Abstract
Many RNA-mediated reactions in transcription, translation, RNA processing, and transport require assembly of RNA complexes, yet assembly pathways remain poorly understood. Assembly mechanisms can be difficult to assess in a biological context because many components interact in complex pathways and individual steps are difficult to isolate experimentally. Our previous studies of self-cleaving hairpin ribozymes showed that kinetic and equilibrium parameters measured in yeast agree well with parameters measured in vitro under ionic conditions that mimic the intracellular environment. We now report studies of intermolecular reactions with ribozyme and target sequences expressed in yeast as separate chimeric U3 snoRNAs. In this system, intracellular cleavage rates reflect the kinetics of ribozyme-substrate complex formation through annealing of base-paired helices. Second-order rate constants increased with increasing helix length for in vitro reactions with 2 mM MgCl2 and 150 mM NaCl and in vivo but not in reactions with 10 mM MgCl2. Thus, efficient RNA complex formation required a larger extent of complementarity in vivo than in vitro under conditions with high concentrations of divalent cations. The most efficient intracellular cleavage reactions exhibited second-order rate constants that were 15- to 30-fold below rate constants for cleavage of oligonucleotides in vitro. Careful analysis of structural features that influence cleavage efficiency points to substrate binding as the rate-determining step in the intracellular cleavage pathway. Second-order rate constants for intermolecular cleavage agree well with diffusion coefficients reported for U3 snoRNPs in vivo suggesting that complex formation between chimeric ribozyme and substrate snoRNPs in yeast nuclei is diffusion limited.
Keywords: hairpin ribozyme, RNA catalysis, RNA assembly, intracellular kinetics, RNA folding, U3 snoRNP
INTRODUCTION
RNA assembly reactions participate in critical steps in gene expression ranging from the transient associations between snRNAs and precursor mRNAs during splicing through the stable assembly of ribonucleoprotein structures that mediate protein synthesis and transport. Recognition typically occurs through the formation of complementary base-paired helices. Well-known examples include mRNA binding to 16S ribosomal RNA through Shine-Dalgarno sequences, U1 snRNA binding to splice sites, the U4/U6 association in the spliceosome, and snoRNA interactions with precursor ribosomal RNA cleavage and modification targets (Shine and Dalgarno 1974; Reich et al. 1992; Staley and Guthrie 1998; Sharma and Tollervey 1999; Kiss 2002; Decatur and Fournier 2003). MicroRNA and siRNA interactions with target RNAs provide further examples of RNA-mediated gene regulation through complementary base-pairing (Carrington and Ambros 2003). These kinds of RNA-mediated reactions involve many components that interact in complex pathways so that individual assembly steps can be difficult to evaluate quantitatively.
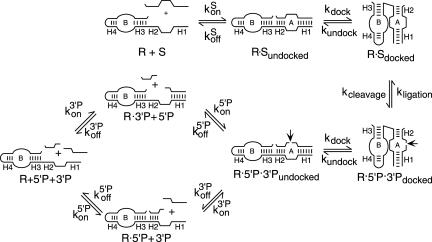
RNA enzymes have been particularly amenable to investigations of RNA recognition and assembly because catalytic activity provides a quantitative signal that a ribozyme has formed a functional complex with its substrate. Application of quantitative physical and enzymological methods has led to detailed insights into some of the structural transitions that define ribozyme assembly pathways in vitro. Intermolecular catalysis by minimal hairpin ribozymes includes substrate binding, cleavage, 5′ and 3′ product dissociation steps, and the reverse steps of product binding, ligation, and substrate dissociation (Fig. 1 ▶). Elemental rate and equilibrium constants for individual steps in the reaction pathway have been determined for several ribozyme variants in “standard” reactions that include 10 mM MgCl2 and 50 mM TrisCl or NaHEPES, pH 7.5 at 25°C (Fedor 2000). The hairpin ribozyme assembles with substrate and product RNAs through complementary base-pairing to form intermolecular helices H1 and H2 (Fig. 1 ▶). Rate constants for substrate and product binding on the order of 107 M−1 min−1 are somewhat slower than those measured for formation of simple base-paired RNA helices (Craig et al. 1971; Pörschke and Eigen 1971; Ravetch et al. 1974; Hegg and Fedor 1995; Esteban et al. 1997; Walter and Burke 1997). Like the kinetics of simple RNA helix formation, substrate binding rates are relatively insensitive to helix length and sequence (Hegg and Fedor 1995). The stability of complexes formed between ribozyme and substrates or products reflects the sum of the free energy of formation of simple helices with the sequences of H1 and H2 and additional stabilizing energy due to extrahelical interactions within and between the loops (Hegg and Fedor 1995; Walter and Burke 1997; Fedor 1999). Because the H1 and H2 sequences determine the magnitude of helical contributions to binding energy, effects of helix sequence changes on dissociation rate and equilibrium constants can be predicted using empirically determined free-energy parameters for simple RNA helix formation (Xia et al. 1998; Mathews et al. 1999). The minimal hairpin ribozyme adopts an extended conformation in the absence of substrate or product RNAs so interdomain docking steps follow binding to complete the assembly process (Walter et al. 1998, 1999).
FIGURE 1.
Kinetic mechanism. The reaction pathway for intermolecular catalysis by minimal hairpin ribozymes includes substrate binding, cleavage, release of 5′ and 3′ cleavage products and the reverse steps of substrate dissociation, ligation, and product binding. Interdomain docking steps follow substrate and product binding steps. H1 and H2 are the intermolecular base-paired helices that ribozymes form with substrate and cleavage product RNAs. (Adapted from Fedor 2000 and reprinted with permission from Elsevier © 2000.)
The intracellular ionic environment, specific and nonspecific RNA binding proteins, and the processes of transcription and translation combine to influence RNA structure and dynamics in vivo, potentially limiting the accuracy with which in vitro reactions can recapitulate RNA assembly reactions in vivo. We have exploited the reversible phosphodiester cleavage reactions mediated by hairpin ribozymes to learn how reaction pathways characterized under standard conditions in vitro translate to the intracellular environment. In previous studies, self-cleaving ribozymes were incorporated into the 3′ untranslated region of the yeast PGK1 mRNA (Donahue and Fedor 1997). Intracellular cleavage kinetics were determined from the acceleration of chimeric ribozyme PGK1 mRNA turnover that resulted from self-cleavage. These experiments revealed that self-cleavage kinetics and the structural requirements for assembly of functional ribozymes are remarkably similar in yeast as in vitro in reactions that approximate intracellular ionic conditions (Donahue and Fedor 1997; Donahue et al. 2000; Fedor et al. 2000; Yadava et al. 2001).
In the current study, hairpin ribozymes and substrate sequences were expressed as separate RNAs so that cleavage depends on substrate recognition, binding, and assembly of a functional complex. We adapted a system previously developed to investigate hammerhead ribozyme activity in which ribozymes and substrates are coexpressed as chimeric U3 snoRNAs (Samarsky et al. 1999). U3 snoRNAs normally mediate precursor rRNA processing in yeast nucleoli (Decatur and Fournier 2003). We investigated how the structures of the intermolecular helices that form between ribozyme and substrate RNAs influence assembly kinetics in vivo. For comparison of results of previous studies that were carried out under standard conditions that include high concentrations of divalent cations, we also determined kinetic parameters in reactions designed to mimic the intracellular ionic environment. Expression of chimeric ribozyme snoRNAs under the control of the GAL1–10 regulatory region allowed us to manipulate intracellular ribozyme concentrations by varying galactose concentrations in yeast-growth media. Quantification of the ribozyme concentration dependence of intermolecular cleavage rates allowed us to calculate second-order rate constants for intracellular ribozyme–substrate complex formation.
RESULTS
We initially tried to investigate ribozyme cleavage of separate RNA substrates using chimeric mRNAs like those that we previously used to evaluate self-cleavage (Donahue and Fedor 1997; Donahue et al. 2000; Yadava et al. 2001). Chimeric PGK1 mRNAs that contained either ribozyme or substrate sequences were coexpressed in yeast under UASGAL control. A chimeric substrate mRNA containing a 42-nucleotide target site was expressed in combination with each of three chimeric ribozyme mRNAs. The chimeric ribozyme variants were designed to recognize chimeric substrate mRNA through complementary base-pairing in intermolecular H2 helices with four base pairs and in intermolecular H1 helices with three (R23), six (R26), or 27 (R227) base pairs. However, none of these ribozyme variants produced detectable amounts of cleavage products in yeast. With chimeric ribozymes that had the potential to form three or six base pairs in H1, the same amounts of uncleaved chimeric substrate mRNAs were found in cells that contained both substrate and ribozyme mRNAs as in cells that expressed substrate mRNAs alone (data not shown). We were able to detect evidence of intermolecular cleavage activity through the depletion of uncleaved substrate only when H1 contained 27 base pairs (data not shown). Even with this extensive complementarity between ribozyme and substrate RNAs, however, intermolecular cleavage activity was too low to allow accurate quantification.
Inefficient intermolecular cleavage could be explained if cytoplasmic chimeric mRNA concentrations are too low to drive ribozyme–substrate complex formation. Hammerhead ribozymes were reported to cleave separate target RNAs almost completely when both sequences were expressed as chimeric U3 snoRNAs in yeast nucleoli (Samarsky et al. 1999). Due to the small volume of the nucleolus and the high abundance of U3 snoRNA at 400 to 1000 copies per cell (Hughes et al. 1987), nucleolar U3 snoRNA concentrations were estimated to be on the order of 1 to 3 μM (Samarsky et al. 1999). PGK1 mRNAs, on the other hand, are expressed at a level of 140 copies per cell (Wang et al. 2002) and are localized in the yeast cytoplasm that has a volume that is 150-fold larger than the volume of the nucleolus (Sherman 1991). Thus, intracellular concentrations of chimeric PGK1 ribozymes and substrate fall in the low nanomolar range. If low cytoplasmic concentrations of chimeric PGK1 mRNAs limit intermolecular cleavage, expressing ribozymes and substrates as chimeric U3 snoRNAs should stimulate cleavage by accelerating complex formation.
Quantification of chimeric snoRNAs self-cleavage kinetics in yeast nuclei
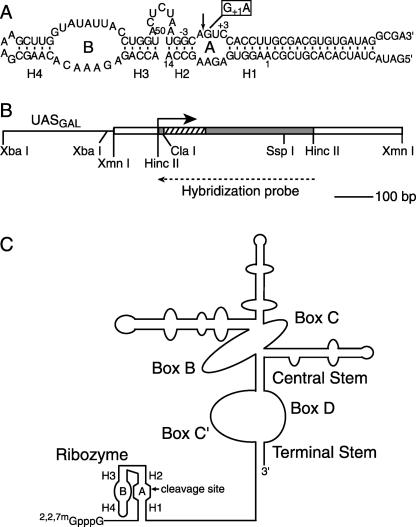
We first evaluated self-cleavage kinetics of chimeric U3 snoRNAs to compare with chimeric PGK1 mRNA self-cleavage rates measured previously (Donahue and Fedor 1997; Donahue et al. 2000; Yadava et al. 2001). Self-cleaving ribozyme coding sequences were substituted for an unstructured region of U3 snoRNA that normally mediates pre-rRNA recognition (Fig. 2 ▶). These chimeric hairpin ribozyme constructions are similar to those previously developed by the Fournier group to examine hammerhead ribozyme activity in yeast (Samarsky et al. 1999). Chimeric U3 genes were fused to GAL1–10 regulatory sequences (Johnston and Davis 1984), and were propagated in yeast using single-copy plasmids (Sikorski and Hieter 1989). Chimeric U3 snoRNAs containing functional ribozymes were compared to chimeric snoRNAs containing an inactivating G+1A mutation to distinguish any effects of the ribozyme insertion on snoRNA expression from effects of self-cleavage. Chimeric HP23, HP26, and HP220 snoRNAs share the same self-cleaving ribozyme sequence (Fig. 2A ▶) except that the length of the H1 helix that forms between 5′ ribozyme and 3′ product RNAs contains three, six, or 20 (shown) base pairs, respectively.
FIGURE 2.
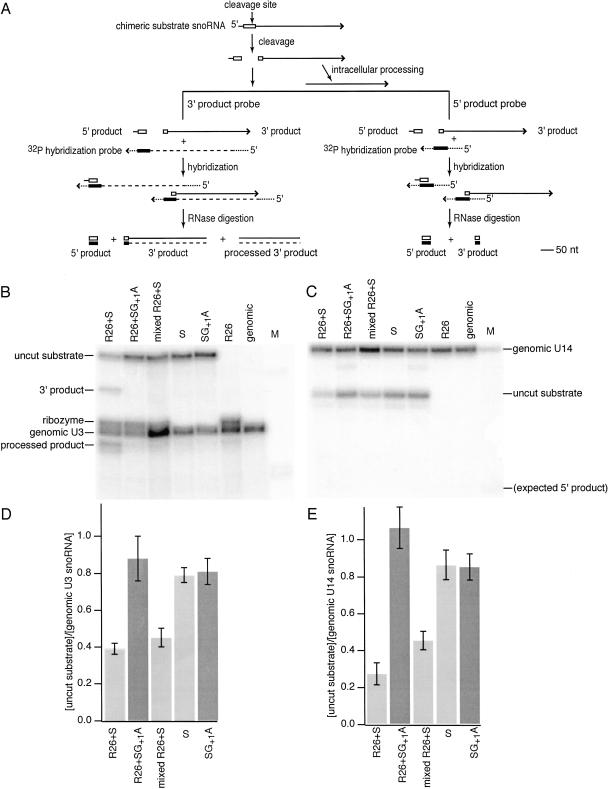
Chimeric self-cleaving snoRNAs. (A) Hairpin ribozyme secondary structure. The essential A and B domains of the minimal hairpin ribozyme are joined by an unstructured oligonucleotide linker. For control experiments, a G+1A mutation (open rectangle) was introduced to inhibit self-cleavage. Base-paired helices H1 through H4 and the conventional numbering system for ribozyme and substrate nucleotide positions are indicated. The variant shown, HP220, has 20 base pairs in H1. Additional self-cleaving variants were examined with three base pairs in H1 (HP23) or six base pairs in H1 (HP26; Donahue et al. 2000). The self-cleaving hairpin ribozyme insertion in the U3 snoRNA gene is the same size in each case, but the 3′ cleavage product sequences flanking the shorter H1 helices are identical rather than complementary to the flanking 5′ ribozyme sequences. (B) Chimeric U3 snoRNA gene. Cassettes encoding self-cleaving hairpin ribozymes (hatched rectangle) were inserted into the 5′ region (shaded rectangle) of the yeast U3 snoRNA gene in place of a U3 sequence that normally participates in precursor rRNA recognition (Samarsky and Fournier 1998). Yeast sequences flanking the U3 coding region are shown as open rectangles. Chimeric snoRNAs were expressed under the control of the GAL1–10 regulatory sequence, UASGAL (solid line). The solid arrow indicates the U3 snoRNA transcription start site. The dashed arrow indicates the sequence of the hybridization probe used to quantify chimeric U3 snoRNAs in RNase protection assays. (C) Chimeric ribozyme–U3 snoRNAs contain hairpin ribozyme sequences (adapted from Samarsky et al. 1999 and reprinted with permission, © 1999 National Academy of Sciences, USA).
Using an established yeast fractionation protocol (Fedor et al. 1988) in combination with RNase protection assays (Fig. 3A ▶), we confirmed that chimeric U3 snoRNAs colocalize with U3 snoRNAs expressed from the unmodified genomic U3 gene, as expected. Chimeric snoRNAs and snoRNAs transcribed from the genomic U3 gene mostly cofractionated with yeast nuclei while unmodified and chimeric PGK1 mRNAs were found primarily in cytoplasmic fractions (data not shown). Thus, consistent with a previous report (Samarsky et al. 1999), insertions of ribozyme sequences into PGK1 mRNA and U3 snoRNA did not perturb intracellular localization.
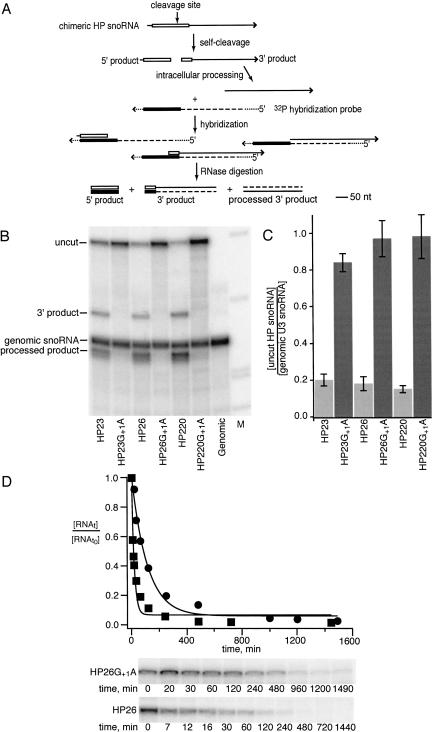
FIGURE 3.
Hairpin ribozyme self-cleavage in nuclei. (A) RNase protection assay of chimeric ribozyme snoRNA self-cleavage. Self-cleavage of the hairpin ribozyme (open rectangle) in U3 RNA (solid line) generates a 140 nt 5′ ribozyme fragment and a 310 nt 3′ cleavage product fragment. Intracellular processing events trim the 3′ cleavage product from the 5′ end to generate shorter processed products (Samarsky et al. 1999). 32P-labeled RNAs hybridized to yeast RNAs in RNase protection assays contain sequences complementary to ribozyme (filled rectangle) and U3 sequences (dashed line) as well as vector sequences (dotted line). (B) RNase protection assays yield fragments corresponding to uncut chimeric snoRNA, 3′ cleavage products and 3′ cleavage products that have undergone intracellular processing as well as sequences flanking the ribozyme insertion site that are derived from the U3 snoRNA that is expressed from the genomic U3 gene. Any small fragments corresponding to 5′ cleavage products are expected to be lost during electrophoresis under these conditions. As expected, chimeric snoRNAs containing ribozymes inactivated by a G+1A mutation yield fragments that correspond to uncut chimeric snoRNA and genomic U3 snoRNA only. (C) The levels of uncut chimeric snoRNAs with functional ribozyme (light shading) or inactive ribozyme (dark shading) insertions are compared to the abundance of unmodified snoRNA expressed from the genomic U3 gene. (D) Kinetics of chimeric snoRNA self-cleavage in yeast. Uncut chimeric snoRNA was quantified at various times after transcription was inhibited by addition of glucose to the growth media for HP26 snoRNA (squares) and HP26G+1A snoRNA (circles). Intracellular decay rates were computed from the fit to [uncut RNAt]/[uncut RNAto = e−kt after normalization to the abundance of genomic U3 RNA. The HP26G+1A snoRNA decay rate reflects turnover of chimeric snoRNA through the endogenous snoRNA degradation pathway. The rate of HP26 snoRNA decay reflects the sum of the intrinsic degradation rate and the intracellular self-cleavage rate (Donahue and Fedor 1997). The self-cleavage rate is calculated from the difference between the HP26 and HP26G+1A decay rates, as described in the text.
RNase protection assays were carried out to examine chimeric snoRNA self-cleavage activity in yeast (Fig. 3 ▶). As expected, fragments corresponding in size to 3′ cleavage products were detected in RNA from yeast expressing self-cleaving chimeric snoRNAs but not in RNA from yeast expressing mutationally inactivated ribozymes (Fig. 3B ▶). Additional chimeric snoRNA fragments were detected that likely result from intracellular processing of 3′ cleavage product RNAs, as observed in previous analyses of chimeric hammerhead ribozyme snoRNAs (Samarsky et al. 1999). Chimeric snoRNAs with inactivating G+1A mutations were nearly as abundant as U3 snoRNA expressed from the genomic U3 gene (Fig. 3B,C ▶). Thus, insertion of ribozyme sequences appeared to have little effect on U3 snoRNA transcription or stability in the absence of self-cleavage. Mutationally inactivated chimeric snoRNAs were four- to fivefold more abundant than their functional counterparts (Fig. 3B,C ▶), evidence that functional chimeric snoRNAs were depleted through self-cleavage, as expected. Yeast expressing chimeric snoRNAs with HP23, HP26, and HP220 insertions contained similar amounts of uncleaved chimeric snoRNAs (Fig. 3B,C ▶). Thus, self-cleaving ribozymes with three, six, or 20 base pairs in H1 appeared to be equally functional in yeast.
The relative depletion of chimeric snoRNAs with functional ribozyme insertions (Fig. 3B,C ▶) shows that self-cleavage occurs within the intracellular lifetime of a chimeric U3 snoRNA. To evaluate self-cleavage kinetics quantitatively, decay kinetics of chimeric snoRNAs with self-cleaving and inactive ribozyme insertions were compared by measuring the time course of chimeric snoRNA disappearance after glucose was added to growth media to inhibit transcription (Donahue and Fedor 1997). HP26G+1A, a chimeric snoRNA with an inactive ribozyme insertion, decayed at a rate of 0.0086 min−1 (circles, Fig. 3D ▶). This decay rate reflects the turnover of this inactive chimeric snoRNA through the endogenous RNA degradation pathway. Similar time course experiments with HP23G+1A and HP220G+1A snoRNAs gave very similar intrinsic degradation rates of 0.0083 min−1 and 0.0092 min−1, respectively (data not shown). These intrinsic degradation rates agree well with the rate of 0.01 min−1 measured for degradation of unmodified U3 snoRNA (Hughes and Ares 1991), confirmation that insertion of ribozyme sequences has little effect on U3 snoRNA expression or stability in the absence of self-cleavage. As expected, chimeric snoRNAs with self-cleaving ribozyme insertions decayed faster, reflecting the combined contributions to decay of self-cleavage and degradation through the endogenous snoRNA turnover pathway. HP26 snoRNA decayed at a rate of 0.068 min−1 (squares, Fig. 3D ▶). The difference between the decay rates self-cleaving HP26 snoRNA of 0.066 ± 0.005 min−1 and inactive HP26G+1A snoRNA of 0.0087 ± 0.0002 min−1 that were obtained by averaging the results of multiple experiments gave an intracellular self-cleavage rate of 0.057 ± 0.005 min−1.
Once intrinsic degradation rates are known for chimeric snoRNAs, intracellular self-cleavage rates also can be calculated from the relative abundance of self-cleaving and inactive chimeric snoRNAs at steady state (Fig. 3C ▶; Donahue and Fedor 1997). Using this approach, an intracellular self-cleavage rate of 0.038 min−1 can be calculated from the average ratio of HP26G+1A and uncut HP26 snoRNAs of 5.4 at steady state (Fig. 3C ▶) and the intrinsic degradation rate of 0.0087 min−1 measured for HP26G+1A snoRNA (Fig. 3D ▶). A rate of 0.05 min−1 can be obtained for HP26 snoRNA self-cleavage in yeast by combining the results of steady-state experiments with the kcleavage value of 0.057 min−1 that was calculated from intracellular decay kinetics. Calculations based on steady-state abundance and decay kinetics gave similar rates of 0.04 min−1 and 0.06 min−1 for intracellular HP23 and HP220 snoRNA self-cleavage, respectively.
Chimeric ribozymes cleave separate RNA targets in yeast
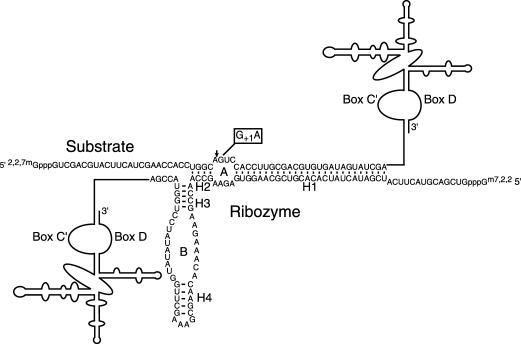
Chimeric snoRNAs with insertions of either ribozyme or target RNA sequences were coexpressed in yeast to evaluate intermolecular cleavage activity that requires assembly of a functional ribozyme–substrate complex (Fig. 4 ▶). When chimeric substrate snoRNA was coexpressed with chimeric R26 snoRNA, the abundance of the chimeric substrate RNA was reduced and fragments corresponding to 3′ cleavage products were detected (Fig. 5 ▶). As in the case of chimeric self-cleaving ribozymes (Fig. 3B ▶), smaller fragments that likely result from intracellular processing of 3′ cleavage product RNAs also were observed (Fig. 5B ▶). Even in RNase protection assays optimized for detection of 5′ cleavage product RNA (Fig. 5A ▶, right); however, no 5′ cleavage products were found (Fig. 5C ▶), suggesting that 5′ product RNAs degrade rapidly after cleavage. The same amounts of chimeric substrate snoRNA were detected in yeast expressing chimeric R26 snoRNA and the noncleavable substrate RNA with an inactivating G+1A mutation as in yeast expressing the cleavable substrate snoRNA alone (Fig. 5D,E ▶). The results of these control experiments suggest that the effect of chimeric ribozyme snoRNAs on substrate RNA levels results exclusively from ribozyme-mediated cleavage. Furthermore, substrate RNA was not depleted when yeast expressing only chimeric ribozymes were combined with yeast expressing only chimeric substrate RNAs before the yeast RNA was extracted and analyzed. This control experiment demonstrates that no cleavage occurs during RNA extraction or RNase protection assays. Finally, we detected no cleavage when chimeric ribozyme mRNAs that should localize in the cytoplasm were coexpressed with chimeric substrate snoRNA that should localize in the nucleus or vice versa. Taken together, these results demonstrate that chimeric hairpin ribozyme snoRNAs can mediate cleavage of separate substrate RNAs in yeast nuclei.
FIGURE 4.
Chimeric ribozyme–substrate complex. Chimeric snoRNAs that contained either ribozyme or substrate sequences were coexpressed in yeast to evaluate intermolecular cleavage activity. Additional ribozyme and substrate variants were examined that formed H1 helices ranging from three base pairs (R23) to six base pairs (R26) in length and with base pair substitutions that enhanced the stability of H2 (Fig. 7 ▶). Substrate analogs with inactivating G+1A mutations were used to distinguish effects of cleavage from effects of complex formation on chimeric substrate snoRNA expression.
FIGURE 5.
Hairpin ribozyme cleavage of separate RNA substrates in yeast. (A) RNase protection assays of chimeric substrates were carried out with 32P-labeled hybridization probes designed to detect 5′ (right) or 3′ (left) cleavage products. Notation is the same as in Figure 3 ▶. (B) RNase protection assays with the 3′ product probe and RNA obtained from yeast at steady state yield fragments corresponding to uncut chimeric substrate snoRNA (327 nucleotides), 3′ cleavage products (298 nt) and 3′ cleavage products that have undergone intracellular processing (262 nt) as well as ribozyme sequences (272 nt) and sequences flanking the ribozyme insertion site derived from U3 snoRNA expressed from the genomic U3 gene (268 nt). A 29-nt protected fragment corresponding to the 5′ cleavage product is expected to be lost during electrophoresis under these conditions. As expected, chimeric snoRNAs containing substrate insertions inactivated by a G+1A mutation yield fragments that correspond to uncut substrate RNA and genomic U3 snoRNA only. The lane labeled “mixed R26 + S” represents results of a control experiment in which equal amounts of yeast that expressed only chimeric substrate RNA were mixed with yeast that expressed only chimeric ribozyme RNA before the RNA was extracted. (C) RNase protection assays with the 5′ product probe and RNA obtained from yeast at steady-state yield fragments corresponding to uncut chimeric substrate snoRNA (47 nt) and to the U14 snoRNA used for normalization (60 nt). No 29-nt fragment corresponding to the size expected for 5′cleavage products was detected. (D) The levels of uncut chimeric snoRNAs with functional ribozyme (light shading) or inactive ribozyme (dark shading) insertions are compared to the abundance of unmodified snoRNA expressed from the genomic U3 gene. (E) The levels of uncut chimeric snoRNAs with functional ribozyme (light bars) or inactive ribozyme (dark shading) insertions are compared to the abundance of unmodified snoRNA expressed from the genomic U14 gene.
An intrinsic degradation rate of 0.0062 ± 0.0004 min−1 was calculated from the abundance of substrate snoRNA at steady state in yeast expressing chimeric substrate alone. Similar values for the intrinsic substrate snoRNA degradation rate were calculated from the abundance of mutation-ally inactivated substrate at steady state in the presence or absence of ribozyme RNA and from the time course of substrate snoRNA disappearance following transcription inhibition. The difference between the substrate snoRNA decay rate of 0.0145 ± 0.006 min−1 measured when chimeric ribozyme and substrate snoRNAs are coexpressed and the intrinsic degradation rate of substrate snoRNA in the absence of ribozyme gave a rate of 0.0083 ± 0.001 min−1 for R26 snoRNA cleavage of substrate snoRNA in yeast.
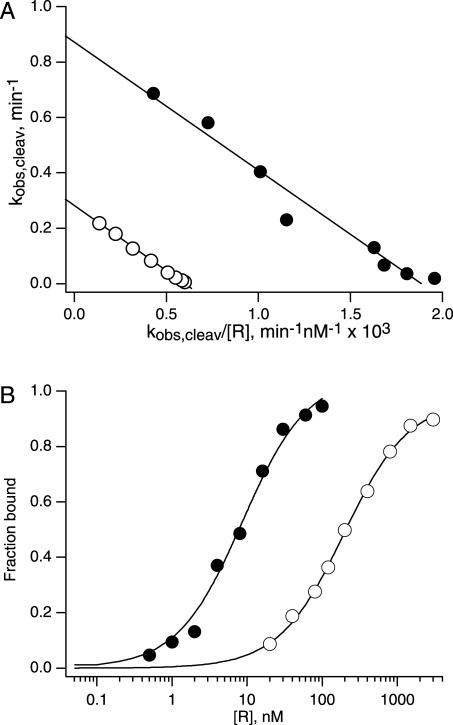
This intermolecular cleavage rate is sixfold below the rate of 0.05 min−1 measured for self-cleavage of chimeric HP26 snoRNA in yeast. Under standard reaction conditions in vitro that include 10 mM MgCl2, a ribozyme with the same sequence as the ribozyme insertion in R26 snoRNA cleaves an oligonucleotide substrate with a much higher cleavage rate constant of 0.87 min−1 (Fig. 6A ▶, filled circles). Even under less favorable reaction conditions that more closely approximate the intracellular ionic environment, the same ribozyme displays a cleavage rate constant of 0.28 min−1 (Fig. 6A ▶, open circles). This rate still is more than 30-fold faster than the intermolecular cleavage rate measured in yeast. We next set out to evaluate what feature(s) of the intermolecular cleavage reaction pathway limits intermolecular cleavage activity in yeast.
FIGURE 6.
Influence of ionic conditions on cleavage kinetics and ribozyme–substrate complex stability. (A) Eadie-Hofstee plots of the R2 concentration dependence of S26 cleavage rates gave a kcleav value of 0.87 min−1 in reactions with 10 mM MgCl2 (filled circles) and a kcleav value of 0.28 min−1 in reactions with 2 mM MgCl2, 150 mM NaCl (open circles). Reactions under both salt conditions exhibited the same KMS value of 460 nM (Table 2 ▶). (B) Equilibrium dissociation constants were measured for the R2 • S26dA complex using nondenaturing gel electrophoresis. The fit to Fraction bound = [R]/([R] + KdS) gave a KdS value of 8.6 nM in buffer with 10 mM MgCl2 (filled circles) and a KdS value of 190 nM in buffer with 2 mM MgCl2, 150 mM NaCl (open circles).
Influence of intermolecular helix structure on cleavage activity
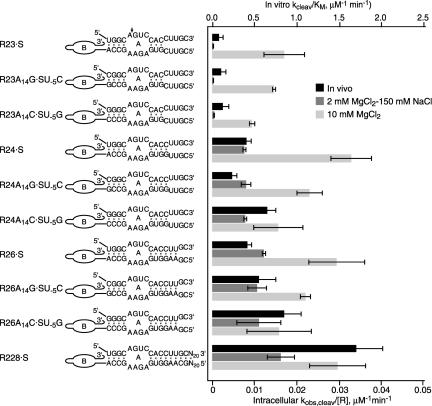
The H2 helix forms within a contiguous RNA sequence in a self-cleaving ribozyme (Fig. 2A ▶) while H2 is stabilized by just four intermolecular base pairs in a ribozyme–substrate complex (Fig. 4 ▶). Ribozyme variants with different base pairs substituted for the U-5:A14 pair in H2 were compared to learn whether enhanced H2 stability might promote intermolecular cleavage by favoring assembly of the functional ribozyme–substrate complex (Fig. 7 ▶; Table 1 ▶). A simple RNA duplex with the same sequence as the H2 helix that forms between R23, R24, or R26 and substrate snoRNA has a calculated stability of -7.2 kcal/mole (Table 1 ▶; Mathews et al. 1999). Variant ribozymes that have H2 helices with calculated stabilities of −7.8 kcal/mole and −8.4 kcal/mole exhibited similar intermolecular cleavage kinetics as R23, R24, or R26 in vitro in reactions with 10 mM MgCl2, or with 2 mM MgCl2 and 150 mM NaCl (Table 2 ▶) and in vivo (Fig. 7 ▶).
FIGURE 7.
Intermolecular cleavage activity of hairpin ribozymes. Intracellular cleavage rates were calculated from the abundance of uncleaved chimeric snoRNA at steady state by assuming an intracellular ribozyme concentration of 1 μM (filled bars). Second-order rate constants (k2/ KMS) were measured for ribozyme cleavage of oligonucleotide substrates that form the same H1 sequences in buffers with 10 mM MgCl2 (open bars) or with 2 mM MgCl2, 150 mM NaCl (shaded bars) except that in vitro kinetic data compared with the intracellular R228•S cleavage rate were obtained with R2 and S28 RNAs that form eight base pairs rather than 28 base pairs in H1. Note that the scales of the X-axes used to plot in vitro and in vivo data differ by 50-fold.
TABLE 1.
Kinetic and equilibrium parameters for ribozyme complex formation under different salt conditions
| 10 mM MgCl2 | 2 mM MgCl2−150 mM NaCl | ||||
| R•S | Δ G30°C,calcH1a kcal/mole | ΔG30°C,calcH2a kcal/mole | Δ G30°C,measE•SdAb kcal/mole | Δ G30°C,measE•SdAb kcal/mole | Δ G30°C,measE•S kcal/mole |
| R2•S23 | −1.4 | −7.2 | — | — | |
| R2A14G•S23U−5C | −1.4 | −7.8 | — | — | |
| R2A14G•S23U−55C | −1.4 | −8.4 | −7.9 | — | −7.0c |
| R2•S24 | −3.7 | −7.2 | −9.4 | — | −8.1d |
| R2A14G•S24U−55C | −3.7 | −7.8 | −10 | — | |
| R2A14G•S24U−55C G | −3.7 | −8.4 | −10 | — | |
| R2•S25 | −5.7 | −7.2 | −11 | −8.3 | |
| R2•S26 | −7.0 | −7.2 | −12 | −9.3 | |
| R2A14G•S26U−55C | −7.0 | −7.8 | — | — | |
| R2A14G•S26U−55C | −7.0 | −8.4 | — | −10 | |
| R2•S28 | −12 | −7.2 | — | — | |
| R2•S228 | −56 | −7.2 | — | — | |
aCalculation based on empirically determined free energy parameters for base-paired RNA helices (Mathews et al. 1999).
bCalculated from ΔGE•S,30°C,meas = −RTln(1/KdS), where R is the gas constant and T is the temperature in Kelvin, with equilibrium dissociation constants measured using nondenaturing gel electrophoresis. Substrate analogs used for nondenaturing gel electrophoresis experiments contained a deoxyadenosine substitution at the −1 position (A−1dA) to prevent cleavage.
cCalculated from the KMS value obtained from the ribozyme concentration dependence of cleavage rates. For this substrate, KMS is equivalent to an equilibrium dissociation constant, or KdS, because substrate dissociation is much faster than cleavage.
dCalculated as above from equilibrium dissociation constants obtained from the ratio of dissociation and binding rate constants. The binding rate constant was calculated from kcleav/ KMS values (Table 2 ▶) and a koff value of 0.53 ± 0.16 min−1 was measured using pulse-chase experiments.
TABLE 2.
Kinetic parameters for ribozyme cleavage of oligonucleotide substrates under different salt conditionsa
| 10 mM MgCl2 | 2 mM MgCl2−150 mM NaCl | ||||||
| R•S | kcleavage min−1 | KMS μM | kcleavage/KMS μM−1 min−1 | kcleavage min−1 | KMS μM | kcleavage/KMS μM−1 min−1 | koffSb min−1 |
| R2•S23 | 1 ± 0.2 | 1.2 ± 0.3 | 0.9 ± 0.2 | 0.11 ± 0.01 | 11 ± 5.0 | 0.011 ± 0.004 | >1 |
| R2A14G•S23U−55C G | 0.63 ± 0.07 | 0.9 ± 0.1 | 0.73 ± 0.02 | 0.18 ± 0.01 | 15 ± 2.0 | 0.012 ± 0.002 | >2 |
| R2A14G•S23U−55C G | 0.51 ± 0.05 | 0.9 ± 0.2 | 0.47 ± 0.03 | 0.19 ± 0.03 | 8.2 ± 0.7 | 0.022 ± 0.005 | >2 |
| R2•S24 | 0.78 ± 0.04 | 0.4 ± 0.1 | 1.6 ± 0.2 | 0.31 ± 0.01 | 0.83 ± 0.02 | 0.38 ± 0.03 | 0.5 ± 0.2 |
| R2A14G•S24U−55C G | 0.79 ± 0.02 | 0.68 ± 0.06 | 1.2 ± 0.15 | 0.24 ± 0.02 | 0.63 ± 0.1 | 0.40 ± 0.06 | — |
| R2A14G•S24U−55C G | 0.5 ± 0.1 | 0.6 ± 0.2 | 0.8 ± 0.3 | 0.25 ± 0.02 | 0.77 ± 0.02 | 0.39 ± 0.02 | 0.11 ± 0.05 |
| R2•S26 | 0.76 ± 0.06 | 0.53 ± 0.07 | 1.5 ± 0.3 | 0.26 ± 0.02 | 0.43 ± 0.02 | 0.6 ± 0.2 | <0.1 |
| R2A14G•S26U−55C G | 0.68 ± 0.07 | 0.61 ± 0.09 | 1.1 ± 0.06 | 0.26 ± 0.01 | 0.49 ± 0.06 | 0.5 ± 0.1 | <0.1 |
| R2A14G•S26U−55C G | 0.47 ± 0.05 | 0.70 ± 0.03 | 0.8 ± 0.4 | 0.25 ± 0.02 | 0.53 ± 0.22 | 0.6 ± 0.3 | — |
| R2•S28 | 0.76 ± 0.01 | 0.6 ± 0.1 | 1.5 ± 0.3 | 0.27 ± 0.02 | 0.33 ± 0.03 | 0.8 ± 0.1 | — |
aMean and range of values obtained from two experiments.
bMeasured using pulse chase experiments as described previously (Hegg and Fedor 1995).
H1 is the intermolecular helix that forms between the ribozyme and 3′ cleavage product RNAs (Fig. 4 ▶). Previous studies have shown that the structure of the H1 helix can influence hairpin ribozyme cleavage activity in vivo and in vitro (Hegg and Fedor 1995; Fedor 1999; Donahue et al. 2000; Yadava et al. 2001). Short H1 helices can be too weak to support assembly of a functional ribozyme, and long H1 helices can be stable enough to inhibit cleavage by promoting religation of bound cleavage products. The influence of H1 stability on self-cleavage activity has been shown to vary with temperature and ionic conditions (Donahue et al. 2000; Yadava et al. 2001). To learn more about how H1 stability influences intermolecular cleavage activity, a series of chimeric ribozyme variants were examined that associate with chimeric substrate RNA to form H1 helices that range in length from three to 28 base pairs (Fig. 7 ▶). Calculations based on empirically determined free energy parameters (Mathews et al. 1999) give ΔG°30°C values ranging from −1.4 kcal/mole to −56 kcal/mole for simple RNA duplexes with the same sequences (Table 1 ▶).
Intracellular cleavage rates increased about sixfold as H1 length increased from three to six base pairs (Fig. 7 ▶). The fastest intracellular cleavage rate was observed for R228 snoRNA, in which H1 contains 28 base pairs. Under standard conditions in vitro that include 10 mM MgCl2, however, ribozymes that form H1 helices ranging from three to eight base pairs in length displayed similar cleavage rate constants and KMS values changed less than twofold (Fig. 7 ▶; Table 2 ▶). Intermolecular cleavage is significantly faster in reactions with 2 mM MgCl2 and 150 mM NaCl than in yeast (Fig. 7 ▶; Table 2 ▶). (Note that axis scales in Fig. 7 ▶ for in vitro and intracellular reactions differ by 50-fold to allow comparison of in vitro cleavage kinetics with intracellular cleavage kinetics, which are much slower.) Nonetheless, cleavage kinetics do show significant dependence on H1 sequence under salt conditions that more closely approximate intracellular ionic conditions (Fig. 7 ▶). In reactions with 2 mM MgCl2 and 150 mM NaCl, cleavage rate constants increase less than twofold when the number of base pairs in H1 increases from three to eight, but KMS values decrease more than 30-fold (Table 2 ▶).
In these single turnover reactions with ribozyme in large excess over substrate RNA, KMS reflects elemental rate constants according to KMS = (koffS + kcleavage)/ konS (Hegg and Fedor 1995). When rate constants for substrate dissociation are fast relative to cleavage rate constants, KMS is virtually the same as an equilibrium dissociation constant, KdS. When substrate dissociation rates are slow relative to cleavage rate constants, however, KMS is not equivalent to KdS, and the ratio of kcleavage and KMS values gives konS, the substrate binding rate constant. Hairpin ribozymes have been shown to display this Briggs-Haldane type of kinetic mechanism under standard reaction conditions that include 10 mM MgCl2 at 25°C (Hegg and Fedor 1995).
Previous analyses of the hairpin ribozyme kinetic mechanism have been carried out exclusively in reactions with high concentrations of divalent cations (Hegg and Fedor 1995; Esteban et al. 1997; Walter and Burke 1997). To compare the kinetic mechanism in reactions with 10 mM MgCl2 to reactions that more closely approximate intracellular ionic conditions, substrate dissociation constants were compared to cleavage rate constants using pulse chase experiments as described previously (Hegg and Fedor 1995). After dilution of ribozyme–substrate complexes into reaction buffer with 10 mM MgCl2, all substrates were found to cleave completely before dissociation could occur (data not shown), as observed previously (Hegg and Fedor 1995). This result indicates that cleavage is faster than substrate S dissociation in reactions with 10 mM MgCl2 and that KM is not the same as KdS.
In contrast, ribozymes with different H1 sequences displayed different kinetic mechanisms in reactions with 2 mM MgCl2 and 150 mM NaCl. Ribozyme complexes with H1 helices that have just three base pairs displayed a Michaelis-Menten type of kinetic mechanism. These variants with three base pairs in H1 dissociated much more rapidly than they cleaved, indicating that KMS values are equivalent to equilibrium dissociation constants in 2 mM MgCl2 and 150 mM NaCl (Table 2 ▶). Ribozyme complexes with more than four base pairs in H1 dissociated more slowly than they cleaved in reactions with 2 mM MgCl2 and 150 mM NaCl, displaying the same Briggs-Haldane type of kinetic mechanism that is characteristic of all hairpin ribozyme reactions in high concentrations of divalent cations. Ribozyme–substrate complexes with four base pairs in H1 displayed intermediate kinetic behavior, by cleaving and dissociating at similar rates under these ionic conditions.
Salt conditions also influenced equilibrium dissociation constants measured for ribozyme complexes with noncleavable substrate analogs. For example, the equilibrium dissociation constant measured for the R2•S26A−1dA complex with six base pairs in H1 was 20-fold higher in 2 mM MgCl2 and 150 mM NaCl than in 10 mM MgCl2 (Fig. 6B ▶). On average, hairpin ribozyme complexes with noncleavable substrate analogs were less stable in 2 mM MgCl2 and 150 mM NaCl than in 10 mM MgCl2 by about 1 to 3 kcal/mole (Fig. 6B ▶; Table 1 ▶).
Intermolecular cleavage rates depend on intracellular ribozyme concentrations
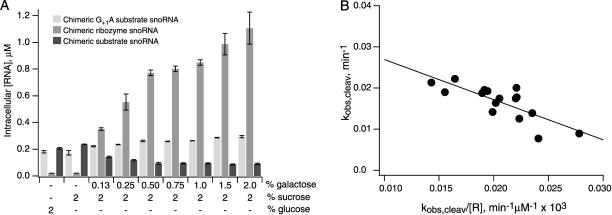
If ribozyme–substrate complex formation is a rate-determining step in the intermolecular cleavage pathway in vivo, cleavage rates are expected to increase with increasing concentrations of chimeric snoRNAs. Chimeric ribozyme snoRNA genes were fused to the GAL1–10 upstream activation sequence so that intracellular concentrations of ribozyme snoRNAs could be modulated by changing the concentration of galactose in yeast growth media (Fig. 8A ▶). Chimeric substrate snoRNAs expressed from the unmodified U3 promoter were not subject to galactose induction. When substrates with an inactivating G+1A mutation were coexpressed with ribozymes under the same growth conditions, intracellular concentrations of the noncleavable chimeric substrate RNAs remained virtually unchanged (Fig. 8A ▶). Thus, changes in galactose concentrations did not affect the abundance of chimeric substrates expressed from an unmodified U3 snoRNA promoter. These control experiments confirm that changes in the levels of chimeric substrates that are observed when ribozymes are coexpressed with cleavable substrate RNAs result from ribozyme-mediated cleavage and not from ribozyme–substrate complex formation alone. When chimeric ribozyme snoRNAs are in excess over chimeric substrate concentrations, intracellular reactions approximate pseudo-first-order conditions, simplifying quantification of the ribozyme concentration dependence of intracellular cleavage kinetics.
FIGURE 8.
Ribozyme concentration dependence of intermolecular cleavage in vivo. (A) Intracellular concentrations of chimeric R228, substrate, and noncleavable substrate snoRNAs with a G+1A mutation in yeast grown in media with 2% glucose or with 2% sucrose and galactose concentrations ranging from 0% to 2%. (B) An Eadie-Hofstee plot of the chimeric R228 snoRNA concentration dependence of rates of chimeric substrate snoRNA cleavage in yeast nuclei gives a kcleav value of 0.037 ± 0.004 min−1 and a KMS value of 1.0 ± 0.2 μM.
At intracellular R26 snoRNA concentrations ranging from 0.3 to 1.2 μM, observed cleavage rates increased slightly from 0.009 to 0.016 min−1, consistent with the idea that complex formation is partially rate determining (not shown). Accurate kinetic parameters could not be determined for intracellular cleavage reactions with R26 snoRNAs, however, due to the small range of observed cleavage rates and the relatively large magnitude of experimental error.
At intracellular R228 snoRNA concentrations ranging from 0.3 to 1.5 μM, observed cleavage rates varied between 0.009 and 0.024 min−1. The narrow range of intracellular cleavage rates that we were able to observe limits the accuracy of kinetic parameters that can be calculated from these rate measurements. However, an Eadie-Hofstee type plot of data obtained from multiple experiments gives an approximate kcleav value of 0.037± 0.004 min−1 and a KMS value of 1.0 ± 0.2 μM for intracellular substrate cleavage by R228 (Fig. 8B ▶). Dissociation of a substrate that forms 28 base pairs in the H1 helix is likely to be much slower than cleavage (Donahue et al. 2000). By assuming that R228-mediated cleavage occurs through a Briggs-Haldane type of kinetic mechanism in vivo, an intracellular substrate binding rate constant of 4 × 104 M−1 min−1 can be calculated from the ratio of kcleav and KMS values.
DISCUSSION
Hairpin ribozymes display similar self-cleavage kinetics in nuclear and cytoplasmic compartments
We began this study by comparing self-cleavage kinetics for chimeric snoRNAs and mRNAs in nuclear and cytoplasmic compartments to learn how intracellular localization influences the hairpin ribozyme reaction pathway. Chimeric hairpin ribozyme snoRNA self-cleavage rates of about 0.05 min−1 were similar to self-cleavage rates measured for the same hairpin ribozyme sequences in vitro in reactions with 2 mM MgCl2 and 150 mM NaCl at 30°C (Donahue et al. 2000). They were about threefold slower than rates ranging from 0.13 to 0.17 min−1 that previously were measured for self-cleavage of the same ribozyme sequences in the context of chimeric PGK1 mRNAs in yeast (Donahue et al. 2000). The similarity of chimeric mRNA and snoRNA self-cleavage kinetics suggests that ribozymes self-cleave through similar reaction pathways in nuclear and cytoplasmic compartments, and that the nuclear environment is not significantly more or less favorable than the cytoplasm for assembly of functional ribozymes.
Hairpin ribozyme self-cleavage rates were somewhat slower than the rate of 0.08 min−1 reported for self-cleavage of chimeric hammerhead U3 snoRNAs in yeast (Samarsky et al. 1999). In contrast to the relatively efficient cleavage reported for chimeric hammerhead ribozyme snoRNAs, little self-cleavage activity has been detected for minimal hammerhead ribozyme insertions in chimeric PGK1 mRNAs (C.P. Donahue and M.J. Fedor, unpubl.) or in a variety of other reporter genes in yeast (Castanotto et al. 1998). Self-cleaving hammerhead ribozymes fused to secreted alkaline phosphatase mRNAs also showed little activity in cultured human embryonic kidney cells unless the minimal catalytic domain was extended to include sequences adjacent to natural hammerhead domains with the potential to form stabilizing tertiary interactions (Khvorova et al. 2003). These differences in hammerhead ribozyme activity in different systems remain to be explained.
H1 helix structure influences intermolecular cleavage activity in yeast and in vitro when reactions mimic intracellular salt conditions
When chimeric ribozymes and substrates both were expressed at maximum levels under fully inducing conditions in yeast, intermolecular cleavage rates varied over a 20-fold range depending on the length and sequence of the intermolecular H1 and H2 helices (Fig. 7 ▶). In self-cleaving ribozymes, H2 forms within a contiguous RNA sequence as part of a bulged two-way helical junction (Fig. 2A ▶). In the intermolecular configuration, H2 forms between separate ribozyme and substrate or 5′ cleavage product RNAs where it is stabilized by just four base pairs (Fig. 4 ▶). The length of H2 is constrained by the requirement to align loops A and B in the folded tertiary structure (Barroso-del Jesus et al. 1999). We considered the possibility that incomplete annealing of H2 in a ribozyme–substrate complex is responsible for the observation that intermolecular cleavage is slower than self-cleavage in vivo. Base pair substitutions that were expected to increase the stability of H2 by 1.2 kcal/mole (Table 1 ▶) did seem to enhance intermolecular cleavage activity somewhat in reactions with 2 mM MgCl2 and 150 mM NaCl (Table 2 ▶). This change in the free energy of H2 formation corresponds to a sevenfold difference in the equilibrium between open and base-paired states. However, stabilizing base pair substitutions enhanced intermolecular cleavage rates by no more than twofold, and only in ribozyme complexes with short, weak H1 helices (Table 2 ▶). When ribozyme variants had four or more base pairs in H1, changes in H2 stability had no detectable effect on cleavage kinetics in vitro or in yeast (Fig. 7 ▶; Table 2 ▶). Thus, H2 formation does not seem to limit intermolecular cleavage activity in vitro or in vivo, although stabilization of H2 could enhance activity by promoting binding of low affinity substrates.
In contrast to the minor influence of H2 sequence on cleavage kinetics, the length of the H1 helix did influence cleavage activity under certain reaction conditions. Extending H1 from three to eight base pairs is expected to decrease the free energy of H1 formation by 10.6 kcal/mole (Table 1 ▶). Changes in the length and stability of H1 had little effect on kinetic parameters of intermolecular cleavage reactions with 10 mM MgCl2 (Fig. 7 ▶; Table 2 ▶), and ribozyme variants that formed as few as three base pairs in H1 exhibited maximum activity. In reactions with 2 mM MgCl2 and 150 mM NaCl, however, kcleav/ KMS values increased more than 70-fold as the number of base pairs in H1 increased from three to eight (Fig. 7 ▶; Table 2 ▶). Most of this increase in catalytic efficiency reflects changes in KMS values, which decreased almost 50-fold.
Previous studies of the hairpin ribozyme kinetic mechanism under standard conditions showed that cleavage is much faster than substrate dissociation (Hegg and Fedor 1995). In reactions with 10 mM MgCl2, virtually all substrates that bind go on to cleave, regardless of substrate size. In this Briggs-Haldane type of kinetic mechanism, KMS is related to elemental rate constants for cleavage and substrate binding according to KMS ≈ kcleav/ konS, and kcleav/ KMS values represent rate constants for ribozyme–substrate complex formation. In contrast, ribozyme complexes with three base pairs in H1 dissociate much more rapidly than they cleave in reactions with 2 mM MgCl2 and 150 mM NaCl (Table 2 ▶). In this Michaelis-Menten type of kinetic mechanism, KMS is related to elemental rate constants for substrate dissociation and binding according to KMS ≈ koffS/ kon S and KMS values are equivalent to equilibrium dissociation constants. Ribozyme complexes with four base pairs in H1 partitioned almost equally between cleavage and dissociation. When H1 contains six or more base pairs, virtually all bound substrate cleaves before it can dissociate and cleavage occurs through the Briggs-Haldane type of kinetic mechanism that is characteristic of all ribozyme variants in reactions with 10 mM MgCl2. For ribozyme variants with six or more base pairs in H1, kcleav/KMS values represent rate constants for substrate binding. For variants with six or more base pairs in H1, kinetic parameters measured in reactions with 2 mM MgCl2 and 150 mM NaCl were virtually the same as those measured under standard conditions with 10 mM MgCl2. Thus, the major difference between in vitro cleavage reactions in 2 mM MgCl2 and 150 mM NaCl and reactions under standard conditions with 10 mM MgCl2 seems to be the extent of complementarity required to promote stable substrate binding.
Cleavage efficiency was 10- to 100-fold lower in vivo than in vitro for similar ribozyme variants. However, intracellular reactions exhibited a similar dependence on H1 structure as in vitro reactions with 2 mM MgCl2 and 150 mM NaCl (Fig. 7 ▶). Little cleavage activity was observed in yeast for ribozymes and substrates with the potential to form weak H1 helices with just three base pairs (Fig. 7 ▶). In vivo, intermolecular cleavage activity increased more than 10-fold as the number of base pairs in H1 increased from three to 28 base pairs. On the other hand, self-cleaving ribozymes with just three base pairs in H1 are no less active than self-cleaving ribozymes with six or 20 base pairs (Fig. 3B,C ▶), evidence that H1 helices with only three base pairs are stable enough to support assembly of fully functional ribozymes. Therefore, the effects of H1 structure on intermolecular cleavage activity in yeast likely reflect an influence of H1 structure on ribozyme–substrate complex formation rather than cleavage of bound substrate. A larger extent of complementarity seems to be required to drive ribozyme–substrate complex formation in vivo and in reactions that approximate the intracellular ionic environment compared to standard conditions with 10 mM MgCl2.
Kinetic parameters can be calculated from the ribozyme concentration dependence of observed cleavage rates in yeast
The chimeric U3 ribozyme self-cleavage rate of 0.05 min−1 may define an upper limit for the rate that can be expected for intermolecular cleavage within a fully assembled ribozyme–substrate complex. The fastest rate observed for intermolecular cleavage was 0.023 min−1 (Fig. 8B ▶), a rate that is less than twofold below rates measured for self-cleavage of ribozymes that do not depend on a binding step to assemble a fully functional RNA structure. The cleavage rate constant of 0.037 min−1 that could be extrapolated from intermolecular cleavage rates observed in vivo at subsaturating ribozyme concentrations (Fig. 8B ▶) is not significantly different from the self-cleavage rate. The similarity in these values demonstrates that hairpin ribozyme complexes that form between two chimeric U3 snoRNAs can be nearly as functional as self-cleaving ribozymes that assemble within a single U3 snoRNA. This is somewhat surprising in view of the fact that U3 snoRNAs normally participate in 18S rRNA biogenesis as part of an 80S ribonucleoprotein particle (Dragon et al. 2002). Unless intermolecular cleavage occurs during some other phase of the U3 snoRNA biological lifetime, before 80S particle assembly or after its dissociation, the complex between chimeric ribozyme and substrate U3 snoRNAs could be a ribonucleoprotein particle with a molecular mass greater than 4,000,000. In these chimeric snoRNAs, ribozyme and substrate sequences are substituted for U3 sequences that normally form complementary base-pairing interactions with the 5′ external spacer region of precursor 18S rRNA (Beltrame et al. 1994; Beltrame and Tollervey 1995). The orientation of these sequences within the 80S particle likely maximizes accessibility and the potential for pre rRNA recognition and binding. It will be interesting to learn whether ribozyme and substrate sequences located at different sites in U3 snoRNA display different intermolecular cleavage kinetics. The formation of functional complexes between chimeric ribozyme and substrate snoRNAs provides direct evidence that U3 snoRNAs can interact with each other as well as with their natural pre rRNA targets.
Intermolecular cleavage kinetics likely reflect slow RNA complex formation in vivo
The KMS value of 1 μM that could be estimated from the ribozyme concentration dependence of observed cleavage rates in vivo is similar to the KMS values measured for similar ribozyme variants in vitro (Fig. 8B ▶; Table 2 ▶). KM S values probably do not correspond to equilibrium dissociation constants in either case. Our previous analyses of chimeric self-cleaving mRNAs suggested that the stability of complexes between ribozymes and cleavage products are about the same in yeast cytoplasm as in reactions with 2 mM MgCl2 and 150 mM NaCl in vitro (Yadava et al. 2001). Unless yeast nuclei contain factors that accelerate dissociation, a ribozyme–substrate complex that is stabilized by 28 base pairs in H1 is likely to cleave much faster than it dissociates. By assuming that chimeric R228 snoRNA mediates substrate cleavage through a Briggs-Haldane type of kinetic mechanism in vivo, a substrate binding rate constant of 4 × 104 M−1 min−1 can be calculated from the ratio of kcleav and KMS values of 0.037 min−1 and 1.0 μM, respectively (Fig. 8B ▶). This estimated intracellular binding rate constant is almost 25-fold slower than binding rate constants on the order of 1 × 106 M−1 min−1 that can be calculated from kinetic parameters measured in vitro (Table 2 ▶).
U3 snoRNA is known to spend at least part of its biological lifetime as part of an 80S ribonucleoprotein particle that accumulates in nucleoli (Kiss 2002). Recent investigations of RNA movements in living cells using fluorescent correlation spectroscopy and fluorescence recovery after photobleaching suggest that some RNAs can move about the nucleus at rates comparable to diffusion in aqueous solution (Pederson 1999; Politz et al. 1999). Photobleaching experiments show that GFP-tagged fibrillarin, which is a U3 snoRNP protein, diffuses freely within mammalian nuclei with a diffusion coefficient of 0.53 μm sec−1 (Phair and Misteli 2000). This diffusion coefficient is 100-fold slower than diffusion coefficients reported for free solutes or GFP alone in the nucleus (Seksek et al. 1997), likely reflecting the large size of the U3 snoRNP. This difference between diffusion coefficients of U3 snoRNP and free solutes corresponds remarkably well with the 25-fold difference we observed between second-order rate constants for in vitro ribozyme–substrate complex formation and complex formation between chimeric ribozyme and substrate snoRNPs in yeast nuclei. This correspondence suggests that intermolecular complex formation between chimeric snoRNPs in yeast nuclei is diffusion limited. It will be important to learn if all RNA assembly reactions in vivo are diffusion limited or if some assembly reactions, such as chimeric snoRNA binding to pre rRNA targets for example, are facilitated.
MATERIALS AND METHODS
Plasmid construction and propagation
Plasmids encoding self-cleaving chimeric U3 snoRNAs under the control of the GAL1–10 regulatory region (Fig. 1 ▶) were prepared in the following way. First, the U3 gene was amplified from genomic DNA of Saccharomyces cerevisiae strain HYF114 (MATα ade2–1 his3–11,15 leu2–3 112 trp1–1 ura3–1 can1–100), a gift of Feng He (University of Massachusetts, Worcester), using U3RNAF and U3RNAR primers (Table 3 ▶). After digestion with HindIII, the PCR product was inserted into the polylinker of pGEM–4Z (Promega) to create pGEM4–32. A unique ClaI site was introduced into the 5′ region of the U3 gene and U3 sequences between 780121 and 780328 (Dujon et al. 1997) were deleted using QuikChange mutagenesis (Stratagene) with PUF and PUR primers (Table 3 ▶) to create pGEM4–U32D2. Sequences encoding the LR26 self-cleaving ribozyme variant (Fig. 2A ▶) were amplified from pTLR26 (Donahue et al. 2000) using RHGLR and NLR26 primers (Table 3 ▶), digested with NarI and ClaI and inserted into the newly created ClaI site in pGEM4–U32D2 to create pGEM4–26U32. Next, an XmnI fragment from pGEM4–26U32 was inserted into the SmaI site of pGAL316, which is a derivative of the single-copy shuttle vector, pRS316 (Sikorski and Hieter 1989), which contains the 270 base pair GAL1–10 regulatory region (Donahue and Fedor 1997; Donahue et al. 2000) and a URA3 selectable marker to create pGULR26. pGULR26 was modified using QuikChange mutagenesis and primers PGULR23F and PGULR23R to create pGULR23, which encodes a self-cleaving ribozyme with three base pairs in H1. QuikChange mutagenesis of pGULR26 with primers PGULR220F and PGULR220R created pGULR220, which encodes a self-cleaving ribozyme with 20 base pairs in H1. For control experiments, an inactivating G+1A mutation was introduced through QuikChange mutagenesis with the appropriate primers (Table 3 ▶) to create pGULR23A, pGULR26A, and pGULR220A.
TABLE 3.
Oligonucleotides used for plasmid constructions
| Plasmids | Sense | Antisense | Primers |
| pGEM4-U32 | 5′-CCGGTAAAGCTTTCTCACAATATACC-3′ | 5′-GTCCTTGAAGCTTCTCGATATAATGAC-3′ | U3RNAF/URRNAR |
| PGEM4-U3D2 | 5′-CTGTGTCGACGTACTTCATCGATGAATCCAACTTGGTTGATGAG-3′ | 5′-CTCATCAACCAAGTTGGATTCATCGATGAAGTACGTCGACACTG-3′ | PUF/PUR |
| pGULR26 | 5′-GCACGGATCGATACTATCACACGTCGCAAGGTGAGAAGCCAACC-3′ | 5′-GATCGCGGCGCCATAGTCTGGAGCGAAGGTGGACTGCCATTAG-3′ | RHGLR/NLR26 |
| pGULR23 | 5′-CTAATGGCAGTCGACGAACGCTCCAGACTATGG-3′ | 5′-CCATAGTCTGGAGCGTTCGTCGACTGCCATTAG-3′ | PGULR23F/PGULR23R |
| pGULR220 | 5′-GGCAGTCGACCTTGCGACGTGTGTAAGGCGATGAATCCAAC-3′ | 5′-GTTGGATTCATCGCCTTACACACGTCGCAAGGTCGACTGCC-3′ | PGULR420F/PGULR420R |
| pGUR228 | 5′-GGTATATTACCTGGTAC/CGATGAATCCAACTTGG-3′ | 5′-CCAAGTTGGATTCATCGGTACCAGGTAATATACC-3′ | PGUR2F/PGUR2R |
| pGUR23 | 5′-CGTACTTCATCGATAGATAGTGTGCAGCGTTCGTGAGAAGCCAACC-3′ | 5′-GGTTGGCTTCTCACGAACGCTGCACACTATCTATCGATGAAGTACG-3′ | PGUR3F/PGUR3R |
| pGUR24 | 5′-GTGTGCAGCGTTGGTGAGAAGCC-3′ | 5′-GGCTTCTCACCAACGCTGCACAC-3′ | PGUR24F/PGUR24R |
| pGUR26 | 5′CGTACTTCATCGATAGATAGTGTGCAGCGAAGGTGAGAAGCCAACC-3′ | 5′GGTTGGCTTCTCACCTTCGCTGCACACTATCTATCGATGAAGTACG-3′ | PGUR6F/PGUR6R |
| pGULR23A | 5′-GTACTCTAATGGCAATCCACGAACGCTCC-3′ | 5′-GGAGCGTTCGTGGATTGCCATTAGAGTAC-3′ | PGULR23AF/PGULR23AR |
| pGULR26A | 5′-GTACTCTAATGGCAATCCACCTTCGCTCC-3′ | 5′-GGAGCGAAGGTGGATTGCCATTAGAGTAC-3′ | PLR26AF/PLR26AR |
| pGULR220A | 5′-GGTACTCTAATGGCAATCCACCTTGCGACG-3′ | 5′-CGTCGCAAGGTGGATTGCCATTAGAGTACC-3′ | PGULR220AF/PGULR220AR |
| pGUS | 5′-CGAACCACCTGGCAGTCCACCTTGCGACGTGTGATAGAT-3′ | 5′-CTATCACACGTCGCAAGGTGGACTGCCAGGTGGTT-3′ | 156/256 |
| pGUSA | 5′-GAACCACCTGGCAATCCACCTTGCGACG-3′ | 5′-CGTCGCAAGGTGGATTGCCAGGTGGTTC-3′ | SGA1/SGA2 |
| pGUSU-5 C | 5′-CATCGAACCACCCGGCAGTCCACCTTGC-3′ | 5′-GCAAGGTGGACTGCCGGGTGGTTCGATG-3′ | PGUSU5CF/PGUSU5CR |
| pGUSU-5 C | 5′-CATCGAACCACCGGGCAGTCCACCTTGC-3′ | 5′-GCAAGGTGGACTGCCCGGTGGTTCGATG-3′ | PGUSU5GF/PGUSU5GR |
| pGUR2A-14 C | 5′-CAAGGTGAGAAGCCCACCAGAGAAACAC-3′ | 5′-GTGTTTCTCTGGTGGGCTTCTCACCTTG-3′ | A14CF/A14CR |
| pGUR2A-14 C | 5′-CAAGGTGAGAAGCCGACCAGAGAAACAC-3′ | 5′-GTGTTTCTCTGGTCGGCTTCTCACCTTG-3′ | A14GF/A14GR |
| pU14 | 5′-GATCGCAGTTTCCACGGTAGGAGTACGCTTACGAACCCATCGTTAGTACTCTCGGTGACC-3′ | 5′-AGCTGCGGTCACCGAGAGTACTAACGATGGGTTCGTAAGCGTACTCCTACCGTGGAAACTGC-3′ | U14F/U14R |
For analysis of ribozyme cleavage of separate RNA targets, plasmids were engineered to encode chimeric U3 snoRNAs with only ribozyme or only substrate sequences. Plasmids encoding chimeric U3 snoRNAs containing the R228 ribozyme sequence (Fig. 7 ▶) were prepared from pGULR220 by deleting the interdomain linker and substrate sequences downstream of ribozyme position A50 using QuikChange mutagenesis and PGUR2F and PGUR2R primers (Table 3 ▶) to create pGUR228. Plasmid derivatives that encode chimeric ribozymes with the potential to form H1 sequences with three base pairs (pGUR23), four base pairs (pGUR24), or six base pairs (pGUR26) were created through QuikChange mutagenesis and the primers shown in Table 3 ▶ (Fig. 7 ▶).
The plasmid encoding the chimeric substrate U3 snoRNA fused to UASGAL was prepared by inserting a synthetic oligonucleotide duplex encoding the substrate sequence (156 and 256, Table 3 ▶) into the ClaI site of U3 in pGEM4–US. For expression in yeast, an XmnI fragment from pGEM4–US and the XbaI fragment from pGAL316 that contains the GAL1–10 regulatory region (Donahue and Fedor 1997) were inserted into pRS313, which is a single-copy number shuttle vector with a HIS3 selectable marker (Sikorski and Hieter 1989) to create pGUS. An inactivating G+1A mutation was introduced into the substrate sequence of pGUS to create pGUSA by QuikChange mutagenesis with SGA1 and SGA2 primers (Table 3 ▶). H2 sequence modifications were introduced into pGUR2 and pGUS derivatives using QuikChange mutagenesis (Fig. 7 ▶; Table 3 ▶).
To facilitate calculation of the intracellular ribozyme concentration of chimeric substrate snoRNA cleavage, experimental conditions were developed to approximate pseudofirst-order conditions, that is, conditions under which the concentration of substrate is low with respect to ribozyme concentrations so that substrate binding does not significantly reduce the concentration of free ribozyme. Under these conditions, the total concentration of chimeric ribozyme snoRNA is a good approximation of the concentration of unbound ribozyme. For these experiments, chimeric ribozyme snoRNAs were expressed under the control of the GAL1–10 regulatory region, as described above, and chimeric substrate snoRNA was expressed from pUS, a plasmid that encodes the chimeric U3 substrate RNA but lacks UASGAL. pUS and the corresponding plasmid encoding a noncleavable substrate analog, pUSA, were constructed from pGUS and pGUSA, respectively, by deletion of the XbaI fragment that contains GAL1–10 sequences.
Plasmid templates for T7 RNA polymerase transcription of hybridization probes complementary to chimeric self-cleaving snoRNAs were prepared by insertion of HincII fragments of the corresponding pRS313 derivatives (Fig. 2B ▶) into the polylinker of pGEM–4Z. In addition to self-cleaving ribozyme sequences, HincII fragments contain 14 base pairs of U3 sequence upstream of the ribozyme insertion site and 268 base pairs of U3 sequence downstream of the ClaI site. Along with the chimeric snoRNA fragments diagrammed in Figure 3A ▶, RNase protection assays with these hybridization probes anneal with unmodified U3 RNA expressed from the genome to give a protected fragment 268 nt in length.
Plasmid templates for transcription of hybridization probes complementary to the chimeric substrate sequences in pUS and pGUS derivatives contained the HincII fragment from pGEM4–U3D2 with the substrate-encoding oligonucleotides (156 and 256, Table 3 ▶) and flanking U3 sequences (Fig. 2B ▶) inserted into the ClaI site (Fig. 5A ▶, left). Plasmids encoding hybridization probes complementary to substrate variants SU−5C and SU−5G were created from pGEM4–U3D2 using QuikChange mutagenesis and primers PGUSU5CF and PGUSU5CR, for SU−5C, and PGUSU5GF and PGUSU5GR for SU−5G (Table 3 ▶). In addition to the chimeric substrate snoRNA fragments diagrammed in Figure 5A ▶ (left), these hybridization probes anneal with chimeric ribozyme snoRNAs and with genomic U3 RNA to give a protected fragment 272 and 268 nt in length, respectively (Fig. 5B ▶). The plasmid template for transcription of a hybridization probe complementary to the ribozyme sequence in chimeric snoRNA, pRK3, was prepared by inserting a duplex oligonucleotide encoding the complement of nucleotides 4–51 of the ribozyme sequence (Fig. 2A ▶) into the KpnI site of pGEM–3Zf.
Templates for transcription of hybridization probes designed to detect 5′ cleavage products (Fig. 5A ▶, right) were prepared by deleting sequences corresponding to the 3′ region of the chimeric substrate RNA through QuikChange mutagenesis with TUSF and TUSR primers.
To construct pTU14, the plasmid template for transcription of a hybridization probe complementary to U14 snoRNA, a synthetic oligonucleotide duplex with nucleotides 747–807 of the SNR128 gene (Zagorski et al. 1988; Dujon et al. 1997) was inserted between the HindIII and EcoRI sites of pGEM–4Z.
The sequence of the U3 gene and all sequence modifications of chimeric ribozyme–snoRNA plasmids were confirmed by dideoxy sequencing. Plasmids were propagated in Escherichia coli strain DH5α (Sambrook et al. 1989) or XL1-Blue (Stratagene). pRS316 derivatives, with the URA3 selectable marker, and pRS313 derivatives, with the HIS3 selectable marker, were transformed into S. cerevisiae strain HYF114 (MATα ade2–1 his3–11,15 leu2–3 112 trp1–1 ura3–1 can1–100). Yeast were grown in minimal medium (0.67% yeast nitrogen base without amino acids, Difco) supplemented with the appropriate amino acids and 2% galactose. For experiments designed to measure the ribozyme concentration dependence of intracellular cleavage rates, galactose was combined with different concentrations of sucrose or glucose to achieve different levels of chimeric ribozyme snoRNA expression (Ronicke et al. 1997).
The ribozyme used for in vitro experiments, R2, was prepared by T7 RNA polymerase transcription of a Bgl II-linearized derivatives of plasmid template, pTR2, as described (Fedor 1999). Variants of R2 with modifications of H2 sequences were created through QuikChange mutagenesis of pTR2 and the A14GF and A14GR primers to create pTR2A14G variants, and with A14CF and A14CR primers to create pTRA14C variants (Fig. 7 ▶; Table 3 ▶).
RNA preparation and quantification
Total RNA was extracted from yeast cultures at mid-log phase and quantified using RNase protection assays as described (Donahue and Fedor 1997) except that all procedures were carried out at pH 4.2 and at 4°C to inhibit ribozyme activity during RNA isolation and analysis. When necessary, yeast cultures were diluted into fresh media to maintain logarithmic growth as samples were collected over the long time courses needed to measure slow chimeric snoRNA decay kinetics. When a 32P-labeled self-cleaving RNA transcript was combined with yeast pellets and subjected to extraction and analysis procedures, less than 10% underwent self-cleavage, confirming that conditions used for RNA isolation and RNase protection assays do not support ribozyme activity. A second type of control experiment was used to test whether ribozyme sequences in chimeric snoRNAs could cleave separate chimeric substrate snoRNAs during yeast RNA extraction and analysis. When yeast expressing chimeric ribozyme snoRNA were combined with yeast expressing chimeric substrate snoRNAs before RNA extraction, no substrate depletion occurred and no cleavage products were detected, evidence that the cleavage activity observed with coexpressed ribozyme and substrate snoRNAs occurred in yeast and not in vitro.
Hybridization probe templates were linearized by digestion with EcoRI and probes were uniformly labeled by including [α −32P]ATP in transcription reactions, as described (Donahue and Fedor 1997). Dharmacon Research, Inc. supplied substrates, deoxynucleotide-substituted substrates, and 3′ cleavage product RNAs. [5′ −32P]RNAs were prepared by reaction with T4 poly-nucleotide kinase and [γ −32P]ATP as described (Fedor and Uhlenbeck 1990).
RNA concentrations were determined by assuming a residue extinction coefficient at 260 nm of 6.6 × 103 M−1 cm−1 or calculated from the specific activity of the [α −32P]ATP used for labeling.
Kinetic and thermodynamic analyses
Methods for determining kinetic parameters from in vitro and in vivo cleavage reactions have been described in detail (Donahue and Fedor 1997; Nesbitt et al. 1997; Fedor 1999; Donahue et al. 2000) and are summarized below. For ribozymes with three or more base pairs in H1, in vitro self-cleavage kinetics were measured in pulse-chase experiments as described (Nesbitt et al. 1999). Briefly, 32P LR was first prepared in a ligation reaction in which a trace amount of 32P5′R was combined with 3′P in standard buffer at a 3′P concentration sufficient to produce half of the maximal amount of 32P LR. Ligation reactions were then diluted 200- or 400-fold into 50 mM NaHEPES, pH 7.5, 10 mM MgCl2, 0.1 mM EDTA (standard buffer) or into 50 mM NaHEPES, pH 7.5, 2 mM MgCl2, 150 mM NaCl, 0.1 mM EDTA at 30°C and the fraction of 32P5′R was measured after various times. Self-cleavage rates were computed from the fit to F = F∞(1−e −kobst), where F is the fraction cleaved at time t.
Kinetic parameters for ribozyme cleavage of oligonucleotide substrates in vitro were determined at 30°C from single-turnover reactions as described (Hegg and Fedor 1995). Briefly, reactions contained ribozyme, at eight or more concentrations ranging from 1 to 10,000 nM, and [32P] substrate, at concentrations that were at least 10-fold lower than the ribozyme concentrations. Plots of kobs,cleav versus kobs,cleav/[R] were used to calculate cleavage rate constants, k2 and KMS values (Eadie 1942; Hofstee 1952).
Equilibrium dissociation constants were measured for complexes between ribozymes and oligonucleotide substrate analogs with deoxynucleotide substitutions at the A−1 position using nondenaturing gel shift assays as described (Fedor and Uhlenbeck 1992; Hegg and Fedor 1995). Briefly, a small amount of [32P] substrate was combined with ribozyme at a range of concentration that were at least 10-fold higher. Solutions were adjusted to a final concentration of 10 mM MgCl2, or 2 mM MgCl2, 150 mM NaCl and 5% glycerol, 0.0002% bromophenol blue, and 0.0002% xylene cyanol and were equilibrated at 30°C for 4 h to 8 h. Bound and unbound ligands were fractionated using 15% acrylamide gels in 50 mM Tris acetate, pH 7.5, 10 mM magnesium acetate or 2 mM magnesium acetate, 150 mM NaCl buffer. The fraction of bound ligand (FB) was plotted as a function of ribozyme concentration, and KdS values were calculated by fitting to FB∞ = [R]/([R] + KdS).
Intracellular chimeric snoRNA self-cleavage and intermolecular cleavage rates were determined from the abundance of uncut chimeric snoRNA at various times after glucose was added to the growth media to inhibit transcription and from the abundance of uncut chimeric snoRNA at a steady state in the experiments similar to those described previously (Parker et al. 1991; Donahue and Fedor 1997). For chimeric snoRNAs with self-cleaving ribozyme or substrate insertions, decay rates were calculated from the amount of uncut chimeric snoRNA remaining after various times by computing the nonlinear least squares fit to [uncut snoRNA]t/[uncut snoRNA]t0 = e−kobst. Intracellular cleavage rates were calculated from the difference between decay rates of self-cleaving snoRNAs and degradation rates of chimeric snoRNAs with an inactivating G+1A mutation at the cleavage site. Intracellular self-cleavage and intermolecular substrate cleavage rates also were calculated from chimeric snoRNA degradation rates and the abundance of chimeric snoRNA at steady state by computing the fit to kobs,cleav = (kdegradation/fraction HP snoRNA) − kdegradation as described (Donahue and Fedor 1997). Virtually the same degradation rate (0.008 ± 0.001 min−1) and the same abundance at steady state ([chimeric U3 snoRNA]/[genomic U3 snoRNA] = 1.0 ± 0.18) were measured for all inactive chimeric snoRNAs regardless of the insertion sequence. Thus, insertion of ribozyme and substrate sequences had no apparent effect on snoRNA turnover in the absence of cleavage. Intracellular cleavage rates obtained from time course and steady-state experiments typically agreed within 40%. The accuracy of time course experiments was limited by our inability to collect yeast samples over short intervals. For optimum accuracy, reported intracellular cleavage rates represent the mean and standard error of four or more rates calculated from HP snoRNA abundance at steady state.
Acknowledgments
This work was supported by NIH Grant GM62277. R.S. Yadava was a Skaggs Postdoctoral Fellow. We thank Elise Calderon, Joseph Cottrell, Carla Da Costa, Jason Harger, and Yaroslav Kuzmin for critical reading of the manuscript.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.rnajournal.org/cgi/doi/10.1261/rna.5234204.
REFERENCES
- Barroso-del Jesus, A., Tabler, M., and Berzal-Herranz, A. 1999. Comparative kinetic analysis of structural variants of the hairpin ribozyme reveals further potential to optimize its catalytic performance. Antisense Nucleic Acid Drug Dev. 9: 433–440. [DOI] [PubMed] [Google Scholar]
- Beltrame, M. and Tollervey, D. 1995. Base pairing between U3 and the pre-ribosomal RNA is required for 18S rRNA synthesis. EMBO J. 14: 4350–4356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beltrame, M., Henry, Y., and Tollervey, D. 1994. Mutational analysis of an essential binding site for the U3 snoRNA in the 5′ external transcribed spacer of yeast pre-rRNA. Nucleic Acids Res. 22: 4057–4065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrington, J.C. and Ambros, V. 2003. Role of microRNAs in plant and animal development. Science 301: 336–338. [DOI] [PubMed] [Google Scholar]
- Castanotto, D., Li, H., Chow, W., Rossi, J.J., and Deshler, J.O. 1998. Structural similarities between hammerhead ribozymes and the spliceosomal RNAs could be responsible for lack of ribozyme cleavage in yeast. Antisense Nucleic Acid Drug Dev. 8: 1–13. [DOI] [PubMed] [Google Scholar]
- Craig, M.E., Crothers, D.M., and Doty, P. 1971. Relaxation kinetics of dimer formation by self complementary oligonucleotides. J. Mol. Biol. 62: 383–401. [DOI] [PubMed] [Google Scholar]
- Decatur, W.A. and Fournier, M.J. 2003. RNA-guided nucleotide modification of ribosomal and other RNAs. J. Biol. Chem. 278: 695–698. [DOI] [PubMed] [Google Scholar]
- Donahue, C.P. and Fedor, M.J. 1997. Kinetics of hairpin ribozyme cleavage in yeast. RNA 3: 961–973. [PMC free article] [PubMed] [Google Scholar]
- Donahue, C.P., Yadava, R.S., Nesbitt, S.M., and Fedor, M.J. 2000. The kinetic mechanism of the hairpin ribozyme in vivo: Influence of RNA helix stability on intracellular cleavage kinetics. J. Mol. Biol. 295: 693–707. [DOI] [PubMed] [Google Scholar]
- Dragon, F., Gallagher, J.E., Compagnone-Post, P.A., Mitchell, B.M., Porwancher, K.A., Wehner, K.A., Wormsley, S., Settlage, R.E., Shabanowitz, J., Osheim, Y., et al. 2002. A large nucleolar U3 ribonucleoprotein required for 18S ribosomal RNA biogenesis. Nature 417: 967–970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dujon, B., Albermann, K., Aldea, M., Alexandraki, D., Ansorge, W., Arino, J., Benes, V., Bohn, C., Bolotin-Fukuhara, M., Bordonne, R., et al. 1997. The nucleotide sequence of Saccharomyces cerevisiae chromosome XV. Nature 387: 98–102. [PubMed] [Google Scholar]
- Eadie, G.S. 1942. The inhibition of cholinesterase by physostigmine and prostigmine. J. Biol. Chem. 146: 85–93. [Google Scholar]
- Esteban, J.A., Banerjee, A.R., and Burke, J.M. 1997. Kinetic mechanism of the hairpin ribozyme. J. Biol. Chem. 272: 13629–13639. [DOI] [PubMed] [Google Scholar]
- Fedor, M.J. 1999. Tertiary structure stabilization promotes hairpin ribozyme ligation. Biochemistry 38: 11040–11050. [DOI] [PubMed] [Google Scholar]
- ———. 2000. Structure and function of the hairpin ribozyme. J. Mol. Biol. 297: 269–291. [DOI] [PubMed] [Google Scholar]
- Fedor, M.J. and Uhlenbeck, O.C. 1990. Substrate sequence effects on “hammerhead” RNA catalytic efficiency. Proc. Natl. Acad. Sci. 87: 1668–1672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ———. 1992. Kinetics of intermolecular cleavage by hammerhead ribozymes. Biochemistry 31: 12042–12054. [DOI] [PubMed] [Google Scholar]
- Fedor, M.J., Lue, N.F., and Kornberg, R.D. 1988. Statistical positioning of nucleosomes by specific protein-binding to an upstream activating sequence in yeast. J. Mol. Biol. 204: 109–127. [DOI] [PubMed] [Google Scholar]
- Fedor, M.J., Donahue, C.P., and Nesbitt, S.M. 2000. Hairpin ribozyme activity in vitro and in vivo. In Ribozyme biochemistry and biotechnology (eds. G. Krupp and R. Gaur), pp. 205–226. Eaton Publishing, Natick, MA.
- Hegg, L.A. and Fedor, M.J. 1995. Kinetics and thermodynamics of intermolecular catalysis by hairpin ribozymes. Biochemistry 34: 15813–15828. [DOI] [PubMed] [Google Scholar]
- Hofstee, B.H.J. 1952. Specificity of esterases. I. Identification of two pancreatic aliesterases. J. Biol. Chem. 199: 357–364. [PubMed] [Google Scholar]
- Hughes, J.M. and Ares Jr., M. 1991. Depletion of U3 small nucleolar RNA inhibits cleavage in the 5′ external transcribed spacer of yeast pre-ribosomal RNA and impairs formation of 18S ribosomal RNA. EMBO J. 10: 4231–4239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes, J.M., Konings, D.A., and Cesareni, G. 1987. The yeast homologue of U3 snRNA. EMBO J. 6: 2145–2155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston, M. and Davis, R.W. 1984. Sequences that regulate the divergent GAL1–GAL10 promoter in Saccharomyces cerevisiae. Mol. Cell Biol. 4: 1440–1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khvorova, A., Lescoute, A., Westhof, E., and Jayasena, S.D. 2003. Sequence elements outside the hammerhead ribozyme catalytic core enable intracellular activity. Nat. Struct. Biol. 10: 708–712. [DOI] [PubMed] [Google Scholar]
- Kiss, T. 2002. Small nucleolar RNAs: An abundant group of noncoding RNAs with diverse cellular functions. Cell 109: 145–148. [DOI] [PubMed] [Google Scholar]
- Mathews, D.H., Sabina, J., Zuker, M., and Turner, D.H. 1999. Expanded sequence dependence of thermodynamic parameters improves prediction of RNA secondary structure. J. Mol. Biol. 288: 911–940. [DOI] [PubMed] [Google Scholar]
- Nesbitt, S., Hegg, L.A., and Fedor, M.J. 1997. An unusual pH-independent and metalion-independent mechanism for hairpin ribozyme catalysis. Chem. Biol. 4: 619–630. [DOI] [PubMed] [Google Scholar]
- Nesbitt, S.M., Erlacher, H.A., and Fedor, M.J. 1999. The internal equilibrium of the hairpin ribozyme: Temperature, ion and pH effects. J. Mol. Biol. 286: 1009–1024. [DOI] [PubMed] [Google Scholar]
- Parker, R., Herrick, D., Peltz, S.W., and Jacobson, A. 1991. Measurement of mRNA decay rates in Saccharomyces cerevisiae. Methods Enzymol. 194: 415–423. [DOI] [PubMed] [Google Scholar]
- Pederson, T. 1999. Movement and localization of RNA in the cell nucleus. FASEB J. 13: S238–S242. [DOI] [PubMed] [Google Scholar]
- Phair, R.D. and Misteli, T. 2000. High mobility of proteins in the mammalian cell nucleus. Nature 404: 604–609. [DOI] [PubMed] [Google Scholar]
- Politz, J.C., Tuft, R.A., Pederson, T., and Singer, R.H. 1999. Movement of nuclear poly(A) RNA throughout the interchromatin space in living cells. Curr. Biol. 9: 285–291. [DOI] [PubMed] [Google Scholar]
- Pörschke, D. and Eigen, M. 1971. Co-operative non-enzymic base recognition. 3. Kinetics of the helix-coil transition of the oligoribouridylic–oligoriboadenylic acid system and of oligoriboadenylic acid alone at acidic pH. J. Mol. Biol. 62: 361–381. [DOI] [PubMed] [Google Scholar]
- Ravetch, J., Gralla, J., and Crothers, D.M. 1974. Thermodynamic and kinetic properties of short RNA helices: The oligomer sequence AnGCUn. Nucleic Acids Res. 1: 109–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reich, C.I., VanHoy, R.W., Porter, G.L., and Wise, J.A. 1992. Mutations at the 3′ splice site can be suppressed by compensatory base changes in U1 snRNA in fission yeast. Cell 69: 1159–1169. [DOI] [PubMed] [Google Scholar]
- Ronicke, V., Graulich, W., Mumberg, D., Muller, R., and Funk, M. 1997. Use of conditional promoters for expression of heterologous proteins in Saccharomyces cerevisiae. Methods Enzymol. 283: 313–322. [DOI] [PubMed] [Google Scholar]
- Samarsky, D.A. and Fournier, M.J. 1998. Functional mapping of the U3 small nucleolar RNA from the yeast Saccharomyces cerevisiae. Mol. Cell Biol. 18: 3431–3444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samarsky, D.A., Ferbeyre, G., Bertrand, E., Singer, R.H., Cedergren, R., and Fournier, M.J. 1999. A small nucleolar RNA:ribozyme hybrid cleaves a nucleolar RNA target in vivo with near-perfect efficiency. Proc. Natl. Acad. Sci. 96: 6609–6614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook, J., Fritsch, E.F., and Maniatis, T. 1989. Molecular cloning: A laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Seksek, O., Biwersi, J., and Verkman, A.S. 1997. Translational diffusion of macromolecule-sized solutes in cytoplasm and nucleus. J. Cell Biol. 138: 131–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman, F. 1991. Getting started with yeast. In Guide to yeast genetics and molecular biology (eds. C. Guthrie and G.R. Fink), pp. 3–21. Academic Press, Inc., New York.
- Shine, J. and Dalgarno, L. 1974. The 3′ -terminal sequence of Escherichia coli 16S ribosomal RNA: Complementarity to nonsense triplets and ribosome binding sites. Proc. Natl. Acad. Sci. 71: 1342–1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorski, R.S. and Hieter, P. 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122: 19–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staley, J.P. and Guthrie, C. 1998. Mechanical devices of the spliceosome: Motors, clocks, springs, and things. Cell 92: 315–326. [DOI] [PubMed] [Google Scholar]
- Walter, N.G. and Burke, J.M. 1997. Real-time monitoring of hairpin ribozyme kinetics through base-specific quenching of fluorescein-labeled substrates. RNA 3: 392–404. [PMC free article] [PubMed] [Google Scholar]
- Walter, N.G., Burke, J.M., and Millar, D.P. 1999. Stability of hairpin ribozyme tertiary structure is governed by the interdomain junction. Nat. Struct. Biol. 6: 544–549. [DOI] [PubMed] [Google Scholar]
- Walter, N.G., Hampel, K.J., Brown, K.M., and Burke, J.M. 1998. Tertiary structure formation in the hairpin ribozyme monitored by fluorescence resonance energy transfer. EMBO J. 17: 2378–2391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Y., Liu, C.L., Storey, J.D., Tibshirani, R.J., Herschlag, D., and Brown, P.O. 2002. Precision and functional specificity in mRNA decay. Proc. Natl. Acad. Sci. 99: 5860–5865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia, T., SantaLucia, J.J., Burkard, M., Kierzek, R., Schroeder, S., Jiao, X., Cox, C., and Turner, D. 1998. Thermodynamic parameters for an expanded nearest-neighbor model for formation of RNA duplexes with Watson-Crick base pairs. Biochemistry 37: 14719–11435. [DOI] [PubMed] [Google Scholar]
- Yadava, R., Choi, A., Lebruska, L., and Fedor, M. 2001. Hairpin ribozymes with four-way helical junctions mediate intracellular RNA ligation. J. Mol. Biol. 309: 893–902. [DOI] [PubMed] [Google Scholar]
- Zagorski, J., Tollervey, D., and Fournier, M.J. 1988. Characterization of an SNR gene locus in Saccharomyces cerevisiae that specifies both dispensible and essential small nuclear RNAs. Mol. Cell Biol. 8:3282–3290. [DOI] [PMC free article] [PubMed] [Google Scholar]