Abstract
U11 and U12 snRNPs bind U12-type pre-mRNAs as a preformed di-snRNP complex, simultaneously recognizing the 5′ splice site and branchpoint sequence. Thus, within the U12-type prespliceosome, U11/U12 components form a molecular bridge connecting both ends of the intron. We have affinity purified human 18S U11/U12 and 12S U11 snRNPs, and identified their protein components by using mass spectrometry. U11/U12 snRNPs lack all known U1 snRNP proteins but contain seven novel proteins (i.e., 65K, 59K, 48K, 35K, 31K, 25K, 20K) not found in the major spliceosome, four of which (59K, 48K, 35K, and 25K) are U11-associated. Thus, protein–protein and protein–RNA interactions contributing to 5′ splice site recognition and/or intron bridging appear to differ significantly in the minor versus major prespliceosome. The majority of U11/U12 proteins are highly conserved in organisms known to contain U12-type introns. However, homologs of those associated with U11 were not detected in Drosophila melanogaster, consistent with the presence of a divergent U11 snRNP in flies. RNAi experiments revealed that several U11/U12 proteins are essential for cell viability, suggesting they play key roles in U12-type splicing. The presence of unique U11/U12 snRNP proteins in the U12-type spliceosome provides insight into potential evolutionary relationships between the major and minor spliceosome.
Keywords: U11/U12 snRNP, pre-mRNA splicing, U12-dependent spliceosome
INTRODUCTION
Pre-mRNA splicing is catalyzed by the spliceosome, a complex dynamic RNP machine. Two types of spliceosomes have been identified to date. The major U2-type spliceo-some is found in all eukaryotes and is responsible for the removal of U2-type introns, which represent the vast majority of pre-mRNA introns. The less abundant minor U12-type spliceosome, which is found in only a subset of eukaryotes, excises U12-type introns, which comprise <1% of all human introns (Levine and Durbin 2001). U12-type introns have distinct 5′ splice site and branch site consensus sequences that are longer and more highly conserved than are those of U2-type introns (Burge et al. 1998). In addition, the region between the U12-type branch site and 3′ splice site lacks a polypyrimidine tract. Splicing of both types of introns proceeds via two apparently identical transesterification reactions (for review, see Patel and Steitz 2003).
The U2-type spliceosome is formed by the interaction of the U1, U2, and U4/U5/U6 snRNPs, as well as non-snRNP splicing factors, with the pre-mRNA (for review, see Reed and Palandjian 1997). The U12-type spliceosome, in contrast, is comprised of the U11, U12, and U4atac/U6atac.U5 snRNPs (Hall and Padgett 1996; Tarn and Steitz 1996a, b). Thus, only U5 is common to both spliceosomes. The U11 and U12 snRNPs are the functional analogs of the U1 and U2 snRNPs, respectively, whereas the U4atac/U6atac snRNP is the functional analog of U4/U6 (Hall and Padgett 1996; Tarn and Steitz 1996a,b; Kolossova and Padgett 1997; Yu and Steitz 1997). Assembly of the U12-dependent spliceosome is analogous to that of the U2-dependent spliceosome, with one major exception. In contrast to the U1 and U2 snRNPs, U11 and U12 bind as a stable, preformed U11/U12 di-snRNP complex. During the first step of U12-type spliceosome formation, the 5′ splice site and branch site are recognized by the U11 and U12 snRNP, respectively, in a cooperative manner, forming the prespliceosome (A complex; Frilander and Steitz 1999). The minor U4atac/U6atac.U5 tri-snRNP subsequently binds, and after major conformational changes, a catalytically active U12-dependent spliceosome is formed (Tarn and Steitz 1996a; Yu and Steitz 1997; Frilander and Steitz 2001).
During the assembly of both spliceosomes, a similar dynamic RNA network is formed. Analogous to U1 and U2, within the U12-type prespliceosome the 5′ ends of the U11 and the U12 snRNA base pair with the 5′ splice site and branch site, respectively (Hall and Padgett 1996; Tarn and Steitz 1996b; Kolossova and Padgett 1997; Yu and Steitz 1997). Similar to U6 and U4, U6atac is first base paired with the U4atac snRNA, and upon integration of the U4atac/U6atac.U5 tri-snRNP into the spliceosome, U4atac is displaced, allowing U6atac to base pair with the 5′ splice site and the 5′ end of the U12 snRNA (Tarn and Steitz 1996a; Yu and Steitz 1997; Incorvaia and Padgett 1998; Frilander and Steitz 2001). Thus, both activated spliceosomes ultimately contain an RNA–RNA network of similar conformation that is thought to catalyze splicing.
Major differences between U2- and U12-type splicing appear to occur mainly during the early stages of spliceosome assembly. These include potential differences in intron bridging interactions, which are responsible for juxtaposing the 5′ splice site and the branch site of the pre-mRNA, as well as differences in 5′ splice site and/or branch site recognition. In the major spliceosome, 5′ splice site recognition is initially mediated by the U1 snRNP; the U1/5′ splice site interaction is facilitated by RNA base-pairing, as well as protein–protein and protein–RNA contacts involving the U1-70K and U1-C proteins (for review, see Will and Lührmann 1997). At this early stage a molecular bridge, likely involving SR proteins, is formed between the U1 snRNP bound to the 5′ splice site and SF1/mBBP bound to the branch site (Reed 1996). Upon stable U2 association with the branch site, SF1/mBBP is displaced and a new set of interactions juxtaposing the reactive groups of the pre-mRNA is formed (for review, see Schwer 2001). However, little is presently known about the nature of the molecular bridge formed between the U1 and U2 snRNPs within the U2-type prespliceosome. Recent studies have implicated the DEAD-box protein Prp5 in bridging the U1 and U2 snRNPs in the major spliceosome (Xu et al. 2004). Due to the concomitant binding of U11 and U12 as a di-snRNP, intron bridging interactions between the 5′ splice site and branch site within the minor prespliceosome must be mediated, at least in part, by U11/U12 proteins.
Mass spectrometry (MS) analyses of the major snRNPs and various splicing complexes have generated an essentially comprehensive list of U2-dependent spliceosomal proteins (Hartmuth et al. 2002; Jurica et al. 2002; Makarov et al. 2002; Rappsilber et al. 2002; Zhou et al. 2002). In contrast, due to the relatively low abundance of the U11, U12, and U4atac/U6atac snRNPs (~1% the level of the major snRNPs; Montzka and Steitz, 1988), knowledge of the protein composition of the minor spliceosome is far from complete. Several studies suggest that many spliceosomal proteins are common to both splicing machineries. For example, five subunits of the U2-associated heteromeric splicing factor SF3b, an essential core component of the major spliceosome, are also associated with purified human U11/U12 snRNPs (Will et al. 1999, 2001, 2002). Immunoprecipitation studies indicate that most proteins associated with the major U4/U6.U5 tri-snRNP are also present in the U4atac/U6atac.U5 tri-snRNP and minor spliceosome (Luo et al. 1999; Schneider et al. 2002). In vitro binding studies also support the presence of U4/U6-associated proteins in the U4atac/U6atac snRNP (Nottrott et al. 2002). Members of the SR protein family are required for the splicing of both U2- and U12-type introns and thus apparently are also shared by both spliceosomes (Hastings and Krainer 2001). On the other hand, the initial characterization of purified human 18S U11/U12 snRNPs revealed the presence of several proteins that did not appear to be present in the U2-type spliceosome (Will et al. 1999).
Here we have characterized proteins associated with human 18S U11/U12 and 12S U11 snRNPs. Both snRNPs were affinity purified, and their protein compositions determined by matrix-assisted laser desorption/ionization (MALDI) MS. Seven novel U11/U12 proteins, not present in major spliceosomes, were identified. A subset of these novel proteins associates with the 12S U11 monoparticle and thus likely contributes to 5′ splice site recognition. These data thus indicate that many protein interactions contributing to prespliceosome formation differ significantly in the U12- versus U2-type spliceosome. Compatible with them playing important roles in U12-type splicing, RNAi-mediated knockdowns demonstrated that several of the newly identified U11/U12-specific proteins are essential for cell viability. Potential evolutionary relationships between the major and minor spliceosomes, based on the identification of unique U12-type spliceosomal proteins, are subsequently discussed.
RESULTS
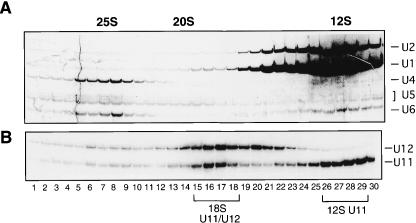
Identification of novel U11/U12 and U11 snRNP proteins
To isolate U11/U12 and U11 snRNPs, human spliceosomal snRNPs were immunoaffinity purified from HeLa nuclear extract with anti-m3G antibodies and then subjected to glycerol gradient centrifugation. The distribution of the major snRNAs (Fig. 1A ▶) or minor U11 and U12 snRNAs across the gradient (Fig. 1B ▶) was determined by silver staining or Northern blot analysis, respectively. Under these conditions, the majority of U1 and U2 sediment as 12S snRNPs (Fig. 1A ▶, lanes 25–28), whereas U4, U5, and U6 are predominantly found in the 25S (U4/U6.U5) tri-snRNP complex (lanes 5–8); a fraction of U5 also sediments as a 20S snRNP (lanes 12–15). Consistent with previous observations (Wassarman and Steitz 1992), immunoaffinity-purified U11 and U12 snRNPs cosediment as an 18S di-snRNP complex (Fig. 1B ▶, lanes 15–18), but are also present as 12S (lanes 26–29) and 15S (lanes 20–22) mono-snRNP particles, respectively.
FIGURE 1.
Sedimentation behavior of human snRNPs containing U11 and U12. Anti-m3G affinity-purified UsnRNPs were separated on a 10%–30% glycerol gradient. RNA was isolated from each fraction and analyzed on a 10% polyacrylamide/7M urea gel. Major snRNAs were visualized by silver staining (A) and U11 and U12 by Northern blotting (B). Sedimentation coefficients of the major snRNPs are indicated at the top. Fractions used to affinity select 18S U11/U12 or 12S U11 snRNPs are bracketed.
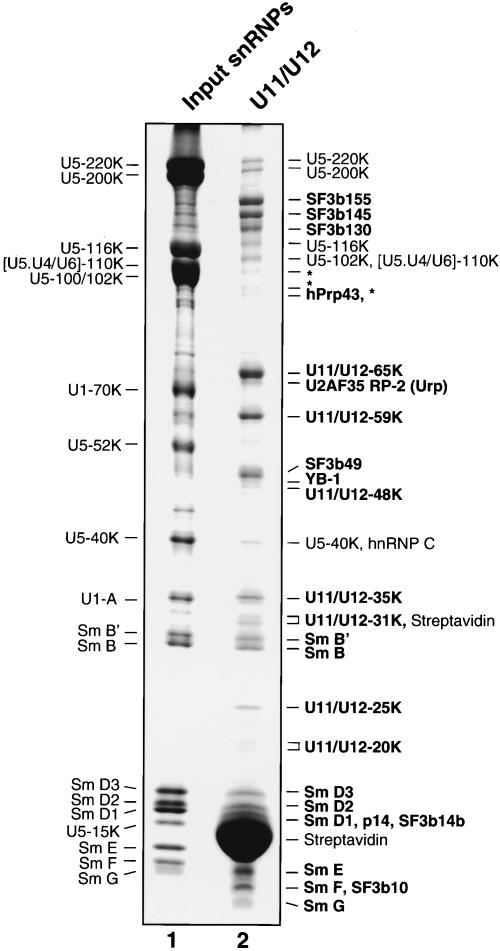
U11/U12 snRNPs were affinity selected from the 18S region of the gradient (which also contained large amounts of U5 snRNPs) by using a biotinylated 2′-O-methyl oligonucleotide complementary to U12 snRNA and streptavidin agarose beads. Analysis of the RNA composition of the affinity-selected snRNPs indicated that predominantly U11/U12 snRNPs were isolated, with only a low level of contaminating U5 snRNP present (data not shown; see Will et al. 1999). Proteins associated with the affinity-selected snRNPs were subjected to SDS-PAGE (Fig. 2 ▶) and identified by MALDI-MS (Table 1 ▶). As previously reported, U11/U12 snRNPs contained all Sm proteins, as well as five subunits of the heteromeric splicing factor SF3b (i.e., SF3b155, SF3b145, SF3b130, SF3b49, and p14; Will et al. 1999, 2001). In addition, two newly identified SF3b subunits, namely, SF3b14b and SF3b10, which are present in 17S U2 snRNPs (Will et al. 2002), were also detected. However, U11/U12 snRNPs, like 17S U2, did not contain the SF3b-associated DEAD-box protein SF3b125. The U2-associated, DEAH-box protein hPrp43, was also found in the 18S U11/U12 di-snRNP, suggesting it may associate with a common component of U2 and U11/U12, such as one of the subunits of SF3b (Will et al. 2002). Purified U11/U12 snRNPs also contained two proteins associated with the major spliceosome, namely, YB-1 and Urp (U2AF35-related protein 2); the latter has been shown to associate with the U2AF65/U2AF35 heterodimer (Tronchere et al. 1997). Due to their apparent association with the U11/U12 snRNP, YB-1, hPrp43p, and Urp are good candidates for factors that also function in U12-type splicing.
FIGURE 2.
Protein composition of affinity-selected 18S U11/U12 snRNPs. Proteins were separated by SDS-PAGE on a 10%/13% poly-acrylamide gel and stained with Coomassie. The identities of the major proteins in the input snRNPs (lane 1) are indicated on the left; proteins in the affinity-selected U11/U12 snRNPs (lane 2), as determined by MS, on the right. U11/U12-associated proteins (as opposed to contaminating U5 proteins) are shown in bold, and the asterisk (*) indicates apparent SF3b145 and/or SF3b130 degradation products.
TABLE 1.
18S U11/U12 snRNP proteins
| Protein | Approximate MW (kDa) | Accession no. | Features |
| SF3b155 | 160 | gil6912654 | HEAT repeats |
| SF3b145 | 150 | gil5803155 | SAP, Pro-rich, Glu-rich |
| SF3b130 | 130 | gil11034823 | CPSF A |
| SF3b49 | 50 | gil2500587 | 2RRMs, His-rich, Pro-rich |
| p14/SF3b14a | 15 | gil15278118 | RRM |
| SF3b14b | 15 | gil10720333 | Cys-rich |
| SF3b10 | 9 | gil13775200 | |
| 65K | 65 | gil16553747 | 2RRM, Pro-rich |
| 59K (ES18) | 59 | gil22027541 | Pro-rich, Arg-rich, Glu-rich |
| 48K | 48 | gil33457355 | Arg-rich |
| 35K | 35 | gil5902144 | RRM, SR |
| 31K (MADP1) | 31 | gil21314767 | RRM, CCHC ZnF |
| 25K | 25 | gil13443018 | |
| 20K | 20 | gil9506863 | C2H2 ZnF, CCCH ZnF, Pro-rich |
| hPrp43 | 90 | gil13124667 | Arg-rich, His-rich, DEAH-box |
| Urp | 64 | gil4827046 | Glu-rich, RRM, 2X CCCH ZnF, SR |
| Y box-1 | 49 | gil6136595 | CSP, Arg-rich |
| SmB/B3 | 28 | gil5870129 | Sm |
| SmD3 | 17 | gil4759160 | Sm |
| SmD2 | 16 | gil29294624 | Sm |
| SmD1 | 15 | gil5902102 | Sm |
| SmE | 10 | gil4507129 | Sm |
| SmF | 9 | gil4507131 | Sm |
| SmG | 8 | gil4507133 | Sm |
Listed features are derived from Prosite Scan (http://hits.isb-sib.ch/cgi-bin/PFSCAN) and Pfam (protein families databases of alignments and HMMs; http://www.sanger.ac.uk/Software/Pfam/index.shtml). Accession number identifies the corresponding protein sequence in the GenBank at the NCBI. U5 snRNP proteins detected by MS are not listed. HEAT is (derived from Huntingtin protein, Elongation factor 3, the Alpha regulatory subunit of protein phosphatase 2A and yeast PI3-kinase TOR1): SAP indicates SAF-A/B, Acinus, and PIAS motif; CPSF A, cleavage and polyadenylation specificity factor A; RRM, RNA recognition motif; SR, Ser/Arg-rich domain; ZnF, zinc finger; and CSP, cold shock protein domain.
Seven additional proteins that are unique components of the U11/U12 snRNP (i.e., they have not been detected in major snRNPs or major spliceosomal complexes; Hartmuth et al. 2002; Jurica et al. 2002; Makarov et al. 2002; Rappsilber et al. 2002; Zhou et al. 2002) were also identified. These include proteins designated 65K, 59K, 48K, 35K, 31K, 25K, and 20K. With the exception of the 59K protein (also known as ES18), which has been implicated in apoptosis (Park et al. 1999), the newly identified U11/U12 proteins have, to date, no known function. Significantly, U11/U12 snRNPs did not contain U1-specific proteins (i.e., U1-A, U1-70K, or U1-C), indicating that protein interactions at U12-type 5′ splice sites are different from those in the major spliceosome.
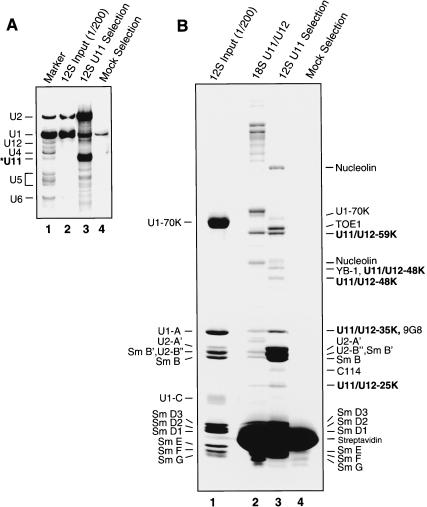
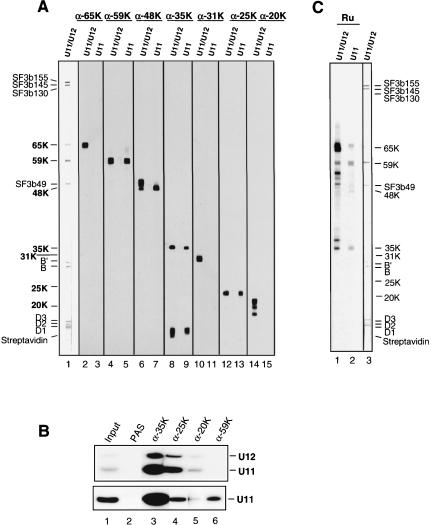
To determine which of the U11/U12 proteins are associated with U11 monoparticles, and thus likely involved in 5′ splice site recognition, the latter were affinity selected from the 12S region of the gradient (Fig. 1B ▶) by using a biotinylated 2′-O-methyl oligonucleotide complementary to U11 snRNA and streptavidin agarose beads. Bound material was eluted from the beads under denaturing conditions, and the RNA and protein composition of the affinity-selected snRNPs were analyzed by denaturing PAGE (Fig. 3A ▶) or SDS-PAGE (Fig. 3B ▶). Compared to the input 12S gradient fractions, which contained very large amounts of U1 and U2, the affinity-selected material was highly enriched in snRNPs containing U11 snRNA (Fig. 3A ▶, cf. lanes 2 and 3). However, due to an apparent cross-reaction of the anti-U11 oligonucleotide, the affinity-selected material also contained a large amount of 12S U2 snRNPs, but only low levels of U1 (Fig. 3A ▶, lane 3). SDS-PAGE analysis of the protein composition of the affinity-selected 12S U2 and U11 snRNPs, in comparison with 18S U11/U12 snRNPs, indicated that several 18S U11/U12 proteins, such as SF3b subunits and the 65K protein, were absent, whereas four U11/U12 proteins (i.e., 59K, 48K, 35K, and 25K) comigrated with proteins in the U11-enriched sample (Fig. 3B ▶, cf. lanes 2 and 3).
FIGURE 3.
Affinity selection of 12S U11 snRNPs. (A) snRNA composition of affinity-selected snRNPs. RNA was isolated from input 12S snRNPs (lane 2), or snRNPs affinity-selected in the presence (lane 3) or absence of anti-U11 oligonucleotide (Mock; lane 4). snRNAs were visualized by silver staining, and the identity of U11 was confirmed by Northern blotting (data not shown). (B) Proteins from 12S input snRNPs (lane 1), 18S U11/U12 snRNPs (lane 2), or 12S snRNPs affinity-selected in the presence (lane 3) or absence of anti-U11 oligonucleotide (lane 4) were analyzed as in Figure 2 ▶. The identities of the major proteins in the input 12S snRNPs are indicated on the left, and proteins in the affinity-selected U11-enriched snRNPs (lane 3), as determined by MS, are indicated on the right. U11-associated proteins (as opposed to U2 or U1 proteins) are shown in bold.
MALDI-MS and liquid chromatography-coupled tandem MS (LC-MSMS) were subsequently performed to precisely identify proteins in the U11-selected material (Table 2 ▶). In addition to known 12S U1 and U2 proteins, MS confirmed the presence of the 59K, 48K, 35K, and 25K U11/U12 proteins, as well as YB-1, the SR protein 9G8, TOE1 (a growth suppressor protein; De Belle et al. 2003), and C114, a double-stranded (ds) RNA-binding protein (Yin et al. 2003). As apparent orthologs of TOE1 and C114 are detected in organisms lacking U12-type introns (not shown) and both proteins localize predominantly in the nucleolus (De Belle et al. 2003; Yin et al. 2003), it is likely that these proteins are not bona fide U11 snRNP proteins. However, at present we cannot exclude that they perform multiple cellular functions, including roles in U12-type splicing. In contrast to the 59K, 48K, 35K, and 25K proteins, 65K, 31K, and 20K were not detected, suggesting that they associate solely with the 18S U11/U12 di-snRNP complex or additionally with the 15S U12 snRNP.
TABLE 2.
Proteins copurifying with 12S U11 snRNPs
| Protein | Approximate MW (kDa) | Accession no. | Features |
| U11/U12 protein | |||
| 59K (ES18) | 59 | gil22027541 | Pro-rich, Arg-rich, Glu-rich |
| 48K | 48 | gil33457355 | Arg-rich |
| 35K | 35 | gil5902144 | RRM, SR |
| 25K | 25 | gil13443018 | |
| SmB/B3 | 28 | gil5870129 | Sm |
| SmD3 | 17 | gil4759160 | Sm |
| SmD2 | 16 | gil29294624 | Sm |
| SmD1 | 15 | gil5902102 | Sm |
| SmE | 10 | gil4507129 | Sm |
| SmF | 9 | gil4507131 | Sm |
| SmG | 8 | gil4507133 | Sm |
| Splicing-related proteins | |||
| YB-1 | 49 | gil6136595 | CSP, Arg-rich |
| 9G8 | 35 | gil24415994 | RRM, CCHC ZnF, SR |
| Other proteins | |||
| TOE-1 | 62 | gil31543815 | CCCH ZnF, Arg-rich |
| C114 | 27 | gil13375901 | Lys-rich |
Listed features are derived from Prosite Scan (http://hits.isb-sib.ch/cgi-bin/PFSCAN) and Pfam (protein families databases of alignments and HMMs; http://www.sanger.ac.uk/Software/Pfam/index.shtml). Accession number identifies the corresponding protein sequence in the GenBank at the NCBI. Nucleolin, which is generally coselected from 12S gradient fractions and is thus a contaminant, and 12S U1 or 12S U2 proteins are not listed. ZnF indicates zinc finger; CSP, cold shock protein domain; RRM, RNA recognition motif; and SR, Ser/Arg-rich domain.
Domain structure of the novel U11/U12 proteins
A search of the nonredundant (nr) protein database at the National Center for Biotechnology Information (NCBI) using the peptide masses obtained by MS allowed the identification of apparently full-length proteins for each of the newly identified U11/U12-specific polypeptides. The predicted molecular weights of the identified proteins generally correlate with their apparent molecular weights. One exception is the 25K protein which, based on the predicted protein identified in the database, is 123 amino acids in length and has a molecular weight of only 14 kDa. However, careful inspection of the human genomic sequence and multiple ESTs, as well as comparisons with putative orthologs, indicated that the identified protein is full-length. Furthermore, in vitro translated protein from the identified 25K cDNA comigrates with the endogenous U11/U12 protein (data not shown).
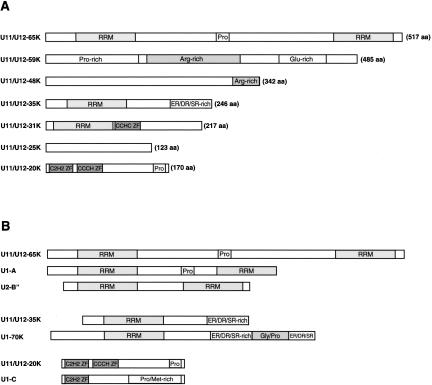
The domain structure of the newly identified proteins is shown in Figure 4A ▶. The presence of certain sequence motifs provides clues to possible functions of some of these proteins. For example, due to the presence of one or more RNA recognition motifs (RRMs), 65K, 35K, and 31K are good candidates for proteins that directly contact the U11 or U12 snRNA. Interestingly, several U11/U12 proteins share general structural features with proteins found in the U1 or U2 snRNP, the functional analogs of U11 and U12, respectively (Fig. 4B ▶). For example, as previously noted, the overall domain structure of the 35K protein is similar to that of the U1-70K protein (Will et al. 1999). Similarly, the general structures of the 20K and 65K proteins are reminiscent of the U1-C and U1-A/U2-B″ proteins, respectively, suggesting that these U11/U12 proteins may be functional analogs of their presumed U1 or U2 counterparts. These proteins also share a moderate level of homology. However, homologous regions are mainly limited to their structural motifs (i.e., RRMs or zinc fingers). Thus, the significance of this observation in terms of a possible evolutionary relationship is presently unclear.
FIGURE 4.
Domain structure of U11/U12-associated proteins. (A) Novel U11/U12 proteins are shown schematically with their number of amino acids (aa) at the right. (B) Comparison of the domain structure of U11/U12-65K with U1-A and U2-B″, U11/U12-35K with U1-70K, and U11/U12-20K with U1-C. RRM indicates RNA recognition motif; ZF, zinc finger; and Pro, proline-rich.
Homologs of the human U11 proteins appear to be missing in Drosophila
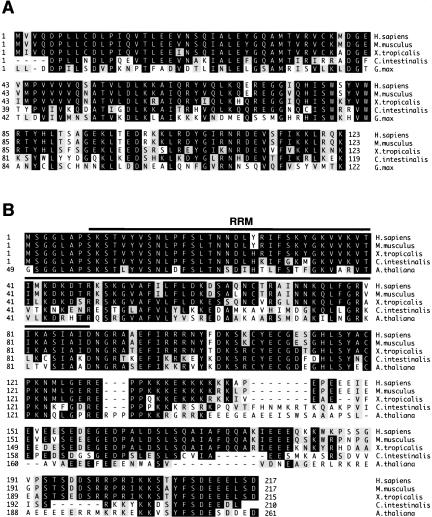
Homologs of most of the novel U11/U12 and/or U11 proteins were detected in a variety of organisms, including vertebrates, insects, and plants (Fig. 5 ▶; data not shown). Consistent with the fact that U12-type introns are not found in Saccharomyces cerevisiae, Schizosaccharomyces pombe, or Caenorhabditis elegans, homologs of the 65K, 59K, 48K, 35K, 31K, 25K, or 20K proteins were not detected by BLAST searches in these organisms. Most of the newly identified proteins are highly evolutionarily conserved. As examples, sequence alignments of two of the human U11/U12 proteins (i.e., 25K and 31K) with a subset of putative orthologs identified in other organisms are shown in Figure 5 ▶. The 25K protein shares 92% and 75% homology with its apparent orthologs in Xenopus tropicalis (frog) and Ciona intestinalis (sea squirt), respectively, and 62% homology with its putative Gycine max (soybean) ortholog. Similarly, the 31K protein shares 89% and 66% homology with its apparent orthologs in X. tropicalis and C. intestinalis, respectively, and 62% homology with its putative Arabidopsis thaliana ortholog. This high level of conservation is consistent with the idea that these proteins perform important cellular functions.
FIGURE 5.
U11/U12 proteins are evolutionarily highly conserved. (A) Amino acid sequence alignment of the Homo sapiens U11/U12-25K protein (gi|13443018) with putative orthologs from Mus musculus (gi|16973675), Xenopus tropicalis (gi|38225467), Ciona intestinalis (gi|24628436) and Glycine max (gi|26044609). The sequences of the X. tropicalis, C. intestinalis and G. max 25K proteins were deduced from EST sequences; note that the 5′ ends of the latter two proteins are apparently not complete. Residues identical in at least three sequences are boxed in black, and conserved residues (grey boxes) are grouped as follows: (D, E), (H, K, R), (A, F, I, L, M, P, V, W), and (C, G, N, Q, S, T, Y). (B) Amino acid sequence alignment of the human U11/U12-31K protein (gi|21314767) with putative orthologs from M. musculus (gi|21313088), X. tropicalis (gi|38394400), C. intestinalis (gi|19440299 and gi|19496343), and Arabidopsis thaliana (gi|15228279). The X. tropicalis and C. intestinalis sequences were generated from EST sequences. Note that the A. thaliana protein contains an N-terminal extension of 48 amino acids that is not included in the alignment. Residues are highlighted and grouped as in A. The position of the RNA recognition motif (RRM) in the 31K protein is indicated by a bar. Sequence alignments were preformed by using the Clustal method.
Intriguingly, we could not detect Drosophila homologs of the human U11-associated proteins (i.e., 59K, 48K, 35K, 25K) using highly sensitive BLAST searches of the Drosophila melanogaster genome; in contrast, homologs of the 35K protein could be identified in other insects and plants (data not shown). Significantly, putative orthologs of the human 65K and 20K proteins, which are found in 18S U11/U12 snRNPs but not in U11 monoparticles, could be identified in Drosophila (data not shown). The Drosophila proteins exhibit only moderate homology (42% for both 65K and 20K) with their human counterparts. Both zinc finger domains are conserved between the Drosophila and human 20K proteins. In contrast, the Drosophila 65K lacks an N-terminal RRM that is present in the human protein, suggesting that this RRM does not carry out a conserved function. These results are consistent with the presence of a highly divergent U11 snRNA in Drosophila (C. Schneider, C.L. Will, J. Brosius, M.J. Frilander, and R. Lührmann, in prep.) and suggest that protein interactions facilitating U12-type 5′ splice site recognition in Drosophila might be different.
Immunoblotting and immunoprecipitation studies
To characterize the novel U11/U12 proteins in more detail, we raised antibodies against each of them. Antibodies against the 65K, 59K, 48K, 35K, 31K, 25K, or 20K proteins reacted with the expected U11/U12 protein on immuno-blots containing U11/U12 proteins (Fig. 6A ▶, even lanes). Anti-35K antibodies also cross-reacted with one or more of the SmD proteins or streptavidin; however, affinity purification of these antibodies abolished this cross-reaction (data not shown). Interestingly, antibodies against the 48K and 20K proteins recognized multiple, slightly higher molecular-weight bands, suggesting that posttranslationally modified forms of these proteins might be present. Consistent with our MS data, immunoblotting confirmed the absence of the 65K, 31K, and 20K proteins in the U11-enriched snRNPs, and the presence of the 59K, 48K, 35K, and 25K proteins (Fig. 6A ▶, odd lanes). In the U11-enriched 12S snRNPs, the majority of the 48K protein recognized by anti-48K antibodies migrated as a single lower-molecular-weight band, suggesting that potentially modified forms of this protein may be present primarily in the 18S U11/U12 snRNP.
FIGURE 6.
The 12S U11 snRNP contains a subset of U11/U12 proteins. (A) Proteins from purified 18S U11/U12 snRNPs (lanes 1,2,4,6,8,10,12,14) or snRNPs enriched in 12S U11 sn-RNPs (lanes 3,5,7,9,11,13,15) were stained with Ponceau S (lane 1) or antibodies against the protein indicated at the top. The positions of the U11/U12 proteins are indicated on the left. (B) Immunoprecipitations were performed with immunoaffinity-purified, glycerol gradient-fractionated snRNPs from the 18S (top) or 12S (bottom) region of the gradient and immuno-affinity-purified antibodies against the 35K (lane 3), 25K (lane 4), 20K (lane 5), 59K (lane 6) protein, or PAS beads alone (lane 2). (Lane 1) 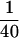 or
or 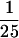 of the input 18S or 12S gradient fractions, respectively. Coprecipitation of the U11 and/or U12 snRNA was determined by Northern blotting. (C) Autoantigenic U11/U12 proteins. Proteins from purified 18S U11/U12 snRNPs (lanes 1,3) or snRNPs enriched in 12S U11 snRNPs (lane 2) were stained with Ponceau S (lane 3) or serum (Ru) from a patient suffering from diffuse systemic sclerosis (lanes 1,2).
of the input 18S or 12S gradient fractions, respectively. Coprecipitation of the U11 and/or U12 snRNA was determined by Northern blotting. (C) Autoantigenic U11/U12 proteins. Proteins from purified 18S U11/U12 snRNPs (lanes 1,3) or snRNPs enriched in 12S U11 snRNPs (lane 2) were stained with Ponceau S (lane 3) or serum (Ru) from a patient suffering from diffuse systemic sclerosis (lanes 1,2).
To provide direct evidence for the physical association of a given protein with the 18S U11/U12 snRNP and/or 12S U11 snRNP, immunoprecipitation studies were performed with affinity-purified antibodies against the 65K, 59K, 48K, 35K, 31K, 25K, or 20K proteins, and 18S or 12S glycerol gradient fractions containing spliceosomal snRNPs (Fig. 1 ▶). Coprecipitated snRNAs were subsequently separated by denaturing PAGE and visualized by Northern blotting (Fig. 6B ▶). Antibodies against the 31K, 48K, or 65K protein failed to precipitate U11/U12 or U11 snRNPs (data not shown), which may indicate that the epitopes recognized by these antibodies are not accessible in these particles. In contrast, precipitation of U11/U12 and U11 snRNPs was observed with antibodies against the 35K and 25K proteins, confirming their association with these snRNPs. Interestingly, anti-59K antibodies clearly precipitated 12S U11 snRNPs, but not 18S U11/U12 particles. One possible explanation for this loss of precipitation is that the region of the 59K protein recognized by this particular antibody (i.e., the C terminus) is no longer accessible upon formation of the U11/U12 di-snRNP, and thus may be located at the U11 and U12 snRNP interface. Only a very low level of U11/U12 and also U11 was precipitated by anti-20K antibodies. The latter is an unexpected result as both MS and immunoblotting failed to detect the 20K protein in the U11-enriched snRNPs; thus, precipitation of the U11 snRNP may be nonspecific or due to a cross-reaction of the anti-20K antibodies. These studies demonstrate a physical association of the 35K and 25K proteins with both U11/U12 and U11 snRNPs, and the 59K protein with the U11 snRNP.
Previous studies with serum from a patient suffering from diffuse systemic sclerosis indicated that one or more proteins associated with the U11 and U11/U12 snRNPs can serve as an autoantigen (Gilliam and Steitz 1993). In particular, this patient serum (designated Ru) contained auto-antibodies against a 65-kDa protein that appeared to be associated with the U11/U12 snRNP. To clearly define which of the U11/U12 proteins are autoantigenic, we performed immunoblotting with U11/U12 or U11-enriched snRNP proteins and Ru serum. The latter reacted very strongly with the 65K protein and, to a lesser extent, with the 59K and 35K proteins (Fig. 6C ▶, lanes 1,2; note that due to its extremely high anti-65K titer, a low level of 65K is detected in the U11-enriched sample). Thus, at least three U11/U12 components can serve as autoantigens in patients suffering from diffuse systemic sclerosis.
RNAi reveals essential cellular functions for novel U11/U12 proteins
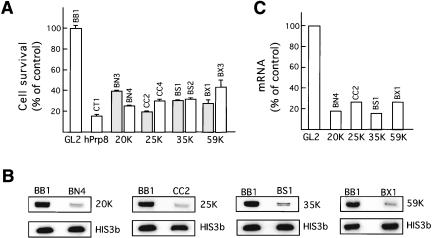
To investigate whether U11/U12-specific proteins carry out essential functions, RNAi experiments were performed with HeLa SS6 cells and 21-nt siRNA duplexes containing a 2-nt 3′ overhang (Elbashir et al. 2001). Multiple siRNA duplexes targeting either the coding region or 3′UTR of mRNAs encoding the 59K, 35K, 25K, or 20K proteins were tested for their ability to inhibit cell growth. Results obtained with two different duplexes that exhibited the greatest effect on cell viability after 72 h are shown in Figure 7A ▶; at the 72-h time point the majority of cells had not undergone apoptosis. siRNA duplexes directed against all of the tested U11/U12 proteins inhibited HeLa cell growth to a significant extent (ranging from ~60% to 80% inhibition) compared with that of the control cells treated with an siRNA duplex against luciferase (GL2). By comparison, knockdown of the essential spliceosomal protein hPrp8, which served as a positive control, led to an ~85% inhibition of cell growth.
FIGURE 7.
RNAi knockdowns reveal essential cellular functions for U11/U12 proteins. (A) Cell viability was determined 72 h after transfection of a given siRNA duplex (indicated above each box) and the average of triple determinations expressed as a percentage of the control knockdown (siRNA BB1, against GL2 luciferase). Results from two different siRNA duplexes are shown for each U11/U12 protein. As positive control, a knockdown was performed with an siRNA duplex against the hPrp8 protein. (B) RT-PCR analysis of 20K, 25K, 35K, and 59K mRNA in control versus knockdown cells. RT-PCR was performed with total cellular RNA and primers specific for the indicated mRNA. PCR products (73–113 base pairs in length) were separated by denaturing PAGE and PCR products were visualized by Southern blotting. (C) mRNA levels after RNAi knockdown were quantitated by using a PhosphorImager and expressed as a percentage of the control (GL2) value.
To demonstrate that the targeted protein had indeed been knocked down, we initially performed Western blotting with antibodies directed against each protein. However, due to the low abundance of the U11/U12 proteins in cellular extract and relatively weak antibodies at our disposal, it was not possible to determine the levels of the 59K, 35K, 25K, or 20K proteins in the control versus knockdown cells. As an alternative, we measured the amount of mRNA of the respective proteins by performing quantitative RT-PCR followed by Southern blotting. As an internal loading control, the level of histone 3B mRNA, which should not be affected by knockdown of U11/U12 proteins, was also determined by RT-PCR. For each protein, we initially determined the linear range of PCR amplification by varying the number of PCR cycles and also the amount of added cDNA. The level of mRNA encoding the 59K, 35K, 25K, or 20K proteins was substantially reduced in the respective knockdown cells, when compared to the control cells and normalized against the level of histone 3B mRNA (Fig. 7B ▶). Quantitation of the amount of each mRNA revealed a 70%–85% reduction in the various mRNAs (Fig. 7C ▶). Taken together, these results indicate that the 59K, 35K, 25K, or 20K proteins play essential roles in the cell, consistent with them functioning in U12-type splicing.
DISCUSSION
MS analyses revealed that the human U11/U12 snRNP lacks all U1-specific proteins but contains seven novel proteins not found in the major spliceosome. These studies thus demonstrate that multiple proteins are not shared by the major and minor spliceosome and suggest that unique molecular interactions facilitate 5′ splice site recognition and intron bridging in the U12-type prespliceosome. Compatible with them playing important roles in U12-type splicing, RNAi-mediated knockdowns demonstrated that several of the novel U11/U12 proteins are required for cell viability.
U11 and U1 snRNPs are structurally distinct
Four of the novel proteins present in U11/U12 snRNPs (i.e., 59K, 48K, 35K, and 25K) were also found in affinity-selected 12S snRNPs enriched in U11. Immunoprecipitation studies confirmed the association of the 59K, 35K, and 25K proteins with U11 monoparticles. Thus, the U11 snRNP is comprised of at least four novel proteins, three of which (59K, 48K, and 25K) bear no resemblance to the 70K, A, and C proteins found in the U1 snRNP. The general secondary structure of the U11 snRNA in vertebrates and plants (but not its sequence) mimics that of U1 (Montzka and Steitz 1988; C. Schneider, C.L. Will, J. Brosius, M.J. Frilander, and R. Lührmann, in prep.). Both can be folded into three stem–loops that form a four-way junction followed by the single-stranded Sm site flanked by a fourth stem–loop. The structure of the human U1 snRNP has been well characterized. The U1-70K and U1-A proteins bind the loop sequences of stem–loops I and II of the U1 snRNA, whereas U1-C associates via protein–protein contacts (for review, see Will and Lührmann 1997). In contrast to U1, the only region of U11 that is conserved among vertebrates and plants is a portion of loop III, making it the best candidate for a protein binding site (C. Schneider, C.L. Will, J. Brosius, M.J. Frilander, and R. Lührmann, in prep.). Furthermore, as only one of the U11-associated proteins (i.e., 35K) contains a motif known to mediate RNA binding, it is likely that only one of the stem–loop structures of the U11 snRNA is bound. These observations suggest that within the U11 snRNP, stem–loop III is bound by the 35K protein and that the remaining U11-proteins associate via protein contacts. Thus, the structural organization of the U11 snRNP is likely very different from that of the U1 snRNP. However, additional analyses of the structure of the U11 snRNP (e.g., chemical/enzymatic structure probing and electron microscopy) are needed to clarify this point.
Differences in 5′ splice site recognition in the minor versus major spliceosome
The identification of novel U11 proteins, coupled with the absence of U1-specific proteins, indicates that protein–protein and protein–RNA interactions at the 5′ splice site are not conserved between both spliceosomes. The association of the U1 snRNP with the 5′ splice site is facilitated by the U1-70K and U1-C proteins, which bind either directly or, in the case of the 70K protein, via interactions with SR proteins, to the 5′ splice site (for review, see Will and Lührmann 1997). Intriguingly, the 35K and 20K U11/U12 proteins exhibit structural similarities with the U1-70K and U1-C proteins, respectively (Fig. 4B ▶). Database searches with the human 35K protein indicate a high degree of homology between its RRM and that of the U1-70K protein. The 35K protein also has an SR-like domain, albeit much less pronounced than that of the U1-70K protein. Aside from suggesting a potential evolutionary relationship, such similarities also point to analogous functional roles for these proteins. In contrast to the 35K protein, several observations suggest that the 20K protein may have a function quite distinct from that of the U1-C protein in the major spliceosome. The N-terminal C2H2 zinc finger of 20K is homologous to that of the U1-C protein, but not significantly more so than C2H2 zinc fingers found in other proteins. 20K also contains a zinc finger of the CCCH type (not found in U1-C), suggesting it may carry out an additional/different function. Furthermore, 20K was not detected in our 12S U11 snRNP preparations, although it cannot be excluded that it was lost during the purification procedure. We are currently investigating which of the U11/U12 proteins contact the U12-type 5′ splice site by using site-specific crosslinking methods. These studies should clarify whether 35K and 20K are functional analogs of the U1-70K and U1-C proteins or whether other U11 proteins, such as 59K, 48K, or 25K, facilitate the interaction of the U11 snRNP with the 5′ splice site.
A unique U11 snRNP in Drosophila
D. melanogaster homologs of the human 59K, 48K, 35K, and 25K U11 snRNP proteins could not be identified by BLAST searches of the Drosophila genome, whereas homologs of the U11/U12-associated 65K and 20K proteins were found. Thus, the aforementioned proteins are either absent or so highly divergent that they escape detection by even highly sensitive BLAST searches. Consistent with these observations, the U11 snRNA in Drosophila shares little sequence similarity (aside from the 5′ splice site interacting region) with U11 from plants and vertebrates (C. Schneider, C.L. Will, J. Brosius, M.J. Frilander, and R. Lührmann, in prep.). Thus, a structurally unique U11 snRNP appears to be present in Drosophila, raising the interesting possibility that, in flies, protein interactions facilitating U12-type 5′ splice site recognition differ from those in most other organisms containing U12-type introns. Intriguingly, this apparent difference may be limited to Drosophila and not generally found in insects; that is, in contrast to Drosophila, homologs of the U11-associated 35K protein were identified in other insects such as A. gambiae (mosquito) and A. mellifera (honey bee).
Conservation of branch site recognition
Many aspects of branch site recognition appear to be conserved in both spliceosomes. The heteromeric complex SF3b, which contacts the pre-mRNA at or in the vicinity of the branch site and thereby stabilizes the U2/branch site interaction (Gozani et al. 1996), is also present in U11/U12 snRNPs. However, SF3a, which also contributes to U2-type branch site recognition, has, to date, not been detected in purified U11/U12 snRNPs. Although it is not clear whether it has dissociated at some step during U11/U12 purification, two observations suggest that this is not the case: (1) U11/U12 snRNPs in nuclear extract exhibit a sedimentation behavior identical to that of immunoaffinity-purified particles (Wassarman and Steitz 1992); and (2) affinity-selected U11/U12 snRNPs, when eluted under native conditions, still sediment as 18S particles (data not shown), suggesting they have not lost SF3a.
Currently, it is not entirely clear which of the novel U11/U12 proteins are present in the 15S U12 monoparticle and thus U12-associated. Based on its sedimentation coefficient, the 15S U12 snRNP likely contains the Sm proteins, as well as SF3b and the 65K protein. The latter proteins were not detected in U11-enriched snRNPs (Table 2 ▶), and in vitro binding studies indicate that 65K directly binds the U12 snRNA (H. Benecke, R. Lührmann, and C.L. Will, in prep). Similarly, U2 snRNPs that have lost SF3a, but contain SF3b, A′/B″ (which analogous to 65K directly contact the U2 snRNA), and the Sm proteins, also sediment as a 15S particle (Krämer et al. 1999). The 31K and 20K proteins were also not detected in purified U11 snRNPs, but whether they additionally associate with U12 monoparticles or solely with the U11/U12 di-snRNP remains to be established. Purification of the 15S U12 monoparticle might help answer these questions, but was not feasible due to the poor separation of 15S and 18S snRNPs on glycerol gradients.
Unique bridging interactions within the minor spliceosome
Previous studies indicated that U11/U12 di-snRNP formation is mediated by protein–protein interactions rather than RNA–RNA base-pairing (Wassarman and Steitz 1992). Thus, aside from facilitating interactions between U11 and the 5′ splice site, and U12 and the branch site, U11/U12 proteins also function in di-snRNP complex formation. Furthermore, due to the simultaneous binding of U11/U12 with the 5′ splice site and branch site, bridging of the reactive groups of the pre-mRNA within the prespliceosome must also be mediated by components of the U11/U12 di-snRNP itself. Although the precise nature of the molecular bridges responsible for juxtaposing the 5′ and 3′ ends of the intron in the major and minor prespliceosomes is not known, the unique composition of the U11 snRNP and presence of additional novel proteins in U11/U12 indicates that at least some (if not all) intron bridging interactions are not conserved. The DEAD-box protein Prp5 appears to bridge the U1 and U2 snRNPs in the major spliceosome (Xu et al. 2004). However, Prp5p was not detected in purified U11/U12 di-snRNPs, and thus, it does not appear to contribute to bridging interactions in the minor spliceosome.
Evolutionary relationships between the major and minor spliceosomes
The identification of several novel proteins in the human U11/U12 snRNP provides additional insight into possible evolutionary relationships between the major and minor spliceosomes. Several models for the evolutionary relationship between both spliceosomes have been proposed (Burge et al. 1998). Previous studies suggested that the majority of spliceosomal proteins are shared between both systems. Indeed, most if not all of the proteins found in the major U4/U6.U5 tri-snRNP are also present in the minor U4atac/U6atac.U5 and all subunits of the heteromeric splicing factor SF3b are found both in U2 and U11/U12 snRNPs (Will et al. 1999, 2001; Schneider et al. 2002). These results strongly argue for a homologous origin (i.e., common ancestry) of both spliceosomes. In contrast, several U11/U12 proteins, in particular those likely involved in 5′ splice site recognition and also di-snRNP formation (and thus intron bridging) are clearly distinct from proteins found in the major spliceosome. Likewise, the sequences of the U11 and U12 snRNAs are not detectably similar to those of U1 and U2, and U4atac and U6atac share only limited sequence homology with U4 and U6, respectively (Tarn and Steitz 1996a; Montzka and Steitz 1988). These results can be explained by a nonhomologous origin of the snRNAs, such as the parasitic invasion model (Burge et al. 1998), or by a homologous origin coupled with a very strong divergence of selected components of each spliceosome. The presence of highly divergent snRNAs and proteins, as well as multiple identical spliceosomal proteins, in both systems is most consistent with the fission/fusion model initially proposed by Burge et al. (1998). In this model, divergence of snRNAs and spliceosomal proteins initially occurred in two separate lineages (presumably after speciation), each with only one type of spliceosome. At some point, both lineages fused (e.g., by endosymbiosis) resulting in an eukaryotic ancestor containing both types of spliceosomes. Those proteins that had not diverged significantly (e.g., SF3b, and many tri-snRNP proteins) became redundant such that only one version was ultimately retained in the genome.
Of proteins known to be associated with the minor snRNPs and/or spliceosome, U11-associated proteins appear to have diverged most significantly. Thus, although the mechanism of branch site recognition appears to be largely conserved, differences in 5′ splice site recognition as well as intron bridging, likely have led to the extensive divergence of proteins involved in these processes. Alternative splicing events are largely determined during 5′ splice site recognition, and the U2-type spliceosome is characterized by its ability to be highly regulated at this stage. The independent binding of U1 and U2 to U2-type introns allows for more flexibility in splice site pairing compared with that in the minor spliceosome. Although apparent examples of alternatively spliced U12-type introns have been described, alternative splicing events involving the U12-type spliceo-some seem to be very limited (Levine and Durbin 2001). Future characterization of those proteins uniquely associated with the U11/U12 snRNP may shed more light on interesting mechanistic differences between the major and minor spliceosome.
MATERIALS AND METHODS
Affinity selection of 18S U11/U12 snRNPs or 12S U11 snRNPs
UsnRNPs were immunoaffinity purified from HeLa nuclear extract with anti-m3G antibodies and subsequently separated on 10%–30% glycerol gradients (Will et al. 1999). 18S or 12S gradient fractions were pooled, and 18S U11/U12 or 12S U11 snRNPs, respectively, were affinity selected with a biotinylated 2′-O-methyl-RNA oligonucleotide complementary to nucleotides 11–28 of human U12 snRNA or to nucleotides 2–18 of U11 snRNA (Will et al. 1999). RNA and protein were recovered under denaturing conditions and analyzed on 10% polyacrylamide/7 M urea gels (RNA) or by 10%/13% SDS-PAGE (proteins).
Immunological techniques
To generate antibodies against the U11/U12 65K, 59K, 48K, 35K, 31K, 25K, or 20K proteins, rabbits were immunized with the following peptides: 65K, CSARPKQDPKEGKR (Pep5.4); 59K, CDWDQYLVPSDHPKGN (Pep7.3); 48K, KIPSITLNKDSQFQIC (Pep9.1); 35K, CEDRKKLRDYGIRNRD (Pep10.2); 31K, CRIKKS TYFSDEEELSD (MADP.2); 25K, CNRDEVSFIKKLRQK (Pep16.3); and 20K, CPVQELPPSLRAPPPG (Pep17.3). For immunoblotting, 18S U11/U12 or 12S U11 proteins were fractionated by SDS-PAGE, transferred electrophoretically to nitrocellulose, and immunostained by using an ECL-Detection Kit (Amersham). For immunoprecipitation, antibodies (with the exception of anti-48K) were affinity purified by using a SulfoLink column (Pierce) containing the respective immobilized peptide. Immunoprecipitations were performed with gradient-fractionated UsnRNPs, and precipitated RNAs were visualized by Northern blotting (Schneider et al. 2002).
MS and database searches
U11/U12 and U11 proteins (separated by SDS-PAGE) were analyzed by MALDI-MS or LC-MSMS and identified in the NCBI nonredundant database by using Mascot as a search engine (Hartmuth et al. 2002). BLAST/BLAT database searches were performed by using the following: http://www.genome.ucsc.edu/, http://www.fruitfly.org/, and http://www.ncbi.nlm.nih.gov/entrez/.
RNAi and RT-PCR
siRNA oligonucleotides were synthesized in house by using 5′-silyl, 2′-ACE phosphoramidites (Dharmacon) and annealed as described (Elbashir et al. 2001). The following 21-nt siRNA duplexes with a 2-nt (dT) 3′ overhang were used (only sense strand is shown):
20K 3′ UTR (BN3): GCCCGGUUCCUGCUACGCC-dTdT;
20K 3′ UTR (BN4): GGCUCCGAGACCAUCUGCC-dTdT;
25K 3′ UTR (CC2): AUCAUCGUGCCUCUUUCAC-dTdT;
25K ORF (CC4): AGGACGUUGUGGUGGCCUC-dTdT;
35K ORF (BS1): GGGUGGAUCCCUCGGCGAC-dTdT;
35K 3′ UTR (BS2): GGCCCAACAGCAGAACCCC-dTdT;
59K ORF (BX1): GCAGCCGCUGAUGGCGUAC-dTdT;
59K 3′ UTR (BX3): GGUGAACUGAGGUUUUUAC-dTdT;
hPrp8 ORF (CT1): GCCCAUCAACGGAGCCAUC-dTdT; and
Luciferase (BB1): CGUACGCGGAAUACUUCGA-dTdT.
The targeted regions were not found via BLAST search of the human genome in any other known genes. Hela SS6 cells were cultured as described previously, and transfections with a given siRNA duplex were performed with Oligofectamine (Life Technologies; Harborth et al. 2001). Cells were harvested after 72 h, and apoptosis was assayed by using a Fluorescein In Situ Cell Death Detection kit (Roche). Total cellular RNA was isolated by using an RNeasy kit (Qiagen), and digested with RQ1 DNase (Promega) to remove contaminating DNA. cDNAs were synthesized by using a SuperScript II Reverse Transcription kit (Invitrogen). Subsequent PCR was performed with Pfu polymerase for 22 cycles (all U11/U12 proteins) or 16 cycles (histone 3B mRNA). The linear range of amplification was determined for each set of primers by varying the cycle number and amount of cDNA. The following PCR primers were used:
20Kfor (exon 1): CTTCCAGGACAACCTCCACAA;
20Krev (exon 1): CTCGGAACATGTCGTACCAGAC;
25Kfor (exon 1): GCAAGAGGAAGATGAGGACGA;
25Krev (exon 1): CTGCACCACCATAGCCAGAC;
35Kfor (exon): GCTACGCCTTCATCGAATACAA;
35Krev (exon): AATAACCAGGCCATCAGCATC;
59Kfor (exon 2): CGCTGATGGCGTACTATCTGAA;
59Krev (exon 2): AAGCCCGTAGAATGTCCACCA;
His3Bfor (exon 3): GCTTCGAGAGATTCGTCGTT; and
His3Brev (exon 3): GAAACCTCAGGTCGGTTTTG.
Southern blots
DNA was fractionated on a 10% polyacrylamide/7 M urea gel and transferred electrophorectically to nylon membrane. Prehybridization, hybridization, and wash conditions were as described previously (Schneider et al. 2002). 32P-labeled probes against histone 3B, 59K, 35K, 25K, and 20K RT-PCR products were generated from PCR fragments of the respective gene, by random priming using a Prime It II kit (Stratagene).
Acknowledgments
We are grateful to G. Heyne for excellent technical assistance, H. Manninga for synthesizing RNA oligonucleotides, and P. Kempkes, B. Hildebrandt, and H. Kohansal for preparing nuclear extract and total UsnRNPs. We thank J. Steitz for kindly providing Ru patient serum, G. Neubauer for initial MS analyses, T. Doerks for help with database searches, and Heike Benecke for critically reading the manuscript. EST clones were kindly provided by the Resource Center of the German Genome Project at the MPI for Molecular Genetics, as well as the IMAGE cDNA Clone Consortium. This work was supported by grants from the DFG (LU294/12-1), BMBF (031U215B) and Fonds der Chemischen Industrie to R.L.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.rnajournal.org/cgi/doi/10.1261/rna.7320604.
REFERENCES
- Burge, C.B., Padgett, R.A., and Sharp, P.A. 1998. Evolutionary fates and origins of U12-type introns. Mol. Cell 2: 773–785. [DOI] [PubMed] [Google Scholar]
- De Belle, I., Wu, J.X., Sperandio, S., Mercola, D., and Adamson, E.D. 2003. In vivo cloning and characterization of a new growth suppressor protein TOE1 as a direct target gene of Egr1. J. Biol. Chem. 278: 14306–14312. [DOI] [PubMed] [Google Scholar]
- Elbashir, S.M., Harborth, J., Lendeckel, W., Yalcin, A., Weber, K., and Tuschl, T. 2001. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature 411: 494–498. [DOI] [PubMed] [Google Scholar]
- Frilander, M.J. and Steitz, J.A. 1999. Initial recognition of U12-dependent introns requires both U11/5′ splice-site and U12/branchpoint interactions. Genes & Dev. 13: 851–863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ———. 2001. Dynamic exchanges of RNA interactions leading to catalytic core formation in the U12-dependent spliceosome. Mol. Cell 7: 217–226. [DOI] [PubMed] [Google Scholar]
- Gilliam, A.C. and Steitz, J.A. 1993. Rare scleroderma autoantibodies to the U11 small nuclear ribonucleoprotein and to the trimethylguanosine cap of U small nuclear RNAs. Proc. Natl. Acad. Sci. 90: 6781–6785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gozani, O., Feld, R., and Reed, R. 1996. Evidence that sequence-independent binding of highly conserved U2 snRNP proteins upstream of the branch site is required for assembly of spliceosomal complex A. Genes & Dev. 10: 233–243. [DOI] [PubMed] [Google Scholar]
- Hall, S.L. and Padgett, R.A. 1996. Requirement of U12 snRNA for in vivo splicing of a minor class of eukaryotic nuclear pre-mRNA introns. Science 271: 1716–1718. [DOI] [PubMed] [Google Scholar]
- Harborth, J., Elbashir, S.M., Bechert, K., Tuschl, T., and Weber, K. 2001. Identification of essential genes in cultured mammalian cells using small interfering RNAs. J. Cell. Sci. 114: 4557–4565. [DOI] [PubMed] [Google Scholar]
- Hartmuth, K., Urlaub, H., Vornlocher, H.P., Will, C.L., Gentzel, M., Wilm, M., and Lührmann, R. 2002. Protein composition of human prespliceosomes isolated by a tobramycin affinity-selection method. Proc. Natl. Acad. Sci. 99: 16719–16724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hastings, M.L. and Krainer, A.R. 2001. Functions of SR proteins in the U12-dependent AT-AC pre-mRNA splicing pathway. RNA 7: 471–482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Incorvaia, R. and Padgett, R.A. 1998. Base pairing with U6atac snRNA is required for 5′ splice site activation of U12-dependent introns in vivo. RNA 4: 709–718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurica, M.S., Licklider, L.J., Gygi, S.R., Grigorieff, N., and Moore, M.J. 2002. Purification and characterization of native spliceosomes suitable for three-dimensional structural analysis. RNA 8: 426–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolossova, I. and Padgett, R.A. 1997. U11 snRNA interacts in vivo with the 5′ splice site of U12-dependent (AU-AC) pre-mRNA introns. RNA 3: 227–233. [PMC free article] [PubMed] [Google Scholar]
- Krämer, A., Gruter, P., Groning, K., and Kastner, B. 1999. Combined biochemical and electron microscopic analyses reveal the architecture of the mammalian U2 snRNP. J. Cell. Biol. 145: 1355–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine, A. and Durbin, R. 2001. A computational scan for U12-dependent introns in the human genome sequence. Nucleic Acids Res. 29: 4006–4013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo, H.R., Moreau, G.A., Levin, N., and Moore, M.J. 1999. The human Prp8 protein is a component of both U2- and U12-dependent spliceosomes. RNA 5: 893–908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarov, E.M., Makarova, O.V., Urlaub, H., Gentzel, M., Will, C.L., Wilm, M., and Lührmann, R. 2002. Small nuclear ribonucleoprotein remodeling during catalytic activation of the spliceosome. Science 298: 2205–2208. [DOI] [PubMed] [Google Scholar]
- Montzka, K.A. and Steitz, J.A. 1988. Additional low-abundance human small nuclear ribonucleoproteins: U11, U12, etc. Proc. Natl. Acad. Sci. 85: 8885–8889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nottrott, S., Urlaub, H., and Lührmann, R. 2002. Hierarchical, clustered protein interactions with U4/U6 snRNA: A biochemical role for U4/U6 proteins. EMBO J. 21: 5527–5538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park, E.J., Kim, J.H., Seong, R.H., Kim, C.G., Park, S.D., and Hong, S.H. 1999. Characterization of a novel mouse cDNA, ES18, involved in apoptotic cell death of T-cells. Nucleic Acids Res. 27: 1524–1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel, A.A. and Steitz, J.A. 2003. Splicing double: Insights from the second spliceosome. Nat. Rev. Mol. Cell. Biol. 4: 960–970. [DOI] [PubMed] [Google Scholar]
- Rappsilber, J., Ryder, U., Lamond, A.I., and Mann, M. 2002. Large-scale proteomic analysis of the human spliceosome. Genome Res. 12: 1231–1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed, R. 1996. Initial splice-site recognition and pairing during pre-mRNA splicing. Curr. Opin. Genet. Dev. 6: 215–220. [DOI] [PubMed] [Google Scholar]
- Reed, R., and Palandjian, L. 1997. Spliceosome assembly. In Eukaryotic mRNA processing (ed. A.R. Krainer), pp. 103–129. IRL Press, Oxford, UK.
- Schneider, C., Will, C.L., Makarova, O.V., Makarov, E.M., and Lührmann, R. 2002. Human U4/U6.U5 and U4atac/U6atac.U5 tri-snRNPs exhibit similar protein compositions. Mol. Cell. Biol. 22: 3219–3229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwer, B. 2001. A new twist on RNA helicases: DExH/D box proteins as RNPases. Nat. Struct. Biol. 8: 113–116. [DOI] [PubMed] [Google Scholar]
- Tarn, W.Y. and Steitz, J.A. 1996a. Highly diverged U4 and U6 small nuclear RNAs required for splicing rare AT-AC introns. Science 273: 1824–1832. [DOI] [PubMed] [Google Scholar]
- ———. 1996b. A novel spliceosome containing U11, U12, and U5 snRNPs excises a minor class (AT-AC) intron in vitro. Cell 84: 801–811. [DOI] [PubMed] [Google Scholar]
- Tronchere, H., Wang, J., and Fu, X.D. 1997. A protein related to splicing factor U2AF35 that interacts with U2AF65 and SR proteins in splicing of pre-mRNA. Nature 388: 397–400. [DOI] [PubMed] [Google Scholar]
- Wassarman, K.M. and Steitz, J.A. 1992. The low-abundance U11 and U12 small nuclear ribonucleoproteins (snRNPs) interact to form a two-snRNP complex. Mol. Cell. Biol. 12: 1276–1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Will, C.L. and Lührmann, R. 1997. Protein functions in pre-mRNA splicing. Curr. Opin. Cell. Biol. 9: 320–328. [DOI] [PubMed] [Google Scholar]
- Will, C.L., Schneider, C., Reed, R., and Lührmann, R. 1999. Identification of both shared and distinct proteins in the major and minor spliceosomes. Science 284: 2003–2005. [DOI] [PubMed] [Google Scholar]
- Will, C.L., Schneider, C., MacMillan, A.M., Katopodis, N.F., Neubauer, G., Wilm, M., Lührmann, R., and Query, C.C. 2001. A novel U2 and U11/U12 snRNP protein that associates with the pre-mRNA branch site. EMBO J. 20: 4536–4546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Will, C.L., Urlaub, H., Achsel, T., Gentzel, M., Wilm, M., and Lührmann, R. 2002. Characterization of novel SF3b and 17S U2 snRNP proteins, including a human Prp5p homolog and an SF3b DEAD-box protein. EMBO J. 21: 4978–4988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, Y.Z., Newnham, C.M., Kameoka, S., Huang, T., Konarska, M.M., and Query, C.C. 2004. Prp5 bridges U1 and U2 snRNPs and enables stable U2 snRNP association with intron RNA. EMBO J. 23: 376–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin, Z., Haynie, J., Williams, B.R., and Yang, Y.C. 2003. C114 is a novel IL-11-inducible nuclear double-stranded RNA-binding protein that inhibits protein kinase R. J. Biol. Chem. 278: 22838–22845. [DOI] [PubMed] [Google Scholar]
- Yu, Y.T. and Steitz, J.A. 1997. Site-specific crosslinking of mammalian U11 and U6atac to the 5′ splice site of an AT-AC intron. Proc. Natl. Acad. Sci. 94: 6030–6035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, Z., Licklider, L.J., Gygi, S.P., and Reed, R. 2002. Comprehensive proteomic analysis of the human spliceosome. Nature 419: 182–185. [DOI] [PubMed] [Google Scholar]