, and in such cases, solid-phase RNA synthesis is the best approach (Earnshaw and Gait 1998; Scaringe et al. 1998; Scaringe 2000,Scaringe 2001). Although solid-phase synthesis of 5′-triphosphorylated RNA has been reported (Ludwig and Eckstein 1989; Gaur et al. 1992), this approach is limited in utility because the 5′-triphosphate must be introduced while the oligonucleotide is still attached to the solid-phase support and before global RNA deprotection. Related m7Gppp-capped RNA may be prepared after solid-phase synthesis using guanyl transferase with 5′-diphosphate RNA (Brownlee et al. 1995), but the 5′-diphosphate RNA must be prepared in a manner analogous to 5′-triphosphate RNA, with the associated limitations. As another option, 5′-activating groups may be appended to RNA using nucleic acid enzymes. For example, an in vitro selected ribozyme was reported with 5′-m7Gppp self-capping activity (Huang and Yarus 1997), and a self-adenylating deoxyribozyme has been identified (Li et al. 2000). However, the range of tolerated substrate sequences is significantly limited in these cases. A sequence-general synthetic route to 5′-activated RNAs that is applicable after the conclusion of solid-phase synthesis (i.e., after both cleavage of the oligonucleotide from the support and global deprotection) would expand the range of available activated RNAs, particularly those with site-specific modifications that are not readily prepared via transcription.
This article reports the general synthesis of 5′-AppRNA of nearly any sequence and indefinite length using T4 RNA ligase and ATP. Chiuman and Li (2002) recently described preparation of 5′-AppDNA using T4 DNA ligase and a complementary DNA oligonucleotide that renders the substrate double-stranded, as required by the ligase enzyme. In preliminary experiments at the outset of this study, it was found that T4 DNA ligase does not 5′-adenylate RNA under any tested conditions (data not shown). This prompted us to investigate T4 RNA ligase for 5′-AppRNA formation. In the approach reported here, the RNA substrate is first 5′- monophosphorylated with T4 polynucleotide kinase and ATP on a large scale and without polyacrylamide gel purification. Alternatively, the 5′-phosphate may be incorporated with standard reagents during solid-phase RNA synthesis, or it may be introduced by in vitro transcription in the presence of excess GMP, which is incorporated only at the 5′-end of the transcript in place of GTP. Subsequently, the T4 RNA ligase-mediated 5′-adenylation reaction follows an interrupted version of the natural ligation mechanism (Fig. 1B ▶). The 5′-AppRNA intermediate is synthesized from the 5′-phosphorylated RNA and ATP, but it does not react further to form ligated RNA (i.e., 1 → 2 but not 2 → 3). In this report, the conditions necessary for 5′-AppRNA synthesis via this strategy have been fully optimized. The 5′-AppRNA product and several side products have been characterized biochemically by a variety of approaches, and the method has been applied on the preparative (nanomole) scale.
RESULTS
A general strategy for 5′-adenylation of RNA
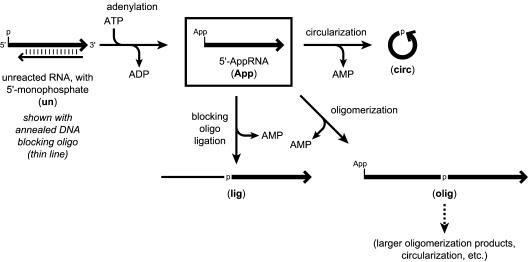
The key 5′-adenylation reaction uses T4 RNA ligase, ATP, and a 5′-monophosphorylated RNA substrate. T4 RNA ligase can circularize single-stranded RNAs of suitable length by intramolecular reaction of the free 3′-hydroxyl with the 5′-terminus of the AppRNA intermediate (see Introduction). Here this undesired side reaction is termed “circularization” (Fig. 2 ▶). T4 RNA ligase is also known to create RNA dimers and larger oligomers by the intermolecular analog of the circularization reaction, here termed “oligomerization.” 5′-Adenylation is strictly required as a preliminary step for both circularization and oligomerization. To enable preparative 5′-adenylation while suppressing any subsequent circularization or oligomerization, we annealed to the RNA substrate a complementary DNA oligonucleotide that we term a “blocking oligo” (Fig. 2 ▶). The key variables in the design of a DNA blocking oligo are the number of overhanging (or recessed) RNA nucleotides at each end of the RNA–DNA duplex, which we denote as n/m for the 5′- and 3′-ends, respectively. Positive values of n or m mean overhanging RNA at the relevant terminus, negative values imply recessed RNA, and zero indicates a blunt-ended duplex.
FIGURE 2.
Possible reaction products from 5′-adenylation of an RNA substrate with T4 RNA ligase and ATP. 5′-monophosphate and 5′-adenyl pyrophosphate termini are abbreviated p and App, respectively. The 5′-to-3′ polarity of each strand is shown by an arrowhead pointing in the 3′-direction. The desired 5′-AppRNA is boxed, and the three possible side reactions starting from 5′-AppRNA are illustrated (circularization, oligomerization, and blocking oligo ligation). The abbreviations used for the other products in the remaining figures of this article are given in boldface within parentheses. For the oligomerization reaction, the RNA substrate that does not provide the reactive 5′-App may itself have either 5′-p or 5′-App. Therefore, two different oligomerization products of any given nucleotide length are possible; only one is shown here.
In a typical 5′-adenylation experiment, a short (17-nt) RNA substrate was first 5′-phosphorylated with T4 polynucleotide kinase. Although this product may be purified by polyacrylamide gel electrophoresis (PAGE) if desired, a simple phenol/chloroform extraction and precipitation was sufficient to enable subsequent manipulations in all cases (see Materials and Methods). The 5′-monophosphorylated RNA was then annealed with a DNA blocking oligo—or no blocking oligo at all—and subsequently treated with T4 RNA ligase and ATP. The reaction progress was monitored by 20% denaturing PAGE, by which unreacted 5′-monophosphorylated RNA, 5′-adenylyated RNA, circularized RNA, and oligomerized RNA are readily resolved (see below). In some cases as noted, a third side product in addition to circularization and oligomerization was formed in significant amounts, by ligation of the 3′-end of excess DNA blocking oligo to the 5′-end of the AppRNA (Fig. 2 ▶). This undesired DNA–RNA chimera (i.e., DNApRNA connectivity) is termed the blocking oligo ligation product.
The product distribution depends strongly on the blocking oligo length
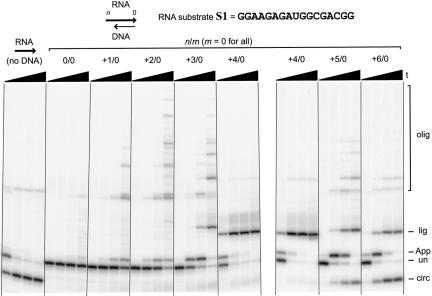
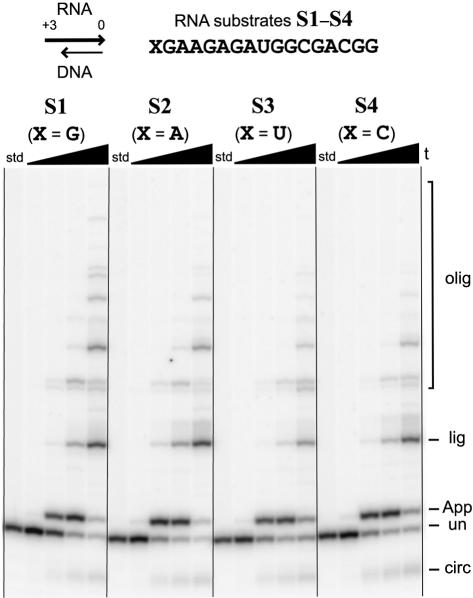
The distribution of T4 RNA ligase products—including the desired product 5′-AppRNA—depends strongly on the number of overhanging RNA nucleotides n at the 5′-end of the RNA substrate:DNA blocking oligo duplex (Fig. 3 ▶). With no blocking oligo at all (i.e., using a single-stranded RNA substrate), circularization of arbitrary 17-mer RNA substrate S1 is rapid (<10 min) and almost quantitative, with only a trace of oligomerization product. Such circularization was anticipated based on previous studies with similar short RNA oligonucleotides, and the circular product migrates slightly faster than its linear precursor, as expected (Silber et al. 1972; Kaufmann et al. 1974).
FIGURE 3.
Survey of 5′-adenylation with 17-mer RNA substrate S1 and various lengths of DNA blocking oligo relative to the RNA 5′-end. Either no blocking oligo was included (far left set of lanes), or the blocking oligo n/0 ranged from n = 0 to +6 (i.e., 0–6 nt RNA 5′-overhang). Gel images from two experiments are shown; the +4/0 experiment is included on both gel images to facilitate comparison. For all sets of lanes, the four time points are at 0.5 min, 10 min, 1 h, and 6 h (20% PAGE). The various species are labeled as follows, bottom to top: circ, circularization; un, unreacted RNA; App, 5′-adenylation; lig, blocking oligo ligation; olig, oligomerization (see Fig. 2 ▶). Similar to the 0/0 experiment (i.e., n = 0), no reaction was observed with blocking oligos for which n is negative (i.e., a recessed RNA 5′-end; data not shown).
The product distribution differs when a blocking oligo was annealed to the substrate before the addition of ATP and T4 RNA ligase. With a blocking oligo n/0 where n is 0 or negative (i.e., a blunt or recessed RNA 5′-terminus and a blunt 3′-terminus), virtually no reaction is observed even after 6 h (Fig. 3 ▶; data not shown). This is consistent with the previous finding that a blunt-ended 5′-terminus is a poor reaction partner (donor) in T4 RNA ligase-mediated ligation reactions (Bruce and Uhlenbeck 1978). As n is increased from +1 to +6 overhanging nucleotides, the RNA 5′-terminus becomes progressively more reactive. With the +1/0 blocking oligo that provides a 1-nt RNA 5′-overhang, adenylation is observable but very slow (hours), and oligomerization following adenylation is a substantial side reaction. With the +2/0 blocking oligo, the same products are observed as with +1/0, but on a somewhat more rapid timescale. With the +3/0 blocking oligo, the 5′-adenylation and oligomerization reactions are even faster, and blocking oligo ligation is now evident as an additional side reaction. For the +4/0 blocking oligo, its undesired ligation to the RNA substrate is the only reaction observed following 5′-adenylation, which is extremely rapid (<10 min). Finally, for +5/0 and +6/0 blocking oligos, both circularization and blocking oligo ligation are found to comparable extents, along with a trace of oligomerization. Curiously, the fastest 5′-adenylation rate is found for the +4/0 blocking oligo, with either longer or shorter RNA overhangs providing slower adenylation.
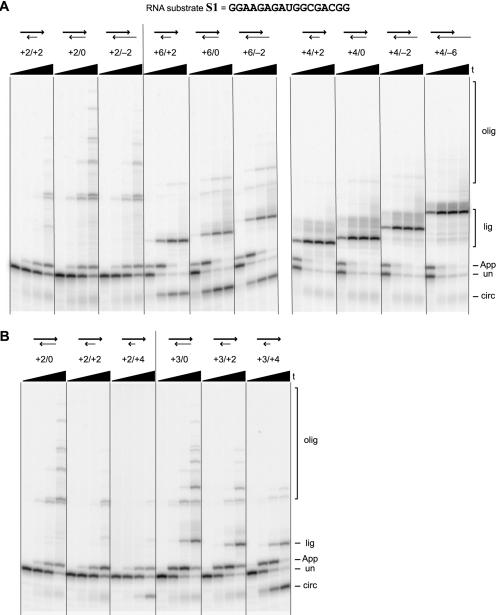
From these data, we conclude that both the rate of 5′-adenylation of S1 and the balance among the four possible subsequent fates (circularization, oligomerization, blocking oligo ligation, and no further reaction) are highly dependent on the value of n, which is determined simply by the choice of blocking oligo sequence. In contrast, other experiments show that the length m of the RNA 3′-overhanging or recessed end plays little role in the product distribution (Fig. 4A ▶). Blocking oligos n/m with n = 2, 4, or 6 were systematically examined. With a +2/m blocking oligo and m either +2, 0, or −2, only oligomerization is observed following 5′-adenylation, regardless of the value of m. Similarly, with blocking oligo +4/m with m either +2, 0, −2, or −6, the only product observed other than 5′-AppRNA is from blocking oligo ligation, independent of m. Note in Figure 4A ▶ that the migration rate of this particular side product tracks with the length of the blocking oligo, providing direct evidence that it arises from ligation of the DNA to the RNA (confirmatory MALDI-TOF mass spectrometry data is given below). Finally, with a +6/m blocking oligo, circularization and blocking oligo ligation occur to comparable extents regardless of the value of m (+2, 0, or −2). Therefore, for all of the tested values of n, the exact value of m is largely irrelevant in terms of determining the product distribution.
FIGURE 4.
Survey of 5′-adenylation with RNA substrate S1 and blocking oligos n/m with systematic variation of m. (A) Variation of m as either positive, zero, or negative, for n = +2 or +6 (left gel image) or n = +4 (right gel image). (B) Variation of m = 0, +2, or +4, with n = +2 or +3. For all sets of lanes, the four time points are at 0.5 min, 10 min, 1 h, and 6 h (20% PAGE).
We also examined various RNA 3′-overhang lengths (i.e., various positive values of m; Fig. 4B ▶). With n = +2 or +3, increasing the length of the RNA 3′-overhang lowers the prevalence of oligomerization side products, although the effect is slight. However, with an m = +4 overhang, circularization becomes a significant problem. Thus m must be chosen to avoid a long RNA 3′-overhang to achieve 5′-adenylation without circularization.
We conclude that for the 17-mer RNA substrate S1, neither circularization, oligomerization, or blocking oligo ligation can be completely suppressed solely by the appropriate length of RNA 3′-overhang (i.e, choice of m). Rather, it is only via judicious selection of both 5′- and 3′-overhang lengths (n and m) that the desired 5′-adenylated product may be formed in good yield, and a small amount of side reaction subsequent to adenylation is inevitable. The most promising value of n for S1 is +3, because circularization is nearly undetectable, and both oligomerization and blocking oligo ligation are much slower than 5′-adenylation itself. For S1, the value of m should be between 0 and +2 for efficient 5′-AppRNA formation. Thus, upon incubation of S1 with the +3/0 blocking oligo for 1 h, a high yield of 5′-AppRNA (~60%) is apparent (Fig. 3 ▶), although the various side products cannot be entirely avoided.
The blocking oligo 5′-adenylation strategy is generalizable to any RNA sequence
Using the +3/0 blocking oligo, we tested the 17-mer RNA substrate S1 alongside three related RNAs S2–S4 for which only the 5′-terminal nucleotide (denoted X in Fig. 1 ▶) was changed from G to any of A, U, or C (Fig. 5 ▶). In all four cases, 5′-adenylation proceeds in high yield, with minimal further reaction in all but the most lengthy incubations. Thus the blocking oligo strategy is general with regard to the 5′-terminal RNA nucleotide, at least for substrates S1–S4.
FIGURE 5.
5′-Adenylation is successful for any 5′-terminal RNA nucleotide. RNA substrates S1–S4 (which differ only in their 5′-terminal nucleotide X) were annealed with the +3/0 blocking oligo and assayed for 5′-adenylation as in Figure 3 ▶. std, RNA substrate incubated without T4 RNA ligase for 6 h. For all sets of lanes, the four time points are at 0.5 min, 10 min, 1 h, and 6 h (20% PAGE).
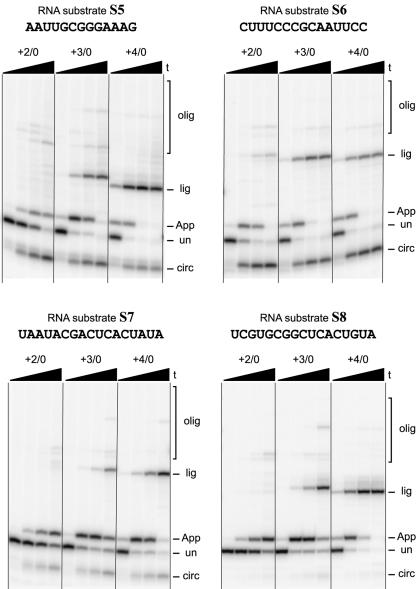
We also performed analogous assays on four unrelated short RNA oligonucleotides S5–S8 that have entirely different nucleotide compositions than S1–S4 but comparable length (Fig. 6 ▶). These assays used the appropriate +2/0, +3/0, or +4/0 blocking oligos for each RNA substrate. In all cases, 5′-adenylation proceeds well using the appropriate +3/0 blocking oligo on a similar timescale as for S1–S4. Furthermore, the +2/0 and +4/0 blocking oligos are inferior for providing solely 5′-adenylation, with the same pattern of side products as for S1–S4. Therefore, the optimal value of n for the blocking oligo is not strongly dependent on the RNA sequence. Instead, n = +3 is optimal for 5′-AppRNA formation for a wide variety of substrate sequences.
FIGURE 6.
Successful 5′-adenylation of a variety of short RNA substrate sequences S5–S8. Each RNA substrate was reacted with the appropriate +2/0, +3/0, or +4/0 blocking oligo, ATP, and T4 RNA ligase as in Figure 3 ▶. For all sets of lanes, the four time points are at 0.5 min, 10 min, 1 h, and 6 h (20% PAGE).
Deactivating (capping) the 3′-end of the DNA blocking oligo prevents formation of a side product, but at the expense of adenylation rate
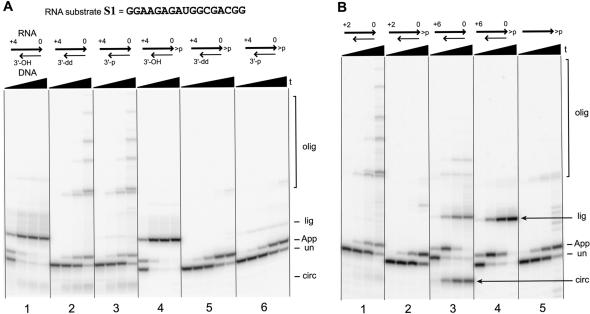
The undesired blocking oligo ligation product is presumably formed by reaction of the blocking oligo 3′-hydroxyl with the 5′-AppRNA. As shown in Figure 3 ▶ for RNA substrate S1, this undesired side reaction is significant for blocking oligos n/0 where n is +3 or greater. It is reasonable to predict that this side reaction would be entirely suppressed by appropriate 3′-capping of the DNA blocking oligo, which in turn may increase the yield of 5′-AppRNA. To explore this possibility, the +4/0 blocking oligo was synthesized with either a 2′,3′-dideoxy terminus (3′-dd) or a 3′-phosphate (3′-p). Either of these modifications removes the 3′-hydroxyl that attacks the 5′-AppRNA during formation of the blocking oligo ligation product. Note in Figure 3 ▶ that the uncapped +4/0 blocking oligo promotes very rapid adenylation of S1 (<10 min) but also extremely rapid blocking oligo ligation on a similar timescale. When the +4/0 blocking oligo was capped as either the 3′-dideoxy or 3′-phosphate and the S1 adenylation assay was performed, formation of the blocking oligo ligation side product was indeed suppressed. Unfortunately, 5′-adenylation itself was slowed markedly as well, allowing oligomerization to become significant (Fig. 7A ▶, compare second and third set of lanes to first set of lanes). An acceptable yield of 5′-AppRNA could still be obtained, but this now required at least 1 h, versus complete reaction in <10 min when the +4/0 DNA blocking oligo was 3′-uncapped. Similar observations were made with RNA substrates S2–S4 (data not shown).
FIGURE 7.
Effects of 3′-capping of the DNA blocking oligo and RNA substrate. (A) 3′-capping of the DNA blocking oligo suppresses formation of the blocking oligo ligation side product, but at the expense of adenylation rate. RNA substrate S1 was reacted with the 3′-uncapped (3′-OH), 2′,3′-dideoxy-capped (3′-dd), or 3′-phosphate-capped (3′-p) +4/0 blocking oligo, ATP, and T4 RNA ligase as in Figure 3 ▶. The RNA substrate was either 2′,3′-diol or 2′,3′-cyclic phosphate as shown. (B) Including a 2′,3′-cyclic phosphate on the RNA substrate prevents the circularization and oligomerization side reactions but not blocking oligo ligation. The DNA blocking oligo was 3′-uncapped. For 5′-adenylation of RNA substrate S1 that has no 2′,3′-cyclic phosphate in the absence of a DNA blocking oligo, see the far left of Figure 3 ▶. The faint set of bands in the middle of the final time point for the fifth set of lanes is of unknown origin. For all sets of lanes in both panels, the four time points are at 0.5 min, 10 min, 1 h, and 6 h (20% PAGE); the fifth time point in A for the final two sets of lanes is at 15 h. The sets of lanes are numbered at the bottom for reference (see text). For the experiments in sets of lanes 4 and 5, the 5′-AppRNA yields at 6 h were 60% and 50%, respectively; the yields at 15 h were 80% and 70%. Note that RNA with a 2′,3′-cyclic phosphate has a slightly faster mobility than 2′,3′-diol RNA due to the additional negative charge.
A 2′,3′-cyclic phosphate on the RNA substrate abolishes both circularization and oligomerization
Both the circularization and oligomerization side reactions are possible only if the 3′ terminus of the RNA substrate has a free 3′-hydroxyl. The inclusion of an appropriate DNA blocking oligo modulates these unwanted reactions but also adds the possibility of undesired ligation of the blocking oligo to the 5′-adenylated RNA substrate (which itself may be suppressed by 3′-capping of the blocking oligo, as described above). If the 3′-end of the RNA substrate could be rendered chemically incompetent for circularization and oligomerization, then a blocking oligo may no longer be necessary to achieve 5′-adenylation. The use of 3′-terminal blocking groups to control T4 RNA ligase-mediated ligation has been reported (Sninsky et al. 1976; Uhlenbeck and Cameron 1977; Hinton et al. 1978). Indeed, we found that 5′-AppRNA formation proceeds on substrate S1 that has a 2′,3′-cyclic phosphate, in the absence of any DNA blocking oligo (Fig. 7B ▶). However, this adenylation reaction is at least an order of magnitude slower than when the blocking oligo is present (compare the fourth and fifth sets of lanes in Fig. 7B ▶). Therefore, although a DNA blocking oligo is not strictly required to achieve 5′-adenylation when the RNA 3′-terminus is chemically blocked as the 2′,3′-cyclic phosphate, a blocking oligo may still be useful in practice for kinetic reasons.
All of the side reactions following 5′-AppRNA formation require a 3′-hydroxyl terminus either of the RNA substrate (for circularization and oligomerization) or of the DNA blocking oligo (for formation of the blocking oligo ligation product). Therefore, capping the 3′-end of the RNA as the 2′,3′-cyclic phosphate as well as the DNA as the 2′,3′-dideoxy or 3′-phosphate should circumvent all side reactions, thereby permitting a higher yield of 5′-adenylated RNA substrate. As anticipated, when both the RNA substrate and the DNA blocking oligo were 3′-capped, the sole product was 5′-AppRNA (right side of Fig. 7A ▶), in up to 80% yield. The trace amount of blocking oligo ligation product with the 3′-phosphate-capped DNA blocking oligo but not the 3′-dideoxy-capped blocking oligo (fifth and sixth set of lanes in Fig. 7A ▶) may arise after 3′-dephosphorylation of the DNA by the T4 RNA ligase, although this requires further study. As observed when the RNA did not have a 2′,3′-cyclic phosphate (left side of Fig. 7A ▶), the 5′-adenylation itself was significantly slowed when the DNA blocking oligo was 3′-capped, requiring at least 6 h of incubation for optimal yield of 5′-AppRNA.
Optimizing the incubation conditions for 5′-adenylation
Using RNA substrate S1 and the optimal uncapped +3/0 blocking oligo, the effects of varying the ATP, ligase, and RNA substrate concentrations were examined. Changing the ATP concentration between 10 and 250 μM (or including 0.05 mg/mL BSA, as recommended by some T4 RNA ligase suppliers) did not significantly affect the product distribution (data not shown). As expected, increasing the ligase concentration increased the reaction rate, and increasing the RNA concentration increased the prevalence of oligomerization products (data not shown; see Materials and Methods). Finally, T4 RNA ligase from two commercial suppliers was compared, with only slight activity differences (data not shown).
Preparative-scale 5′-adenylation and MALDI-TOF mass spectrometry product characterization
Any 5′-adenylation method would be most useful if it may be applied on the preparative rather than analytical scale (e.g., with nanomoles of RNA substrate rather than merely tens of picomoles). Relatively large-scale 5′-adenylation reactions were performed using 2 nmoles of 5′-phosphorylated RNA substrates S1–S4 and the uncapped +3/0 DNA blocking oligo, with incubation times from 20 to 90 min at 37°C. Isolated yields were 0.5–0.8 nmoles (25%–40%) after polyacrylamide gel electrophoresis and quantitation by UV absorbance (see Materials and Methods). Undesired side products from oligomerization and blocking oligo ligation were observed, consistent with the analytical-scale results (Fig. 3 ▶). In separate reactions, some of the normally undesired side products were deliberately prepared to permit their further characterization. The circularization product from RNA substrate S1 was obtained as essentially the only product by omitting the blocking oligo altogether, as expected based on the analytical-scale experiment (Fig. 3 ▶). When the uncapped +4/0 blocking oligo was included with S1, the blocking oligo ligation product predominated, also as expected. Finally, upon relatively lengthy incubation with the uncapped +3/0 blocking oligo, two specific oligomerization products from S1 were isolated in small amounts (~0.05 nmoles from 2.0 nmoles S1).
MALDI-TOF mass spectrometry characterization data were obtained for four of the 5′-AppRNAs, the circularized RNA from S1, and the oligomerization side products. The masses of the 5′-AppRNAs from S1–S4 (which collectively have all four possible 5′-terminal nucleotides) matched the expected values. The found (expected) masses were as follows: 5′-AppG (S1) 6018 ± 6 (6018); 5′-AppA (S2) 6002 ± 6 (6002); 5′-AppU (S3) 5979 ± 6 (5979); 5′-AppC (S4) 5978 ± 6 (5975).
Similarly, all of the assayed side products had the expected masses. The found (expected) masses were as follows: circularized S1 5669 ± 6 (5670); ligation product of S1 with the +4/0 blocking oligo 9526 ± 10 (9517); 5′-adenylated S1–S1 dimer 11,691 ± 12 (11,686); 5′-adenylated S1–S1–S1 trimer 17,377 ± 34 (17,356). In the latter two cases, the masses of the side products demonstrate that they are themselves 5′-adenylyated, as expected if 5′-AppRNA formation is rapid relative to subsequent oligomerization.
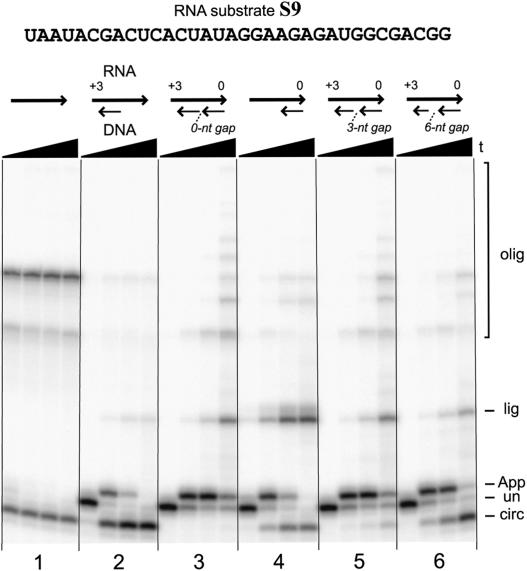
Longer RNA substrates require a blocking oligo for each terminus
Each of the RNA substrates S1–S8 is merely 13 to 17 nt long. Therefore, just one DNA blocking oligo is easily able to span the length of each substrate. In practice, one may wish to 5′-adenylate a longer RNA of arbitrary length, for which a single blocking oligo may be impractical to prepare. We therefore explored 5′-adenylation of longer RNAs. With the 34-nt substrate S9 (Fig. 8 ▶), two separate blocking oligos may anneal to the same RNA molecule without interference between the DNA strands. Using S9, blocking oligos at both the 5′- and 3′-termini are simultaneously required for 5′-adenylation without ensuing side reactions (Fig. 8 ▶; note in the third set of lanes that 1 h incubation gives predominantly 5′-AppRNA). Significantly, if either or both blocking oligos are omitted, then rapid circularization of S9 follows 5′-adenylation (first, second, and fourth sets of lanes; oligomerization is also observed without any blocking oligo). In addition, when only the 5′-blocking oligo is omitted, ligation of the RNA to the 3′-blocking oligo occurs to a significant extent (fourth set of lanes).
FIGURE 8.
5′-adenylation of the medium-length 34-mer RNA substrate S9. Up to two blocking oligos were used, one for each terminus of the RNA. The 5′-blocking oligo was n = +3, and the 3′-blocking oligo was m = 0 (each was 3′-uncapped). For all sets of lanes, the four time points are at 0.5 min, 10 min, 1 h, and 6 h (12% PAGE). The six sets of lanes are numbered at the bottom for reference (see text).
When the two blocking oligos shown in Figure 8 ▶ anneal to the RNA substrate, they form what is equivalent to a nicked RNA–DNA duplex that has no missing nucleotides along the DNA complement (i.e., a 0-nt gap). We considered the possibility that this may suppress circularization in a fashion not readily applicable to an even longer RNA substrate. Such a longer substrate would have a flexible single-stranded RNA region in the middle (Cohen and Cech 1998), and this flexibility could permit circularization despite the blocking DNA oligos annealed to each end. To model this situation without increasing the length of the RNA, we examined S9 using two blocking oligos that leave either a 3-nt region or a 6-nt stretch of the RNA substrate as a single-stranded flexible gap between the annealed DNA oligos. The relevant experiments are in the final two sets of lanes in Figure 8 ▶. As anticipated, increasing the size of this gap permits circularization. This is particularly evident for the 6-nt gap, where circularization is almost as rapid as when the 5′-blocking oligo is omitted altogether (compare the sixth and fourth sets of lanes). Nevertheless, even with the 6-nt gap, at short incubation times (e.g., 10 min) a substantial portion of the RNA substrate is 5′-adenylated but not yet circularized.
5′-adenylation of long RNAs
Finally, we examined 5′-adenylation of long RNAs (>100-mers), for which two special challenges arise. First, 5′-AppRNA formation does not lead to a measurable gel shift, unlike the situation for the relatively short RNA substrates S1–S8 and S9. Therefore it is impossible to assess directly by PAGE whether or not adenylation of the long RNA has occurred without resorting to a functional (rather than structural) assay. Second, if the 3′-terminus of the long RNA substrate cannot be protected as a 2′,3′-cyclic phosphate, then circularization and oligomerization can occur, prompting the use of blocking oligos. However, it may be impractical to prepare blocking oligos long enough to span the entire length of the RNA, as was successful with the two blocking oligos used together for 34-mer RNA substrate S9 (Fig. 8 ▶). It is not clear whether very long RNAs can be 5′- adenylated in such cases.
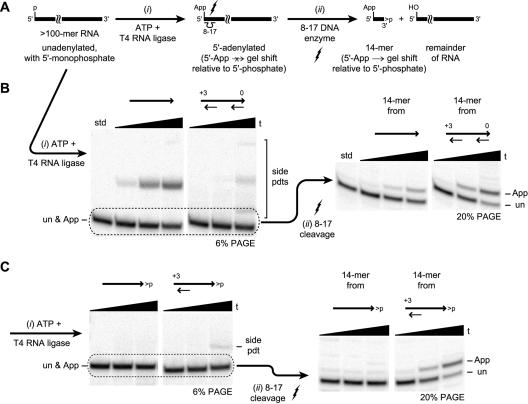
These challenges were addressed experimentally. The problem of directly observing 5′-adenylation of a long RNA substrate was solved with the aid of an RNA-cleaving 8–17 deoxyribozyme (Santoro and Joyce 1997) that excises the 5′-terminal portion of the long RNA. In such a case, 5′-adenylation of the RNA followed by deoxyribozyme cleavage near the 5′-terminus should lead to an easily detected gel shift when the 32P-radiolabeled products are cleaved with the deoxyribozyme and subsequently analyzed by PAGE (Fig. 9A ▶).
FIGURE 9.
5′-adenylation of long RNA substrates. (A) Schematic diagram of the experimental strategy. The >100-mer RNA substrate is too long for 5′-AppRNA formation to induce a measurable gel shift relative to a 5′-monophosphate. Therefore, an appropriate 8–17 deoxyribozyme is used to cleave the 5′-portion of the RNA substrate, leaving a small fragment for which 5′-AppRNA formation does cause a gel shift. (B) The strategy in A applied to the 160-nt P4–P6 domain of the Tetrahymena group I intron RNA. Blocking oligos were uncapped. The three time points are at 0.5 min, 10 min, and 1 h (6% PAGE). The RNA substrate was internally radiolabeled by transcription incorporating α-32P-ATP; the 5′-monophosphate was provided by performing the transcription in the presence of excess GMP (see Materials and Methods). Although the side products have not been studied in great detail, the side product formed in the first experiment (P4–P6 with no DNA blocking oligo) is tentatively assigned as circularized P4–P6 on the basis of attempted 5′-32P-radiolabeling with T4 polynucleotide kinase and γ-32P-ATP; no reaction was observed alongside a positive control. Only the lower band (a mixture of 5′-monophosphate and 5′-AppRNA) was carried to the 8–17 deoxyribozyme cleavage experiment. std, P4–P6 standard RNA carried through all reactions with no blocking oligo, except that T4 RNA ligase was omitted. (C) The strategy in A applied to the 136-nt P4–P6–24 RNA, which is missing the final 24 nt of P4–P6 and terminates with a 2′,3′-cyclic phosphate (Golden et al. 1996).
Using this approach, it was shown directly that two blocking oligos—one for each end of the long RNA substrate—permit a significant yield of 5′-adenylated RNA (Fig. 9B ▶). For these experiments, the substrate was the 160-nt P4–P6 domain of the Tetrahymena group I intron RNA, a representative RNA domain that is structurally well characterized (Murphy and Cech 1993; Cate et al. 1996; Juneau et al. 2001). P4–P6 presents a particular challenge for 5′-adenylation, for two reasons. First, the extensive secondary structure within P4–P6 renders binding of blocking oligos relatively difficult. The experiments in Figure 9B ▶ used two 3′-uncapped 50-mer blocking oligos, whereas shorter 15-mer blocking oligos failed to bind (data not shown). Second, in the folded secondary structure, the 5′- and 3′-termini of P4–P6 are fairly close in space, which is expected to promote unwanted circularization (although the influence of tertiary structure on circularization is difficult to predict). Despite these challenges, the data in Figure 9B ▶ demonstrate that a good yield of 5′-adenylated RNA may be obtained for this particular long RNA substrate. These data also demonstrate that even a very long (~60 nt) flexible single-stranded region between the blocked RNA 5′- and 3′-termini does not automatically lead to rapid circularization. Because a mere 6-nt gap in S9 led to significant circularization (Fig. 8 ▶), the relative lack of circularization for P4–P6 when blocking oligos are annealed may be enforced in part by residual secondary or tertiary structure.
Finally, a long RNA substrate with a 2′,3′-cyclic phosphate was examined. For this purpose we used a 136-nt derivative of P4–P6 in which the final 24 nt (out of 160) have been removed, and the remaining 136-nt RNA (named P4–P6–24) terminates with a 2′,3′-cyclic phosphate (Golden et al. 1996). 5′-AppRNA formation with P4–P6–24 as the substrate was successful when a 50-mer 5′-blocking oligo was used, but not in its absence (Fig. 9C ▶). A shorter 15-mer blocking oligo failed to bind (data not shown), as observed for the 160-nt P4–P6 RNA itself. In all of these assays, no 3′-blocking oligo was used, because the 3′-terminus of the RNA was already blocked as the cyclic phosphate.
Experiments to confirm the 5′-AppRNA structure and its mechanism of formation
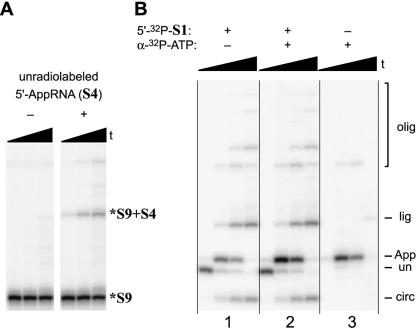
As additional confirmation of the structure of both the 5′-adenylation product and its mechanism of formation, two further experiments were performed. First, the unradiolabeled 5′-AppRNA product from the preparative adenylation reaction using 17-mer substrate S4 was incubated with a 5′-32P-radiolabeled 2′,3′-diol RNA (34-mer substrate S9) and T4 RNA ligase, all in the absence of ATP. Both RNAs were annealed with an appropriate blocking oligo before addition of T4 RNA ligase (see Materials and Methods). Only in the presence of the 5′-AppRNA (S4) was a higher molecular weight ligation product formed; this product was assigned as S9 + S4 (Fig. 10A ▶). This is consistent with 5′-AppRNA entering the T4 RNA ligase mechanistic pathway after the ATP-dependent step, as shown in Figure 1B ▶. In this experiment, the 5′-AppRNA (S4) and radiolabeled 5′-monophosphate RNA (S9) were deliberately chosen to have different lengths, such that any dimerization of S9 would not complicate interpretation of the results. However, no S9 dimerization was observed, regardless of the presence of 5′-adenylated S4.
FIGURE 10.
Experiments to confirm the 5′-AppRNA structure and its mechanism of formation. (A) Reaction of 5′-32P-radiolabeled 34-mer RNA substrate S9 with T4 RNA ligase in the absence of ATP and in the absence or presence of excess unradiolabeled 5′-AppRNA derived from 17-mer RNA substrate S4. The asterisk on *S9 denotes a 5′-radiolabel. As expected, only when unradiolabeled 5′-App-S4 is included is a higher molecular weight radiolabeled ligation product formed (*S9 + S4). For each set of lanes, the three time points are at 0.5 min, 10 min, and 1 h (12% PAGE). An appropriate blocking oligo was included in excess for both of the RNA substrates (see Materials and Methods). (B) 5′-adenylation of 17-mer RNA substrate S1 using either 5′-32P-S1, α-32P-ATP, or both. The +5/0 blocking oligo promotes formation of all three side products (see Fig. 3 ▶), therefore providing a stringent survey of the products into which the radiolabel becomes incorporated. The band patterns are as expected for the 5′-adenylation mechanism of Figure 1B ▶. For all sets of lanes, the four time points are at 0.5 min, 10 min, 1 h, and 6 h (20% PAGE). The three sets of lanes are numbered at the bottom for reference (see text).
Second, we noted that in the overall 5′-adenylation reaction, the β-phosphorus atom (5′-AppRNA) of the product originates as the α-phosphate of the ATP that is consumed in its formation (Fig. 1B ▶). This was exploited to provide further evidence of the 5′-AppRNA structure (Fig. 10B ▶). Using all three combinations of unradiolabeled and 32P-radiolabeled S1 RNA and ATP that include 32P in at least one source, the labeling patterns of the various products were as expected for the assigned mechanism. Specifically, when the RNA is 5′-32P-radiolabeled, all of the various reaction products incorporate 32P, regardless of whether the ATP is α-32P-radiolabeled or not (first and second set of lanes). However, when the sole source of 32P in the reaction solution is α-32P-ATP, only the 5′AppRNA product becomes radiolabeled (third set of lanes), as predicted. Furthermore, the 5′-AppRNA product appears only transiently on precisely the timescale anticipated from the experiments with the 5′-32P-labeled RNA substrate (compare the third set of lanes with the other two sets of lanes).
Testing 5′-AppRNA in place of 5′-pppRNA in functional assays
The nucleotide sequence of S1 corresponds to that of a key substrate for several in vitro selected RNA ligase deoxyribozymes that were previously discovered in our laboratory using the 5′-triphosphorylated version of S1 (Wang and Silverman 2003a,b; Coppins and Silverman 2004). As a preliminary survey of the mechanistic utility of 5′-AppRNA, we determined whether these deoxyribozymes can utilize 5′-AppRNA in place of 5′-pppRNA. The 7S11 deoxyribozyme that creates 2′,5′-branched RNA uses 5′-AppRNA in place of 5′-pppRNA with almost no decrease in ligation rate or yield (Coppins and Silverman 2004). In contrast, the branch-forming 9F7 deoxyribozyme (Wang and Silverman 2003a,b) is intolerant of 5′-AppRNA, whereas the related 9F13 deoxyribozyme maintains high ligation yield but with an approximately 10-fold lower rate (Y. Wang and S.K. Silverman, unpubl. results).
DISCUSSION
Using the blocking oligo strategy (Fig. 2 ▶), 5′-adenylation of RNA may be achieved on a reasonable timescale in good yield for a wide range of substrate sequences and lengths. The scope of potential side reactions following 5′-adenylation has been established, and the results indicate several ways to suppress formation of these side products to the greatest possible extent.
Demonstration of the 5′-AppRNA product structure
Multiple lines of evidence support assignment of the 5′-AppRNA structure of Figure 1A ▶ to the products formed by the new method. First, the gel mobilities of the products as well as the general reactivity patterns (Figs. 3 ▶–10 ▶) are consistent with the structural assignments. Second, in all tested cases, the MALDI-TOF mass spectrometry data of the preparative-scale 5′-adenylation products are as calculated for 5′-AppRNA. Third, the 5′-AppRNA product reacts as anticipated with a 2′,3′-diol RNA in the presence of T4 RNA ligase but in the absence of ATP (Fig. 10A ▶). Fourth, the labeling experiments of Figure 10B ▶ are consistent with the assigned product structure as well as its known mechanism of formation as shown in Figure 1B ▶. Overall, we have successfully interrupted the T4 RNA ligase adenylation–ligation mechanism of Figure 1B ▶ after the adenylation step to achieve synthesis of 5′-AppRNA in a universal fashion.
General observations on optimal 5′-adenylation of short RNA substrates
The data allow us to draw conclusions about optimal 5′-adenylation procedures using the blocking oligo strategy of Figure 2 ▶. For short RNA substrates such as S1–S8, a +3/0 DNA blocking oligo is optimal if it is 3′-uncapped (Figs. 3 ▶–6 ▶). That is, the blocking oligo should leave 3 nt of RNA as a 5′-overhang, and it should provide a blunt 3′ RNA terminus. Including a 2′,3′-cyclic phosphate on the RNA completely suppresses circularization and oligomerization, but at the expense of adenylation rate unless a blocking oligo is used, which then leads to blocking oligo ligation (Fig. 7 ▶). For the most favorable 5′-adenylation yield for each RNA substrate, the exact combination of substrate concentration, T4 RNA ligase concentration, and incubation time needs to be optimized. This is particularly true because the detailed time course of product formation varies with the RNA substrate sequence (Figs. 5 ▶, 6 ▶).
When a +4/0 DNA blocking oligo that is 3′-capped is employed, formation of the undesired blocking oligo ligation product is suppressed, but the 5′-adenylation itself is substantially slowed as well (Fig. 7 ▶). As a consequence, for an RNA substrate with a free 2′,3′-diol, oligomerization becomes an important side reaction, and there may be little advantage to 3′-capping the DNA blocking oligo. In contrast, if the RNA substrate has a 2′,3′-cyclic phosphate, then no side products at all are formed when a +4/0 3′-capped DNA blocking oligo is used. The 5′-adenylation is again slowed significantly, but yields are high. This latter approach would be particularly suitable if the RNA may be blocked as its 2′,3′-cyclic phosphate and an incubation time of several hours for the 5′-adenylation reaction is tolerable.
Requirement for a blocking oligonucleotide and/or a 2′,3′-cyclic phosphate on the RNA substrate
The major design element of the new synthetic approach to 5′-AppRNA is the use of at least one blocking oligo to modulate the RNA substrate’s reactivity (Fig. 2 ▶). A blocking oligo is not strictly required for 5′-adenylation, as shown by rapid circularization following adenylation of single-stranded RNA substrates that have a free 2′,3′-diol terminus (Fig. 3 ▶, first set of lanes). When the RNA 3′-terminus is blocked with a 2′,3′-cyclic phosphate, circularization is abolished (Fig. 7B ▶, fifth set of lanes), but the adenylation reaction is significantly slowed as well. Including a suitable blocking oligo increases the adenylation rate appreciably (Fig. 7B ▶, fourth set of lanes). Based on such observations, for practical purposes it may therefore be necessary to include a blocking oligo merely to enhance the adenylation rate and not solely to suppress subsequent side reactions. If introduction of a 2′,3′-cyclic phosphate into the RNA substrate is difficult or the cyclic phosphate is unwanted for subsequent experiments, then the blocking oligo approach provides a convenient alternative for satisfactory 5′-adenylation.
The relationship between 5′-adenylation rate and RNA 3′-terminus structure suggests that the 5′- and 3′-termini of an individual RNA substrate molecule may interact simultaneously with the same T4 RNA ligase molecule during the adenylation reaction, regardless of whether circularization subsequently occurs. This hypothesis is consistent with previous findings (Sugino et al. 1977), although it is also possible that trimolecular associations—two RNA substrates plus an enzyme molecule—are involved. In reactions that include a DNA blocking oligo, the excess blocking oligo may serve the role of acceptor nucleic acid to activate the adenylation step (Sugino et al. 1977), even though the blocking oligo does not always become ligated to the RNA. The observation that 3′-capping of the DNA blocking oligo significantly slows 5′-adenylation of the RNA substrate (Fig. 7A ▶) supports this role of the DNA.
The detailed distribution of side products differs for various RNA substrates
For reasons that we have not investigated in detail, the distribution of side products depends on the precise RNA substrate sequence. For example, substrates S1 and S8 shows very little circularization compared with S5–S7 (Figs. 5 ▶, 6 ▶). The lack of circularization with S8 is particularly unexpected because adenosine as the acceptor nucleoside (i.e., as the nucleoside whose 3′-hydroxyl reacts with the 5′-adenylated terminus) is known to provide the fastest ligation rate (England and Uhlenbeck 1978a), and on this basis, S8 was expected to have especially rapid circularization. Observations such as this imply that the particular array of side products and their relative rates of formation will depend idiosyncractically on the RNA substrate sequence. This may be particularly true for long RNAs (see below), where secondary and tertiary structure can influence the product distribution. Therefore, incubation conditions and blocking oligo design must be optimized experimentally for each new RNA adenylation substrate.
5′-adenylation of long RNA substrates
Some additional considerations arise for relatively long RNA substrates. For a medium-length substrate such as the 34-mer S9, two shorter blocking oligos may successfully be annealed to promote 5′-adenylation (Fig. 8 ▶). Presumably a single long DNA blocking oligo would work as well. For longer RNAs such as the 160-nt P4–P6 (Fig. 9 ▶), the situation is more complicated. Long RNAs are likely to have secondary and perhaps tertiary structure under the incubation conditions; the T4 RNA ligase buffer includes 10 mM Mg2+ at 1× strength, and RNA tertiary structure generally requires Mg2+ (Tinoco and Bustamante 1999). In the case of P4–P6, relatively lengthy blocking oligos (50-mers) are required, because shorter blocking oligos fail to bind. This is almost certainly due to secondary structure formation within P4–P6 that prevents the binding of short blocking oligos. In contrast to the results with S9 (Fig. 8 ▶), 5′AppRNA formation on P4–P6 is successful even with a large nominally single-stranded gap in the middle of the RNA (Fig. 9B ▶). This region of the RNA may adopt internal secondary structure that discourages any side reactions (particularly circularization) following 5′-adenylation, although we have not investigated this directly. Every long RNA substrate will probably have its own peculiarities that can only be discovered by direct experimentation.
When the 3′-terminus of the 136-nt P4–P6–24 is blocked as the 2′,3′-cyclic phosphate, only the 5′-blocking oligo is required for its successful 5′-adenylation (Fig. 9C ▶). This is consistent with the results for the shorter RNA substrate S1 (Fig. 7B ▶). Both sets of results agree that the inclusion of the 5′-blocking oligo significantly speeds adenylation relative to the single-stranded RNA substrate alone. However, for P4–P6–24, no adenylation at all was observed without the 5′- blocking oligo. Residual secondary structure within P4–P6– 24 in the absence of a 5′-blocking oligo may prevent 5′-adenylation altogether. Again, such considerations are highly specific to the individual long RNA substrate and must be determined on a case-by-case basis.
A practical concern for 5′-adenylation of long RNA substrates is that the 5′-monophosphate RNA and 5′-AppRNA are inherently inseparable by PAGE, because 5′-adenylation does not lead to a measureable gel shift even on the analytical scale. The deoxyribozyme-based cleavage assay of Figure 9 ▶ provides a straightforward means to assay the extent of 5′-adenylation for any particular RNA substrate. However, there appears to be no simple escape from the preparative impossibility of separating the 5′-AppRNA product from unadenylated RNA by conventional PAGE. For many purposes, having a fraction of the RNA molecules unadenylated should be tolerable. These unactivated molecules will simply fail to react with any nucleophile, and this lack of reactivity can be taken into account in the experimental design. For example, in a selection procedure, the maximum theoretical activity of a pool using a partially adenylated RNA substrate will be lower than 100%, but pool activities are usually well below 100% even in successful selection procedures.
Anticipated utility of 5′-AppRNA
The straightforward synthetic approach described here for 5′-AppRNA will be valuable for preparing activated RNA substrates for in vitro selected ribozymes and deoxyribozymes. In particular, the ability to activate the 5′-terminus of an RNA without regard to its sequence or length greatly expands the range of useful RNA substrates. For example, 5′-AppRNA formation may be the best practical way to activate the 5′-end of pyrimidine-rich RNAs that are difficult or impossible to transcribe using a phage polymerase. In addition, when the RNA incorporates site-specific modifications that require its preparation by solid-phase synthesis, adenylation by the method reported here may be the only straightforward way to activate the 5′-end. In all cases, for short RNA substrates, the 5′-AppRNA product may be separated cleanly by PAGE. For long RNAs, the 5′-AppRNA product will be an inseparable mixture with unadenylated RNA, as described above.
In the functional assays that used 5′-AppRNA in place of 5′-triphosphorylated RNA as a substrate for RNA ligase deoxyribozymes, a range of outcomes was observed with the 7S11, 9F7, and 9F13 deoxyribozymes that form 2′,5′-branched RNA. These diverse results support the utility of 5′-AppRNA for examining the mechanisms of already-identified nucleic acid enzymes, as well as for providing alternative activated RNA substrates for ongoing selection efforts.
MATERIALS AND METHODS
RNA substrates and DNA blocking oligos
RNA substrate oligonucleotides S1–S9 each terminating in a 2′,3′-diol were prepared by solid-phase synthesis (Dharmacon, Inc. or HHMI-Keck Laboratory, Yale University). Their nucleotide sequences are given in the figures. The RNA substrate S1 terminating in a 2′,3′-cyclic phosphate was prepared by in vitro transcription of a precursor RNA using an annealed DNA oligonucleotides template (Milligan et al. 1987; Milligan and Uhlenbeck 1989) followed by cleavage with a 10–23 deoxyribozyme (Santoro and Joyce 1997; Pyle et al. 2000). All of these RNAs were purified by 20% denaturing PAGE followed by ethanol precipitation.
The RNA substrates were 5′-phosphorylated in 2-nmole portions (1 mM ATP, 10 U T4 polynucleotide kinase from New England Biolabs, 1× PNK buffer, 50 μL reaction volume; 37°C, 2–4 h), followed by standard phenol-chloroform extraction and ethanol precipitation. The resulting 5′-monophosphorylated RNAs were used directly in subsequent 5′-adenylation experiments. PAGE purification of the unradiolabeled monophosphorylated RNA did not change the observed adenylation product distributions (data not shown). Radiolabeled RNAs were prepared with γ-32P-ATP (PerkinElmer) and T4 PNK (New England Biolabs) and purified by 20% denaturing PAGE followed by ethanol precipitation.
Short DNA blocking oligos (<20-mers) were obtained from IDT. Uncapped blocking oligos were purified by conventional phenol-chloroform extraction and ethanol precipitation. PAGE purification did not change the observed adenylation product distributions (data not shown). The 3′-capped blocking oligos were purified by 20% denaturing PAGE followed by ethanol precipitation; they were not tested before PAGE purification.
For experiments with long RNA substrates, the P4–P6 RNA (Silverman and Cech 1999) or P4–P6–24 RNA (Golden et al. 1996) was prepared by T7 RNA polymerase transcription from linearized plasmid DNA templates. The transcription solutions included excess GMP (10 mM GMP, 0.5 mM GTP) to provide >95% 5′-monophosphate RNA. The 50-mer DNA blocking oligos from IDT were purified by 8% denaturing PAGE.
T4 RNA ligase
T4 RNA ligase was obtained from either MBI Fermentas or New England Biolabs. Enzyme from either source gave comparable results (data not shown). The data shown in the figures were obtained with the MBI Fermentas enzyme.
5′-adenylation assays for short and medium-length RNA substrates
A typical analytical-scale 5′-adenylation procedure is as follows. A sample containing 0.25 pmoles of 5′-32P-radiolabeled RNA substrate, 10 pmoles of unradiolabeled 5′-monophosphorylated RNA substrate, and 30 pmoles of DNA blocking oligo in 8 μL of 5 mM Tris (pH 7.5), 15 mM NaCl, and 0.1 mM EDTA was annealed by heating to 95°C for 2 min and cooling on ice for 5 min. The solution was brought to 10 μL total volume containing 1× T4 RNA ligase buffer (50 mM HEPES at pH 8.0, 10 mM MgCl2, 10 mM DTT), 50 μM ATP, and 10 units of T4 RNA ligase (MBI Fermentas). Final concentrations were 1 μM RNA substrate, 3 μM DNA blocking oligo, and 1 U/μL T4 RNA ligase. The sample was fully immersed in a 37°C water bath. At appropriate time points, 2-μL aliquots were quenched into 8 μL of stop solution (80% formamide, 1× TB, 50 mM EDTA, 0.25% each bromophenol blue and xylene cyanol, where 1× TB contains 89 mM each Tris and boric acid at pH 8.3). Time points were generally taken at 0.5 min, 10 min, 1 h, and 6 h. Quenched aliquots were electrophoresed on 20% denaturing PAGE and imaged with a PhosphorImager.
In some analytical-scale experiments (not shown), the concentration of T4 RNA ligase was varied between 0.1 and 2 U/μL for 1 μM RNA substrate and 3 μM DNA blocking oligo. Separately, the concentration of T4 RNA ligase was varied between 0.4 and 4 U/μL for 5 μM RNA substrate and 15 μM DNA blocking oligo. In all cases, successful 5′-AppRNA formation was observed; the optimal incubation time varied considerably (10 min to 6 h) depending on the exact conditions.
In all experiments, the DNA blocking oligo sequences were checked to verify that the computed ΔG values for binding to the relevant RNA substrate were adequate. Values of ΔG were computed using published parameters (Sugimoto et al. 1995). We and others have previously concluded that ΔG values of >12–14 kcal/mole are reasonable in such experiments that require DNA–RNA hybridization (Pyle et al. 2000; Flynn-Charlebois et al. 2003). For RNA substrate S1 and the 0/0 blocking oligo (Fig. 3 ▶), the computed ΔG is −26.5 kcal/mole, and the optimal +3/0 blocking oligo has a computed ΔG of −21.3 kcal/mole. For the +6/+2 blocking oligo (Fig. 4A ▶) the computed ΔG is −11.4 kcal/mole. This latter blocking oligo is the shortest used for S1, and its results are comparable to those obtained with longer blocking oligos that have more favorable ΔG (compare the +6/0 and +6/−2 experiments in Fig. 4A ▶).
Several of the assays for the medium-length RNA substrate S9 used two blocking oligos (Fig. 8 ▶). The sequence of S9 is shown in Figure 8 ▶. The sequence of the 5′ blocking oligo for all of these experiments was 5′-TATAGTGAGTCGTA-3′. The sequence of the 3′ blocking oligo for the 0-nt gap experiment was 5′-CCGTCGC CATCTCTTCC-3′, which is the same as the +0/0 blocking oligo for substrate S1. For the 3-nt and 6-nt gap experiments, 3 and 6 nt, respectively, were omitted from the 3′ terminus of the latter oligo (equivalent to the +3/0 and +6/0 blocking oligos for S1). For the experiment shown in the sixth set of lanes in Figure 8 ▶ (6-nt gap), the 3′-blocking oligo has a computed ΔG of −16.4 kcal/mole. It is therefore unlikely that the increase in circularization side product is due to poor annealing of either blocking oligo.
For the experiment of Figure 10A ▶, an appropriate DNA blocking oligo was used for both the S9 and 5′-App-S4 RNA substrates. The blocking oligo for S9 was n = 0 to prevent any action by T4 RNA ligase on its 5′-terminus, yet a long 17-nt RNA 3′-overhang was left such that the 3′-terminus could react with 5′-App-S4 if present. The blocking oligo for 5′-App-S4 was +3/0 such that the 5′-terminus was free for T4 RNA ligase-mediated reaction but the 3′-terminus was unavailable for circularization or self-dimerization.
5′-adenylation assays for long RNA substrates
Each long RNA substrate (P4–P6 or P4–P6–24) was assayed for 5′-adenylation in a two-step procedure as shown in Figure 9A ▶. First, the internally 32P-radiolabeled RNA transcript was 5′-adenylated using the general procedure described above for the shorter RNA substrates (~20 pmoles RNA, 80 pmoles each blocking oligo as appropriate, and 20 units of T4 RNA ligase in 20 μL; 4 μL aliquots were quenched into 15 μL of stop solution). The products were separated by 6% denaturing PAGE (Fig. 9B,C ▶, left) in preparation for the cleavage step. Second, the products were cleaved with an appropriate 8–17 deoxyribozyme (5′-GCTGTT GACCCTCCGAGCCGGACGATTCCCGCAATTC-3′, where the underlined regions bind to the RNA), which excises 14 nt from the 5′-end of the RNA. The RNA product from the first step (~2–10 pmoles) was incubated with 50 pmoles of deoxyribozyme in 50 mM Tris (pH 7.5) and 60 mM MgCl2 at 37°C for 6 h. The products were assayed by 20% denaturing PAGE (Fig. 9B,C ▶, right), which revealed 5′-adenylation as a gel shift comparable to that seen for the shorter RNA substrates.
Because the long RNA substrates were prepared with internal 32P radiolabels, the remaining 146 nt of P4–P6 (remaining 122 nt of P4–P6–24) were observed as expected at the top of the 6% PAGE image; they are not shown in Figure 9 ▶. For practical application of this deoxyribozyme-based assay strategy to other long RNAs, it may be more expedient to 5′-32P-radiolabel the RNA before adenylation, which will provide RNA of higher specific activity. However, the 3′-terminus of such 5′-radiolabeled RNAs will not be observable using 32P imaging.
Preparative 5′-adenylation reactions
Preparative 5′-adenylation reactions were performed using 2 nmoles of 5′-monophosphorylated RNA substrate as follows. RNA substrate (2.0 nmoles) and excess +3/0 3′-uncapped DNA blocking oligo (4.0 nmoles) were annealed in 80 μL of 5 mM Tris (pH 7.5), 15 mM NaCl, and 0.1 mM EDTA by heating to 95°C for 2 min and cooling on ice for 5 min. The solution was brought to 100 μL total volume containing 1× T4 RNA ligase buffer (see above), 50 μM ATP, and 40–200 units of T4 RNA ligase (MBI Fermentas). Final concentrations were 20 μM RNA substrate, 40 μM DNA blocking oligo, and 0.4–2.0 U/μL T4 RNA ligase. After incubation at 37°C for 20–90 min, the solution was quenched with 200 μL of stop solution (see above) and purified by 20% denaturing PAGE. The RNA products were eluted from the preparative gel and precipitated with ethanol as described (Wang and Silverman 2003b), with quantification by UV absorbance (A260). Between 0.5 and 0.8 nmoles of 5′-adenylated RNA product (25%–40% isolated yield) was obtained, depending on the RNA substrate sequence, ligase concentration, and incubation time. MALDI-TOF mass spectra were obtained on an Applied Biosystems Voyager-DE STR instrument in the Mass Spectrometry Laboratory of the University of Illinois at Urbana–Champaign (UIUC) School of Chemical Sciences.
Acknowledgments
This research was supported by the Burroughs Wellcome Fund (New Investigator Award in the Basic Pharmacological Sciences), the March of Dimes Birth Defects Foundation (5-FY02-271), the National Institutes of Health (GM-65966), and the UIUC Department of Chemistry. Acknowledgement is made to the donors of The Petroleum Research Fund, administered by the American Chemical Society, for partial support of this research (38803-G4). S.K.S. is the recipient of a fellowship from The David and Lucile Packard Foundation. Rebecca Coppins, Chandra Miduturu, and Richard Gumport are thanked for valuable discussions.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.rnajournal.org/cgi/doi/10.1261/rna.5247704.
Footnotes
In this article, the term “adenylated” is used for simplicity instead of “adenylylated.” Both terms have been used elsewhere.
REFERENCES
- Bain, J.D. and Switzer, C. 1992. Regioselective ligation of oligoribonucleotides using DNA splints. Nucleic Acids Res. 20: 4372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrio, J.R., Barrio, M.C., Leonard, N.J., England, T.E., and Uhlenbeck, O.C. 1978. Synthesis of modified nucleoside 3′,5′-bisphosphates and their incorporation into oligoribonucleotides with T4 RNA ligase. Biochemistry 17: 2077–2081. [DOI] [PubMed] [Google Scholar]
- Bartel, D.P. and Szostak, J.W. 1993. Isolation of new ribozymes from a large pool of random sequences. Science 261: 1411–1418. [DOI] [PubMed] [Google Scholar]
- Brennan, C.A., Manthey, A.E., and Gumport, R.I. 1983. Using T4 RNA ligase with DNA substrates. Methods Enzymol. 100: 38–52. [DOI] [PubMed] [Google Scholar]
- Brownlee, G.G., Fodor, E., Pritlove, D.C., Gould, K.G., and Dalluge, J.J. 1995. Solid phase synthesis of 5′-diphosphorylated oligoribonucleotides and their conversion to capped m7Gppp-oligoribonucleotides for use as primers for influenza A virus RNA polymerase in vitro. Nucleic Acids Res. 23: 2641–2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruce, A.G. and Uhlenbeck, O.C. 1978. Reactions at the termini of tRNA with T4 RNA ligase. Nucleic Acids Res. 5: 3665–3677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cate, J.H., Gooding, A.R., Podell, E., Zhou, K., Golden, B.L., Kundrot, C.E., Cech, T.R., and Doudna, J.A. 1996. Crystal structure of a group I ribozyme domain: Principles of RNA packing. Science 273: 1678–1685. [DOI] [PubMed] [Google Scholar]
- Chiuman, W. and Li, Y. 2002. Making AppDNA using T4 DNA ligase. Bioorg. Chem. 30: 332–349. [DOI] [PubMed] [Google Scholar]
- Cohen, S.B. and Cech, T.R. 1998. A quantitative study of the flexibility contributed to RNA structures by nicks and single-stranded gaps. RNA 4: 1179–1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coppins, R.L. and Silverman, S.K. 2004. A DNA enzyme that mimics the first step of RNA splicing. Nature Struct. Mol. Biol. 11 (in press). [DOI] [PubMed]
- Earnshaw, D.J. and Gait, M.J. 1998. Modified oligoribonucleotides as site-specific probes of RNA structure and function. Biopolymers 48: 39–55. [Google Scholar]
- Ekland, E.H. and Bartel, D.P. 1996. RNA-catalysed RNA polymerization using nucleoside triphosphates. Nature 382: 373–376. [DOI] [PubMed] [Google Scholar]
- Ekland, E.H., Szostak, J.W., and Bartel, D.P. 1995. Structurally complex and highly active RNA ligases derived from random RNA sequences. Science 269: 364–370. [DOI] [PubMed] [Google Scholar]
- England, T.E. and Uhlenbeck, O.C. 1978a. Enzymatic oligoribonucleotide synthesis with T4 RNA ligase. Biochemistry 17: 2069–2076. [DOI] [PubMed] [Google Scholar]
- ———. 1978b. 3′-terminal labelling of RNA with T4 RNA ligase. Nature 275: 560–561. [DOI] [PubMed] [Google Scholar]
- Flynn-Charlebois, A., Wang, Y., Prior, T.K., Rashid, I., Hoadley, K.A., Coppins, R.L., Wolf, A.C., and Silverman, S.K. 2003. Deoxyribozymes with 2′-5′ RNA ligase activity. J. Am. Chem. Soc. 125: 2444–2454. [DOI] [PubMed] [Google Scholar]
- Gaur, R.K., Sproat, B.S., and Krupp, G. 1992. Novel solid-phase synthesis of 2′-O-methylribonucleoside 5′-triphosphates and their α-thio analogues. Tet. Lett. 33: 3301–3304. [Google Scholar]
- Glasner, M.E., Yen, C.C., Ekland, E.H., and Bartel, D.P. 2000. Recognition of nucleoside triphosphates during RNA-catalyzed primer extension. Biochemistry 39: 15556–15562. [DOI] [PubMed] [Google Scholar]
- Golden, B.L., Gooding, A.R., Podell, E.R., and Cech, T.R. 1996. X-ray crystallography of large RNAs: Heavy-atom derivatives by RNA engineering. RNA 2: 1295–1305. [PMC free article] [PubMed] [Google Scholar]
- Hager, A.J. and Szostak, J.W. 1997. Isolation of novel ribozymes that ligate AMP-activated RNA substrates. Chem. Biol. 4: 607–617. [DOI] [PubMed] [Google Scholar]
- Harvey, C.L., Gabriel, T.F., Wilt, E.M., and Richardson, C.C. 1971. Enzymatic breakage and joining of deoxyribonucleic acid. IX. Synthesis and properties of the deoxyribonucleic acid adenylate in the phage T4 ligase reaction. J. Biol. Chem. 246: 4523–4530. [PubMed] [Google Scholar]
- Higgins, N.P. and Cozzarelli, N.R. 1979. DNA-joining enzymes: A review. Methods Enzymol. 68: 50–71. [DOI] [PubMed] [Google Scholar]
- Hinton, D.M. and Gumport, R.I. 1979. The synthesis of oligodeoxyribonucleotides using RNA ligase. Nucleic Acids Res. 7: 453–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinton, D.M., Baez, J.A., and Gumport, R.I. 1978. T4 RNA Ligase joins 2′-deoxyribonucleoside 3′,5′-bisphosphates to oligodeoxyribonucleotides. Biochemistry 17: 5091–5097. [DOI] [PubMed] [Google Scholar]
- Huang, F. and Yarus, M. 1997. 5′-RNA self-capping from guanosine diphosphate. Biochemistry 36: 6557–6563. [DOI] [PubMed] [Google Scholar]
- Jaeger, L., Wright, M.C., and Joyce, G.F. 1999. A complex ligase ribozyme evolved in vitro from a group I ribozyme domain. Proc. Natl. Acad. Sci. 96: 14712–14717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston, W.K., Unrau, P.J., Lawrence, M.S., Glasner, M.E., and Bartel, D.P. 2001. RNA-catalyzed RNA polymerization: Accurate and general RNA-templated primer extension. Science 292: 1319– 1325. [DOI] [PubMed] [Google Scholar]
- Juneau, K., Podell, E., Harrington, D.J., and Cech, T.R. 2001. Structural basis of the enhanced stability of a mutant ribozyme domain and a detailed view of RNA–solvent interactions. Structure 9: 221–231. [DOI] [PubMed] [Google Scholar]
- Kaufmann, G. and Littauer, U.Z. 1974. Covalent joining of phenylalanine transfer ribonucleic acid half-molecules by T4 RNA ligase. Proc. Natl. Acad. Sci. 71: 3741–3745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufmann, G., Klein, T., and Littauer, U.Z. 1974. T4 RNA ligase: Substrate chain length requirements. FEBS Lett. 46: 271–275. [DOI] [PubMed] [Google Scholar]
- Krieg, P.A. and Melton, D.A. 1987. In vitro RNA synthesis with SP6 RNA polymerase. Methods Enzymol. 155: 397–415. [DOI] [PubMed] [Google Scholar]
- Lehman, I.R. 1974. DNA ligase: Structure, mechanism, and function. Science 186: 790–797. [DOI] [PubMed] [Google Scholar]
- Li, Y., Liu, Y., and Breaker, R.R. 2000. Capping DNA with DNA. Biochemistry 39: 3106–3114. [DOI] [PubMed] [Google Scholar]
- Ludwig, J. and Eckstein, F. 1989. Rapid and efficient synthesis of nucleoside 5′-O-(1-thiotriphosphates), 5′-triphosphates and 2′,3′-cyclophosphorothioates using 2-chloro-4H-1,3,2-benzodioxaphosphorin-4-one. J. Org. Chem. 54: 631–635. [Google Scholar]
- Milligan, J.F. and Uhlenbeck, O.C. 1989. Synthesis of small RNAs using T7 RNA polymerase. Methods Enzymol. 180: 51–62. [DOI] [PubMed] [Google Scholar]
- Milligan, J.F., Groebe, D.R., Witherell, G.W., and Uhlenbeck, O.C. 1987. Oligoribonucleotide synthesis using T7 RNA polymerase and synthetic DNA templates. Nucleic Acids Res. 15: 8783–8798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mörl, M., Niemer, I., and Schmelzer, C. 1992. New reactions catalyzed by a group II intron ribozyme with RNA and DNA substrates. Cell 70: 803–810. [DOI] [PubMed] [Google Scholar]
- Moseman McCoy, M.I. and Gumport, R.I. 1980. T4 ribonucleic acid ligase joins single-strand oligo(deoxyribonucleotides). Biochemistry 19: 635–642. [DOI] [PubMed] [Google Scholar]
- Murphy, F.L. and Cech, T.R. 1993. An independently folding domain of RNA tertiary structure within the Tetrahymena ribozyme. Biochemistry 32: 5291–5300. [DOI] [PubMed] [Google Scholar]
- Ohtsuka, E., Nishikawa, S., Sugiura, M., and Ikehara, M. 1976. Joining of ribooligonucleotides with T4 RNA ligase and identification of the oligonucleotide-adenylate intermediate. Nucleic Acids Res. 3: 1613–1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan, T., Gutell, R.R., and Uhlenbeck, O.C. 1991. Folding of circularly permuted transfer RNAs. Science 254: 1361–1364. [DOI] [PubMed] [Google Scholar]
- Pokrovskaya, I.D. and Gurevich, V.V. 1994. In vitro transcription: Preparative RNA yields in analytical scale reactions. Anal. Biochem. 220: 420–423. [DOI] [PubMed] [Google Scholar]
- Pyle, A.M., Chu, V.T., Jankowsky, E., and Boudvillain, M. 2000. Using DNAzymes to cut, process, and map RNA molecules for structural studies or modification. Methods Enzymol. 317: 140–146. [DOI] [PubMed] [Google Scholar]
- Reader, J.S. and Joyce, G.F. 2002. A ribozyme composed of only two different nucleotides. Nature 420: 841–844. [DOI] [PubMed] [Google Scholar]
- Rogers, J. and Joyce, G.F. 1999. A ribozyme that lacks cytidine. Nature 402: 323–325. [DOI] [PubMed] [Google Scholar]
- Romaniuk, P.J. and Uhlenbeck, O.C. 1983. Joining of RNA molecules with RNA ligase. Methods Enzymol. 100: 52–59. [DOI] [PubMed] [Google Scholar]
- Santoro, S.W. and Joyce, G.F. 1997. A general purpose RNA-cleaving DNA enzyme. Proc. Natl. Acad. Sci. 94: 4262–4266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scaringe, S.A. 2000. Advanced 5′-silyl-2′-orthoester approach to RNA oligonucleotide synthesis. Methods Enzymol. 317: 3–18. [DOI] [PubMed] [Google Scholar]
- ———. 2001. RNA oligonucleotide synthesis via 5′-silyl-2′-orthoester chemistry. Methods 23: 206–217. [DOI] [PubMed] [Google Scholar]
- Scaringe, S.A., Wincott, F.E., and Caruthers, M.H. 1998. Novel RNA synthesis method using 5′-O-silyl-2′-O-orthoester protecting groups. J. Am. Chem. Soc. 120: 11820–11821. [Google Scholar]
- Sherlin, L.D., Bullock, T.L., Nissan, T.A., Perona, J.J., Lariviere, F.J., Uhlenbeck, O.C., and Scaringe, S.A. 2001. Chemical and enzymatic synthesis of tRNAs for high-throughput crystallization. RNA 7: 1671–1678. [PMC free article] [PubMed] [Google Scholar]
- Silber, R., Malathi, V.G., and Hurwitz, J. 1972. Purification and properties of bacteriophage T4-induced RNA ligase. Proc. Natl. Acad. Sci. 69: 3009–3013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverman, S.K. and Cech, T.R. 1999. Energetics and cooperativity of tertiary hydrogen bonds in RNA structure. Biochemistry 38: 8691–8702. [DOI] [PubMed] [Google Scholar]
- Sninsky, J.J., Last, J.A., and Gilham, P.T. 1976. The use of terminal blocking groups for the specific joining of oligonucleotides in RNA ligase reactions containing equimolar concentrations of acceptor and donor molecules. Nucleic Acids Res. 3: 3157–3166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stump, W.T. and Hall, K.B. 1993. SP6 RNA polymerase efficiently synthesizes RNA from short double-stranded DNA templates. Nucleic Acids Res. 21: 5480–5484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugimoto, N., Nakano, S., Katoh, M., Matsumura, A., Nakamuta, H., Ohmichi, T., Yoneyama, M., and Sasaki, M. 1995. Thermodynamic parameters to predict stability of RNA/DNA hybrid duplexes. Biochemistry 34: 11211–11216. [DOI] [PubMed] [Google Scholar]
- Sugino, A., Snoper, T.J., and Cozzarelli, N.R. 1977. Bacteriophage T4 RNA ligase. Reaction intermediates and interaction of substrates. J. Biol. Chem. 252: 1732–1738. [PubMed] [Google Scholar]
- Tinoco Jr., I. and Bustamante, C. 1999. How RNA folds. J. Mol. Biol. 293: 271–281. [DOI] [PubMed] [Google Scholar]
- Tuschl, T., Sharp, P.A., and Bartel, D.P. 2001. A ribozyme selected from variants of U6 snRNA promotes 2′,5′-branch formation. RNA 7: 29–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlenbeck, O.C. and Cameron, V. 1977. Equimolar addition of oligoribonucleotides with T4 RNA ligase. Nucleic Acids Res. 4: 85–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlenbeck, O.C. and Gumport, R.I. 1982. T4 RNA Ligase. In The Enzymes (ed. P.D. Boyer), pp. 31–58. Academic Press, New York.
- Walker, G.C. and Uhlenbeck, O.C. 1975. Stepwise enzymatic oligoribonucleotide synthesis including modified nucleotides. Biochemistry 14: 817–824. [DOI] [PubMed] [Google Scholar]
- Wang, Y. and Silverman, S.K. 2003a. Deoxyribozymes that synthesize branched and lariat RNA. J. Am. Chem. Soc. 125: 6880–6881. [DOI] [PubMed] [Google Scholar]
- ———. 2003b. Characterization of deoxyribozymes that synthesize branched RNA. Biochemistry 42: 15252–15263. [DOI] [PubMed] [Google Scholar]
- Weiss, B., Jacquemin-Sablon, A., Live, T.R., Fareed, G.C., and Richardson, C.C. 1968. Enzymatic breakage and joining of deoxyribonucleic acid. VI. Further purification and properties of polynucleotide ligase from Escherichia coli infected with bacteriophage T4. J. Biol. Chem. 243: 4543–4555. [PubMed] [Google Scholar]