Abstract
In bacteria, translation of all the ribosomal protein cistrons in the spc operon mRNA is repressed by the binding of the product of one of them, S8, to an internal sequence at the 5′ end of the L5 cistron. The way in which the first two genes of the spc operon are regulated, retroregulation, is mechanistically distinct from translational repression by S8 of the genes from L5 onward. A 2.8 Å resolution crystal structure has been obtained of Escherichia coli S8 bound to this site. Despite sequence differences, the structure of this complex is almost identical to that of the S8/helix 21 complex seen in the small ribosomal subunit, consistent with the hypothesis that autogenous regulation of ribosomal protein synthesis results from conformational similarities between mRNAs and rRNAs. S8 binding must repress the translation of its own mRNA by inhibiting the formation of a ribosomal initiation complex at the start of the L5 cistron.
Keywords: RNA-protein complex, X-ray crystallography, ribosomal, autoregulation
INTRODUCTION
The synthesis of many bacterial ribosomal proteins is regulated at the translational level by systems that prevent the accumulation of ribosomal proteins in the cytoplasm un-associated with rRNA (Zengel and Lindahl 1994). In bacteria, the genes for many ribosomal proteins are components of operons that contain the genes of several other ribosomal proteins. Translation of all the ribosomal proteins encoded by the mRNA produced from each such operon is inhibited by mRNA interactions with just one of them (Dean et al. 1981).
In the late 1970s, it was proposed that autoregulatory ribosomal proteins bind their own mRNAs the same way they bind to rRNA in the ribosome because their mRNAs and rRNAs have locally similar three-dimensional structure (Fallon et al. 1979; Lindahl and Zengel 1979; Fiil et al. 1980; Nomura et al. 1980). This idea was attractive because if the RNA-binding activity that enables these proteins to participate in ribosome assembly also causes translational regulation, competition between rRNAs and ribosomal protein mRNAs will guarantee that the production of ribosomal proteins decreases when supply exceeds demand. Biochemical and genetic data obtained subsequently support this proposal for some ribosomal protein operons, but not all of them (Zengel and Lindahl 1994), and now that crystal structures have been obtained for both ribosomal subunits, it can be tested by direct comparison of the structures of the complexes formed by regulatory ribosomal proteins with their mRNAs and their rRNAs.
In Escherichia coli, the spc operon encodes 11 ribosomal proteins and sec Y, a protein involved in secretion (Zengel and Lindahl 1994). The translation of its mRNA is regulated by ribosomal protein S8, which is encoded by its fifth cistron. Genetic experiments have demonstrated that the regulatory sequence in the spc operon includes the 5′ end of the L5 cistron (Cerretti et al. 1988; Gregory et al. 1988), which is the third cistron in the operon. In the absence of S8, the stem/loop this sequence forms is compatible with normal translation, but when S8 binds, translation of L5 and the other ribosomal protein cistrons in the spc operon mRNA is repressed. The genes on the 5′ side of the regulatory site are retroregulated, and those on its 3′ side are repressed by translational coupling (Dean et al. 1981; Mattheakis and Nomura 1988; Mattheakis et al. 1989). It is not known whether S8 regulates the translation of the two most distal genes in the operon, secY and L36 (Zengel and Lindahl 1994).
Crystal structures are available for Bacillus stearothermophilus S8 and for Thermus thermophilus S8, and the structure of Methanococcus jannaschii S8 has been solved bound to a fragment of 16S rRNA (Davies et al. 1996; Nevskaya et al. 1998; Tishchenko et al. 2001). In addition, the structure of S8 is known in the intact 30S ribosomal subunit from T. thermophilus (Wimberly et al. 2000). However, there are no direct structural data available on the conformation of the mRNA sequence to which S8 binds, either in isolation or with S8 bound.
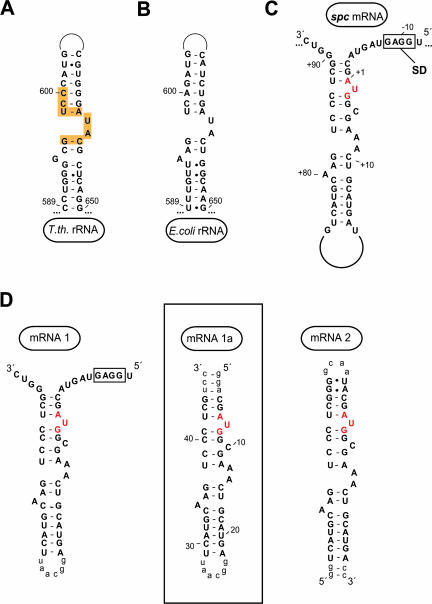
Biochemical data suggest that S8 binds to the spc operon mRNA the way it binds to 16S rRNA, consistent with the standard model for autogenous regulation (Cerretti et al. 1988; Gregory et al. 1988; Wu et al. 1994). In the ribosome, S8 binds to a site that includes sequences from helices 19, 20, 21, 25, and 26a of 16S rRNA (Brodersen et al. 2002), but biochemical experiments have shown that the interaction with helix 21 is energetically dominant (Mougel et al. 1986). The secondary structures of the relevant region of helix 21 in the 16S rRNAs from T. thermophilus (Wimberly et al. 2000) and E. coli (Kalurachchi et al. 1997) are compared in Figure 1, A and 1B ▶. The S8-binding sequence in the spc operon mRNA (Gregory et al. 1988; Mattheakis and Nomura 1988) appears capable of adopting a similar secondary structure (Fig. 1C ▶), but the conformational implications of the sequence differences between helix 21 of 16S rRNA and the region of the spc operon mRNA hypothesized to be its mimic are unknown.
FIGURE 1.
Secondary structure of RNA molecules to which ribosomal protein S8 binds. (A,B) Helix 21 of 16S rRNA from T. thermophilus and E. coli, respectively. Yellow regions indicate S8 contacts within the T. thermophilus 30S (Brodersen et al. 2002). (C) Wild-type regulatory site in the spc operon mRNA from E. coli. The start codon for the L5 gene is shown in red, and the Shine-Dalgarno region is boxed. (D) mRNA constructs used in this study. Lowercase nucleotides were added for stability or transcriptional purposes and are not part of the wild-type sequence. The boxed construct was crystallized in complex with E. coli S8.
Here we present a crystal structure for E. coli S8 bound to an RNA representing the autogenous regulation site within the E. coli spc operon mRNA. It demonstrates that for the S8/spc operon system, the site to which the regulatory ribosomal protein binds in its mRNA is indeed similar to the site that it binds in 16S rRNA, despite its sequence differences. Unexpectedly, the two “extra” nucleotides in the stem of the mRNA-binding site, which are known to reduce its affinity for S8 (Fig. 1D ▶; U7, C10; Wu et al. 1994), have virtually no effect on the conformation of the parts of the RNA that interact with S8. Modeling studies suggest that the 30S ribosomal subunit is unlikely to form a productive initiation complex with the spc operon message when S8 is bound.
RESULTS
Identification of a spc mRNA–S8 complex suitable for structure determination
Several RNA constructs that included the spc mRNA stem were synthesized and tested for their S8-binding activity by using both gel shift and filter binding assays. mRNA2 (Fig. 1D ▶) was used to bench mark these experiments because its S8-binding properties had been reported previously (Wu et al. 1994). mRNA1 is an extended version of mRNA2 that includes the L5 Shine-Dalgarno sequence, but although its stem sequence is the same as that of mRNA2, its closing loop is necessarily at the other end of the molecule, and thus, mRNA1 is closer to the intact spc operon mRNA sequence topologically (Fig. 1C ▶) than is mRNA2. mRNA1a is a truncated version of mRNA1.
As Table 1 ▶ shows, mRNA1 and mRNA1a bind S8 with similar affinity, confirming that the L5 Shine-Dalgarno sequence does not contribute to the interaction of S8 with spc operon mRNA (Cerretti et al. 1988; Wu et al. 1994). Unexpectedly, however, both mRNA1 and mRNA1a bind S8 about an order of magnitude more tightly than that of mRNA2, indicating that the placement of the terminal loop does influence binding. In addition, binding affinities of mRNA1 and mRNA1a for S8 are only about fivefold less than that of 16S rRNA or fragments of 16S rRNA large enough to include all the sequences that interact with S8 in the ribosome. mRNA1a was used for the crystallographic studies described below because gel shift, filter binding, gel filtration, and dynamic light scattering experiments all demonstrated that it forms stable 1:1 complexes with S8 (data not shown).
TABLE 1.
S8 Binding by rRNA and mRNA constructs
| RNA | Kd(μM) | Reference |
| spc operon | 0.2 | Gregory et al. 1988 |
| mRNA2 | 4.2 ± 1.1 | This study |
| mRNA2 | 1.0 | Wu et al. 1994 |
| mRNA1 | 0.10 ± 0.1 | This study |
| mRNA1a | 0.16 ± 0.1 | This study |
| 16S rRNA | 0.02–0.04 | Schwarzenbauer and Craven 1981; Mougel et al. 1986; Gregory et al. 1988 |
| 16S rRNA frag | 0.04 | Gregory et al. 1988 |
Determination of the crystal structure of the S8/mRNA1a complex
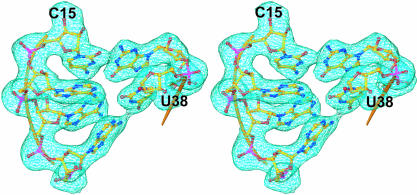
Crystals of the complex of S8 and mRNA1a were grown at 23°C in hanging drops with a well solution consisting of 100 mM Zn acetate, 100 mM Na acetate buffer, and 7% PEG 8000 (pH 5.2). The crystals belong to the space group P3221 and have unit cell dimensions of a = b = 61.2 Å, c = 332.5 Å, and γ = 120. Although these crystals were twinned, as could be demonstrated by analyzing the distribution of diffraction intensities (Yeates 1997), the diffraction data they yielded could be detwinned computationally. Phases were determined by molecular replacement using a model for the S8/mRNA1a complex derived from the crystal structure of the T. thermophilus 30S ribosomal subunit (Wimberly et al. 2000). Omit-map electron density for a few of the RNA bases in the middle of the S8-binding site in mRNA1a is shown in Figure 2 ▶. The E. coli sequence was built into electron-density maps and refined to a final free R-factor of 27.3% (Table 2 ▶). 〈I/σ〉decreases to 2.0 at a resolution of 2.58 Å in the merged data set, but the nominal resolution of the resulting structure is lower than that, 2.79 Å, because of the way error propagates during detwinning.
FIGURE 2.
Typical electron-density. Stereoview depicting typical electron density from a composite omit map contoured to 1.0σ at a resolution limit of 2.7 Å. The region of mRNA1a shown is a portion of the S8-binding site.
TABLE 2.
Data collection and refinement statistics for detwinned data
| Space group | P3221 |
| Unit cell | |
| a = b | 61.2 |
| c | 332.5 |
| Resolution (Å)a | 50–2.7 (2.8–2.7) |
| Reflectionsa | 832,569 |
| Unique reflectionsa | 38,349 (3907) |
| Completeness (%)a | 99.9 (100) |
| Average l/σ (l)a,b | 37.5 (2.1) |
| Redundancya | 21.7f |
| Rsym (%)c | 8.7 |
| Effect of detwinning on l/σ (l) for l/σ (l) = 2.0 | |
| Highest resolution bin detwinned data | 2.82–2.79 Å |
| Refinement statistics | |
| Resolution (Å) | 50–2.7 (2.85–2.7) |
| Reflections (free)a | 19,878 (2817) |
| Rcrystal (%)d | 23.7 (42.1) |
| Rfree (%)e | 27.3 (45.1) |
| Rms | |
| Bonds (Å) | 0.016 |
| Angles (°) | 2.331 |
| Non-hydrogen atoms | 3,951 |
| Average B-factor (Å2) | 44.6 |
aNumbers in parentheses for the highest resolution shell.
bl/σ (l) is the mean reflection intensity/estimated error.
cRsym = ∑|l-〈l〉|∑l, where l is the intensity of an individual reflection and 〈l〉is the average intensity over symmetry equivalents.
dRcrystal = ∑||Fo| − |Fc||/∑|Fo, where Fo and Fc are the observed and calculated structure factor amplitudes.
eRfree is equivalent to Rcrystal but calculated for a randomly chosen set of reflections that were omitted from the refinement process.
fHigh redundancy due to merging of three data sets at different wavelengths with full anamolous: λpeak = 1.0049 Å, λinflection = 1.0089 Å, λremote = 0.9928 Å.
Overview of the complex
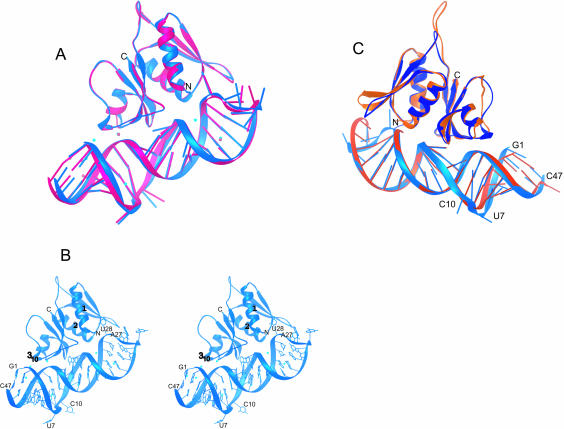
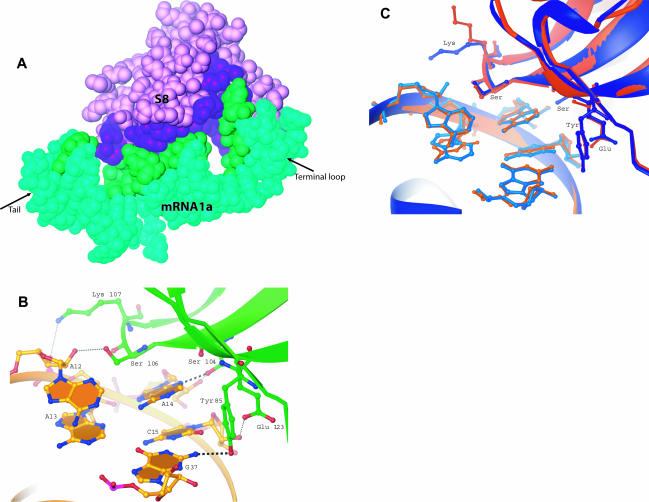
The crystals of S8/mRNA1a contain two similar, but nonidentical copies of the complex (copies A and B, Fig. 3A ▶). In both, S8 is a globular protein interacting with one side of the largely helical mRNA1a (Fig. 3A,B ▶). There are 17 Zn2+ ions in the asymmetric unit: nine associated with complex A and eight with complex B. Most of the ions make only second shell interactions with atoms of the complex, mostly with RNA. The differences between copies A and B are small. When copies A and B are superimposed by using binding site phosphorus atoms (G17–A12/C34–U38) and the Cα atoms of S8, the root mean square deviation (rmsd) of Cα atoms is 0.48 Å; for phosphorus atoms in the section of the RNA to which S8 binds, only 0.2 Å. The most notable differences between the two copies are at the ends of the RNA (rmsd of 1.58 Å) and are a result of differences in crystal packing.
FIGURE 3.
The structure of E. coli S8/mRNA1a and its comparison with the T. thermophilus S8/helix 21 structure. (A) By using the phosphorus atoms of the S8-binding site (A12–G17/C34–U38) and the Cαs of S8, Complex A (blue) was superimposed on complex B (pink). (B) Stereoview of the structure of the E. coli S8–mRNA complex shown in blue (complex A), with the Zn2+ ions shown in cyan. (C) A comparison of E. coli complex A (blue) with T. thermophilus S8/helix 21 (orange; Wimberly et al 2000). Superposition was done by using the phosphorus atoms of the S8-binding site (A12–G17/C34–U38) and the Cα atoms of S8. Note that the complexes have been rotated 180° relative to those shown in A and B.
The structure of E. coli S8
E. coli ribosomal protein S8 folds into an N- and a C-terminal domain (Fig. 3B ▶). The N-terminal domain consists of a three-stranded antiparallel β-sheet that contacts a pair of α-helices on one side. The C-terminal domain is a four-stranded antiparallel β-sheet that closely contacts the two α-helices of the N-terminal domain across a hydrophobic interface. The antiparallel β-sheet of the C-terminal domain contains most of the residues that interact with mRNA1a.
The structure of mRNA1a
mRNA1a is an A-form RNA helical stem/loop made somewhat irregular by imperfections in base-pairing in its central region (U7–A14, A35–U42). Its terminal loop has a sequence that suggests it should form a GNRA tetraloop closed by a G–U base pair (G23–U28), but it does not. Instead, G23 and U28 are not paired, which allows U28 to contact the N-terminal β-sheet of S8 (Fig. 3B ▶), and the bases of the loop (G24–A27) protrude into solution. The conformation of the loop, which is not the same in the A and B versions of the complex (Fig. 3A ▶), is thus almost certainly a consequence of its interactions with S8 (discussed below). The B-factors of the loop nucleotides, similar to those of the amino acids that interact with them, are much higher than the average for the whole complex, suggesting that this region of the structure is relatively flexible.
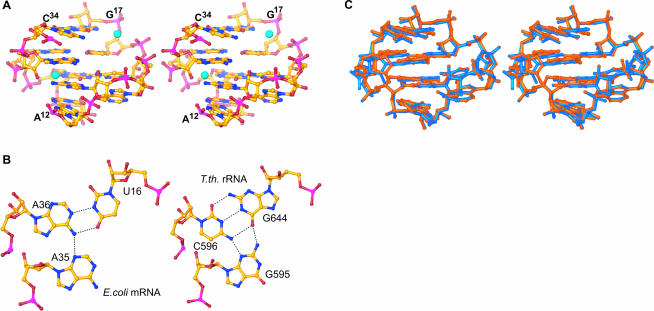
S8 binds to a site in mRNA1a centered on the internal loop formed by bases A12–G17, C34–U38 (Fig. 4A ▶). Two Watson-Crick base pairs form the center of the motif: G37–C15, and A36–U16. They are separated from the two Watson-Crick pairs at its termini (U38–A12, C34–G17) by bulged bases: A13, and A14 in the 5′ strand, and A35 in the 3′ strand. A35 is inserted into the major groove side of A36–U16, where it makes an A-platform with A36 (Fig. 4B ▶, left). On the 5′ strand, bulged bases A13 and A14 form a similar structure with Watson-Crick base pair G37–C15. A14 stacks underneath C15, and the Watson-Crick face of A13 inserts into the major groove side of G37–C15 between the bases of A12 and A35. The resulting A13–G37–C15 structure looks like a base triple but is not because A13 hydrogen bonds with neither G37 nor C15. The end result is a structure that looks like two base triples sandwiched between two Watson-Crick base pairs.
FIGURE 4.
Structure of S8s mRNA and rRNA-binding sites. (A) Stereoview of E. coli mRNA-binding site (A12–G17/C34–U38). Zn ions, shown in cyan, are tetrahedrally coordinated and participate in water-mediated interactions with the mRNA. (B) The U16–A35–A36 base triple from E. coli mRNA1a (left) and the corresponding base triple from T. thermophilus G644–C596–G595 (right). (C) Stereoview of the superposition of the E. coli mRNA-binding site (blue) with the T. thermophilus rRNA-binding site (orange). The C1′ atoms of the conserved residues, which interact with S8 (C15, G37, A14, A12), were used for the superposition.
The stem of the spc operon mRNA includes two bulged nucleotides (U7, C10) that have no counterparts in helix 21 but are known to modulate the affinity of that mRNA for S8 (Wu et al. 1994). Surprisingly, neither contacts S8. The bases protrude from the helix, locally perturbing the backbone geometry of the strand to which they belong but causing almost no change in the conformation of other strand, which is the strand contacting S8, relative to what it is in helix 21 (Fig. 3C ▶). The conformations of these two nucleotides are not the same in the A and B copies of the complex (Fig. 3A ▶), and their B-factors are almost double those of the average for rest of the RNA (100 Å2 versus ~55 Å2), which again suggests local flexibility.
mRNA–S8 interactions
Ribosomal protein S8 binds to one face of the stem-loop of mRNA1a (Fig. 5A ▶). Three parts of mRNA1a are involved: G2 at the open end of its stem, its internal loop (A12–G17/ C34–C41), and its terminal loop region (U28–A31). These contacts bury 868 Å2 of RNA surface (Fig. 5A ▶). Interactions with the terminal loop account for 19.8% of the RNA surface buried, the G2 interaction accounts for 4%, and the internal loop accounts for the rest. It should be noted that the spc operon mRNA does not include equivalents for either G2 or the terminal loop of mRNA1a (Fig. 1 ▶).
FIGURE 5.
S8–RNA interactions. (A) Surface contacts between E. coli S8 (purple) and mRNA1a (green). Buried surface area on the mRNA is shown in dark green and buried surface from S8 is shown in dark purple. (B) Hydrogen bonding between E. coli S8 and its spc operon mRNA-binding site. Two base-specific hydrogen bonds occur (Y85:G37, and S104:A14). All other protein hydrogen bonds involve phosphates or sugars. (C) A comparison of RNA–protein interactions between E. coli S8/mRNA (blue) and T. thermophilus S8/rRNA (orange). Phosphorus atoms and Cα atoms shown were used for the superposition.
The N-terminal α-helix of S8 interacts with the terminal loop region of the mRNA (U28–A31) but most extensively with U28, which protrudes into the gap located in the N-terminal domain of S8 between its β-sheet and the N-terminal portions of its two α-helices (Figs. 3B ▶, 5A ▶). The β-strand of the N-terminal domain of S8 that links α-helix one with α-helix two (Pro27–Lys32) packs against the major groove side of residues 28–31 in the terminal loop, and Lys32 makes a base-specific hydrogen bond with U28 (Lys NH2 with O2 of U28). Polar and basic amino acids in this region interact with the backbone of mRNA1a electrostatically. For example, the backbone NH group from Lys30 hydrogen bonds to the phosphate of C30, and its side-chain NH2 group hydrogen bonds to the phosphate of A31. Interaction of S8 with G2 is also largely electrostatic. It involves the phosphate of G2 and the NH2 group of Arg87.
The most important interactions between mRNA1a and S8 result from the packing of an antiparallel β-sheet in the C-terminal domain of S8 against the minor groove of the internal loop in the mRNA (Figs. 3B ▶, 5B ▶). Side-chains belonging to this β-sheet make the only base-specific hydrogen bonds in the entire complex, and there are only two of them: G37N2–Tyr85OH, and A14N3–Ser104OH. There are four other protein–RNA hydrogen bonds in this region, but they involve RNA backbone atoms only. The OH from Ser104, which makes the hydrogen bond with the N3 of A14, also hydrogen binds to the O2′ of the same nucleotide. Glu123 and Ser106 make hydrogen bonds with the sugars of C15 and A12, respectively, whereas Lys107 makes a hydrogen bond with the phosphate of A13. Hydrophobic interactions are also important in this region. The S8 backbone from residues 119–123 makes hydrophobic contacts with the sugars of U38 and C39. In addition, the β-turn in the C-terminal β-sheet (Thr105–Gly108) inserts into the minor groove of the binding site, where it stacks on what would otherwise be the solvent-exposed face of A14.
DISCUSSION
Comparison of E. coli S8 with the S8s from other species
Not surprisingly, the structure of E. coli S8 is similar to that of the S8s from other species. For example, the rmsd between the structure of the A version of S8 described above and that of S8 in the intact 30S ribosomal subunit from T. thermophilus (Wimberly et al. 2000) is only 0.88 Å for the 122 Cα atoms that can be aligned in the sequence, which is only modestly larger than the rmsd between the A and B copies of E. coli S8 reported here, 0.48 Å for all Cαs. The structure of E. coli S8 is comparably similar to the structures reported for B. stearothermophilus S8 (Davies et al. 1996), and T. thermophilus S8 in isolation (Nevskaya et al. 1998). Even the structure of M. jannaschii S8, which is from another kingdom and is much less homologous in sequence, superimposes quite well on that of E. coli S8 structure. The rmsd is 1.3 Å for the 114 Cα atoms that can be aligned by sequence (Tishchenko et al. 2001).
Comparison of RNA-binding ligands of ribosomal protein S8
The rmsd between mRNA1a and helix 21 of T. thermophilus 16S rRNA is larger than is the rmsd for the two S8s: 2.0 Å for phosphorus atom superposition over the entire RNA, versus 0.88 Å for the Cα atoms of the two proteins, but this is still only ~0.5 Å greater than the rmsd between the two copies of mRNA1a in the asymmetric unit of these crystals. If just the heart of the S8-binding site in mRNA1a (A12–G17/U38–C34, phosphorus atoms only) is superimposed on the corresponding region of helix 21 in T. thermophilus (A640–C645/U598–G594) an rmsd of 0.5 Å is obtained, but when the comparison is limited to nucleotides that contact S8 directly, the rmsd is much smaller: 0.13 Å (Fig. 4C ▶).
Comparisons of helix 21 from M. jannaschii 16S rRNA and mRNA1a produced similar results. Superposition of phosphorus atoms of the two RNA molecules in their entirety gave an rmsd of 0.63 Å, but when the only the phosphorus atoms in the S8-binding site were compared, an rmsd of 0.52 Å was obtained. Restriction of the comparison to those nucleotides that contact S8 reduced the rmsd to 0.31 Å.
Aside from the terminal loop in mRNA1a, which helix 21 lacks, the biggest differences between helix 21 and the S8-binding region of the spc operon mRNA is the two nucleotides in the mRNA stem that helix 21 lacks. When the S8/helix 21 complex is superimposed on the S8/mRNA1a complex, it becomes apparent that RNA strands on the S8 side of the mRNA1a duplex are almost identical (rmsd = 0.30 Å), but the opposite strands, which in E. coli contain both “extra” nucleotides, are not (rmsd = 2.2 Å). The bases of the extra nucleotides bulge into the solvent and do not disrupt the base-pairing interactions of their neighbors in any major way. Only a modest increase in the interstrand spacing of the mRNA1a helix is required to compensate for the unequal length of the two strands.
The conformation of the four-layered internal loop in the S8/mRNA1a complex is almost identical to that of the corresponding internal loop in helix 21, despite sequence differences (Fig. 4C ▶). The A35–A36–U16 base triple in mRNA1a is replaced in helix 21 by a base triple composed of G595–C596–G644, which is surprisingly isosteric (Fig. 4B ▶, right). The triple in the small subunit could thus be thought of as providing an example of a “G–C platform”. The pseudo-triple in mRNA1a (G37–C15–A13) is replaced by an alternative in 16S rRNA (G597–C643–U641), which is again essentially isosteric. The difference in sequence orientation of the Watson-Crick G–C pair that closes the motif (G17–C34 versus C645–G594) has no structural effect, as expected.
The internal loop from the M. jannaschii S8/rRNA complex is intermediate in sequence between the corresponding loops in E. coli spc operon mRNA and T. thermophilus 16S rRNA. The internal loop of M. jannaschii contains the same A35–A36–U16 base triple as does mRNA1a, but its second base triple is the same as that seen in T. thermophilus 16S rRNA (G597–C643–U641). The closing base pair is a G–U wobble pair, rather than a Watson-Crick base pair, but this does not affect the architecture of the internal loop, and thus, the internal loops in all three RNAs are essentially isosteric.
A solution structure has been obtained by NMR for an RNA oligonucleotide containing the S8-binding portion of helix 21 from E. coli 16S rRNA in the absence of S8 (Kalurachchi et al. 1997). The family of structures reported has six members, which have an rmsd of 1.15 Å to the average, and within the S8-binding site, the average NMR structure superimposes on the crystal structure of mRNA1a with an rmsd of 1.5 Å (phosphorus atoms). The architecture of the NMR structure is the same as that of mRNA1a in its internal loop region. The similarity of this solution structure to that of the RNA portion of all the S8/RNA complex structures known indicates that S8 binding has little effect on the conformation of the internal loop.
Comparisons of protein–RNA interactions
The similarities between the E. coli S8/mRNA1a complex and the T. thermophilus S8/helix 21 complex extend to their protein–RNA interactions. The amino acids, which hydrogen bond to mRNA1a (Fig. 5 ▶), are all highly conserved, and with the exception of one residue, those in T. ther-mophilus S8 interact the same way with RNA as do the corresponding residues in E. coli S8 (Fig. 5C ▶). The exception is K107 (K116 in T. thermophilus S8). The side-chain rotamer of that lysine is not the same in the two molecules, and thus in T. thermophilus, K116 of S8 does not interact with the phosphate of U641. In B. stearothermophilus, the residue in this position is a glutamine not a lysine, which suggests that the interaction observed in E. coli is likely to be electrostatic rather than a hydrogen bond critical for recognition. The four bases in the binding site that interact directly with the protein are as highly conserved as are the amino acids that interact with them. Despite sequence differences in the M. jannaschii S8–rRNA complex, the RNA–protein interactions in that complex are similar to those discussed above.
The interactions of the terminal loop of mRNA1a with S8 mimic the interactions S8 makes with the 21/20/22 helix junction and helix 25 in the 30S subunit of T. thermophilus (Brodersen et al. 2002). U28 protrudes out to interact with the N-terminal α-helix of S8, which in the 30S subunit flanks the minor groove of helix 25 of 16S rRNA. Thus, the terminal loop of mRNA1a shelters some of the same amino acids in N-terminal helix that are buried in the minor groove of helix 25 when S8 is part of the small subunit. The remainder of the terminal loop of mRNA1a is in approximately the same position as the junction of helices 21, 20, and 22 in 16S rRNA, and mimics the interactions of this junction with S8. A27 folds back into the minor groove of mRNA1a just the way A653 does in the three-helix junction in 16S rRNA, but its position is intermediate between those of A27 and U28 in mRNA1a, rather than that of A27 exactly. The fact that the three-helix junction in question, which includes an irregular loop-like structure not unlike the terminal loop of mRNA1a, is in almost the same location as the 60-nucleotide loop, which connects the two strands of the stem in the spc operon mRNA that binds S8, suggests that a re-examination of the importance of that mRNA loop for S8 binding and spc operon regulation might be in order. Although wild-type S8 affinity is observed for mRNA constructs lacking that large loop (Gregory et al. 1988; Wu et al. 1994), S8 interactions with it could nevertheless influence translational regulation by stabilizing a specific conformation of spc operon mRNA.
Binding affinities
As pointed out above, roughly one-fourth of the surface area of mRNA1a buried when S8 binds involves nucleotides that have no equivalents in either 16S rRNA or spc operon mRNA. However, by using the structure for S8/mRNA1a, it is straightforward to construct a model for the S8/mRNA2 complex. In this model, as expected, the RNA surface area buried by S8 binding is significantly less than that buried when the S8/mRNA1a complex forms (768 Å2 versus 868 Å2). This difference in buried surface area undoubtedly explains why mRNA1a has a higher affinity for S8 than does mRNA2 or longer spc operon mRNA constructs.
Binding constant data provide additional reasons for wondering whether S8/terminal loop interactions might be important for the regulation. As a general rule, the energy of intramolecular interactions is proportional to the surface area they bury (Chothia and Janin 1975). The RNA surface covered by S8 in the T. thermophilus 30S subunit is 1887 Å2, but yet the dissociation constant of the S8/16S rRNA complex is only ~100 times smaller than that of the complexes S8 forms with RNAs that include helix 21 sequences only, like mRNA2, in which the amount of RNA surface buried is much less, 768 Å2 (Table 1 ▶). If energies were strictly proportional to surface area, one would predict that the Kd for 16S rRNA would be <10−12 M. Presumably, the extra interaction energy provided by the additional surface buried in the S8/16S rRNA complex pays for the entropy change required to fix the conformation of 16S rRNA (see D.J. Klein, P.B. Moore, and T.A. Steitz, unpubl.). The same kind of conformational freezing could be important for regulation too.
The mechanism of translational repression by ribosomal protein S8
It is not understood why the formation of the S8/spc operon mRNA complex described here represses the translation of all the ribosomal protein cistrons in that mRNA in a coordinate manner. The S8-binding site is just 3′ of the Shine-Dalgarno sequence of the cistron that encodes L5. The repressive effect of S8 on the expression of the two cistrons 5′ of the L5 cistron is described as retroregulation and has been attributed to destabilization of the mRNA (Mattheakis et al. 1989). The effects of S8 on downstream cistrons is described as translational coupling (Mattheakis and Nomura 1988). We will limit our discussion here to the latter phenomenon.
It would be easier to understand translational coupling in the spc operon system if the sequence of its mRNA were such that translation could begin only at its 5′ end, but this is not the case. Only three of the 10 ribosomal protein cistrons in that mRNA lack a Shine-Dalgarno sequence, and some of them are 3′ to the site where S8 binds. Nevertheless, even though the sequence of the mRNA appears to support internal initiation, one could imagine all these internal Shine-Dalgarno being cryptic because of the conformation of the mRNA, and that its translation begins always at its 5′ end. The UAA that terminates the synthesis of L24, the cistron immediately 5′ of the L5 cistron, is only 15 nucleotides 5′ of the AUG where L5 synthesis begins, and it is known that mRNAs enter the ribosome ~15 nucleotides 3′ of the 5′-most nucleotide in the ribosomal P site (Yusupova et al. 2001). Thus binding of S8 to this mRNA could arrest the passage of the ribosome along this mRNA just as the last amino acid or two was being added to the nascent L24 chain and thus inhibit all further translation of the message. If this is what happens, one might expect to find incomplete L24 chains in cells in which S8 concentrations are artificially raised.
The model for translational inhibition just described is not entirely satisfactory. Experiments done with spc operon deletions lacking almost the entire region 5′ of the L5 cistron have shown that prior translation of the cistrons upstream of L5 is not required for regulation of the translation of L5 and cistrons downstream of L5 (Mattheakis and Nomura 1988). Clearly, protein synthesis can begin at the L5 cistron, at least in a truncated mRNA, and thus, S8 regulation can and may always be caused by an inability of ribosomes to initiate L5 synthesis when S8 is bound to its target site. We have done extensive modeling studies to see if the sequences where L5 synthesis begins can bind to the ribosome properly when S8 is bound, and we have concluded that it cannot. However, there are still large uncertainties. It is not known how mRNAs get threaded into the mRNA-binding site of the ribosome during normal initiation, and the structures we have examined suggest that the stem to which S8 binds is likely to impede initiation whether S8 is bound or not. Thus, until the mechanics of initiation are better understood, the explanatory power of modeling exercises like these will be limited.
MATERIALS AND METHODS
Protein and RNA preparation
E. coli S8 was overexpressed from the pETrpswt plasmid (Wower et al. 1992) in BL21(DE3)/pLysS cells (Studier et al. 1990), isolated in the form of inclusion bodies, and solubilized with 6 M urea as described elsewhere (Wu et al. 1993). Solubilized material was purified on a 250 mL SP-Sephadex Fast Flow column (Pharmacia Biotech) using a salt gradient of 0 to 0.3 M LiCl. It eluted at ~0.15 M LiCl, and its identity was confirmed using Phastgels (Pharmacia Biotech) and molecular weight markers. S8 was renatured using a procedure described elsewhere (Muto et al. 1974; Mougel et al. 1986; Wu et al. 1993). The concentration was determined using UV absorbance, assuming an extinction coefficient at 280 nm of 5762 M−1 cm−1 (Wu et al. 1993).
The mRNA constructs shown in Figure 1D ▶ were synthesized by in vitro transcription using T7 RNA polymerase and linearized plasmid templates containing the appropriate sequences downstream of T7 promoters (Dallas and Moore 1997). RNA transcripts were purified on polyacrylamide gels containing 8 M urea and were extracted from the gel by electroelution.
Filter binding assay
The binding of S8 to mRNA constructs was measured using a nitrocellulose filter binding assay (Wu et al. 1993, 1994). The binding buffer used was 50 mM Tris-OAc (pH 7.6), 20 mM Mg(OAc)2, 350 mM KOAc, and 5 mM DTT (TMK-OAc; Wu et al. 1994). Each Kd value reported is the average of three independent measurements.
Crystallization
mRNA1a and E. coli S8 samples dialyzed into TMK-OAc were mixed in a 1:1 molar ratio on ice to yield a final complex concentration of 7.2 mg/mL; 600 μL of this material was sent to the Hauptman-Woodward Institute (HWI) in Buffalo, New York, where 1536 crystallization trials were conducted at 23°C using microbatch techniques. Of the several conditions found to generate crystals at HWI, the one that ultimately yielded useful crystals involved precipitation using 100 mM zinc acetate, 100 mM Na acetate buffer, and 20% PEG 8000 (pH 5.0). Optimization using hanging drop methods at 23°C revealed that crystals of ~250 × 250 × 100 microns could be obtained by mixing 100 mM Zn acetate, 100 mM Na acetate buffer, and 7% PEG 8000 (pH 5.2) in equal parts with complex solution at a concentration of 7.2 mg/mL. Crystals were stabilized in 25% PEG 8000 overnight, transferred to Paratone-N oil (Hampton Research) briefly, and frozen in liquid N2.
Structure determination
The data used to solve the structure were collected at beam line X25 at the National Synchrotron Light Source (Brookhaven National Laboratory) using a crystal that had been soaked in stabilization buffer that included ethyl mercury phosphate prior to freezing. Full anomalous data sets were measured at three wavelengths (0.9928 Å, 1.0049 Å, 1.008998 Å) in hopes of obtaining MAD phase information from the Hg that had been introduced into the crystals, but subsequent analysis showed that no Hg had bound. Data were therefore integrated, scaled, and merged using HKL2000 (Otwinowski and Minor 1997). The result of merging the three complete data sets was an unusually high redundancy (Table 2 ▶). The merged data were detwinned using the CCP4 DETWIN program (CCP4) following determination of the twin fraction using the Web server interface (http://www.doe-mbi.ucla.edu/Services/twinning; Yeates 1997). Self-rotation computations using the program GLRF with the detwinned data provided evidence for the existence of what ultimately proved to be an approximate, noncrystallographic, two-fold screw axis almost parallel to the c-axis (Tong and Rossman 1990). Thus, the unit cell contains 12 S8/mRNA1a complexes and has a solvent content of ~ 60%.
A molecular replacement solution was obtained using the program AMoRe (Navaza 2001) with a search model derived from the T. thermophilus 30S subunit (S8 [chain H]/A590–A609/A629–A649; Wimberly et al. 2000). The initial molecular replacement solution had a correlation coefficient four standard deviations (SDs) above the average for all solutions and had an R-factor of 51% at 2.7 Å resolution following rigid body refinement. Electron-density maps improved dramatically with very little bias toward initial 2Fo-Fc maps after using “prime and switch”, noncrystallographic symmetry (NCS) averaging and solvent-flattening as implemented in the program RESOLVE (Terwilliger 2000). Subsequent maps were calculated using real-space averaging with RAVE (Kleywegt and Read 1997).
The model was built using Ono8 (Jones and Kjeldgard 1997) and refined using Refmac (Murshudov et al. 1997) from the CCP4 suite. During the early stages of refinement, the two complexes were restrained to obey NCS. This restraint was gradually relaxed and removed entirely at the end of the refinement because the two copies of the complex were not identical. Composite omit maps were computed using CNS (Brunger et al. 1998). Figures were generated by RIBBONS (Carson 1991) and by PyMol (DeLano 2002). Seventeen Zn2+ ions were added to the map late in the refinement following their identification in anomalous difference Fourier maps computed using the combined data set.
Coordinates
The coordinates and structure factors have been deposited with the Protein Data Bank with accession number 1S03.
Acknowledgments
We thank the Hauptman-Woodward Institute for setting up initial crystal screening conditions, S.A. Strobel for assistance with data collection, Y. Xiong for valuable suggestions and discussions regarding structural calculations, and the staff at the Yale Center for Structural Biology for their help. We also thank R.A. Zimmer-mann for his gift of the pETrpswt plasmid, and T.M. Schmeing for his assistance with figures and reading of the manuscript. This work was supported by a grant from National Institutes of Health (GM 61258).
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.rnajournal.org/cgi/doi/10.1261/rna.7030704.
REFERENCES
- Brodersen, D.E., Clemons Jr., W.M., Carter, A.P., Wimberly, B.T., and Ramakrishnan, V. 2002. Crystal structure of the 30S ribosomal subunit from Thermus thermophilus: Structure of the proteins and their interactions with 16S RNA. J. Mol. Biol. 316: 725–768. [DOI] [PubMed] [Google Scholar]
- Brunger, A.T., Adams, P.D., Clore, G.M., DeLano, W.L., Gros, P., Grosse-Kunstleve, R.W., Jiang, J.S., Kuszewski, J., Nilges, N., Pannu, N.S., et al. 1998. Crystallography and NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 54: 905–921. [DOI] [PubMed] [Google Scholar]
- Carson, M. 1991. Ribbons 2.0. J. Appl. Crystallogr. 24: 958–961. [Google Scholar]
- Cerretti, D.P., Mattheakis, L.C., Kearney, K.R., Vu, L., and Nomura, M. 1988. Translational regulation of the spc operon in Escherichia coli: Identification and structural analysis of the target site for S8 repressor protein. J. Mol. Biol. 204: 309–329. [DOI] [PubMed] [Google Scholar]
- Chothia, C. and Janin, J. 1975. Principles of protein–protein recognition. Nature 256: 705–708. [DOI] [PubMed] [Google Scholar]
- Dallas, A. and Moore, P.B. 1997. The loop E–loop D region of Escherichia coli 5S rRNA: The solution structure reveals an unusual loop that may be important for binding ribosomal proteins. Structure 5: 1639–1653. [DOI] [PubMed] [Google Scholar]
- Davies, C., Ramakrishnan, V., and White, S.W. 1996. Structural evidence for specific S8–RNA and S8–protein interactions within the 30S ribosomal subunit: ribosomal protein S8 from Bacillus stearothermophilus at 1.9 Å resolution. Structure 4: 1093–1104. [DOI] [PubMed] [Google Scholar]
- Dean, D., Yates, J.L., and Nomura, M. 1981. Escherichia coli ribosomal protein S8 feedback regulates part of the spc operon. Nature 289: 89–91. [DOI] [PubMed] [Google Scholar]
- DeLano, W.L. 2002. The PyMOL molecular graphics system. DeLano Scientific, San Carlos, CA.
- Fallon, A.M., Jinks, C.S., Strycharz, G.D., and Nomura, M. 1979. Regulation of ribosomal protein synthesis in Escherichia coli by selective mRNA inactivation. Proc. Natl. Acad. Sci. 76: 3411–3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiil, N.P., Friesen, J.D., Downing, W.L., and Dennis, P.P. 1980. Post-transcriptional regulatory mutants in a ribosomal protein–RNA polymerase operon of E. coli. Cell 19: 837–844. [DOI] [PubMed] [Google Scholar]
- Gregory, R.R., Cahill, P.B.F., Thurlow, D.L., and Zimmermann, R.A. 1988. Interaction of Escherichia coli ribosomal protein S8 with its binding sites in ribosomal RNA and messenger RNA. J. Mol. Biol. 204: 295–307. [DOI] [PubMed] [Google Scholar]
- Jones, T.A. and Kjeldgard, M. 1997. Electron-density map interpretation. Methods Enzymol. 277: 173–208. [DOI] [PubMed] [Google Scholar]
- Kalurachchi, K., Uma, K., Zimmermann, R.A., and Nikonowicz, E.P. 1997. Structural features of the binding site for ribosomal protein S8 in Escherichia coli 16S rRNA defined using NMR spectroscopy. Proc. Natl. Acad. Sci. 94: 2139–2144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleywegt, G.J. and Read, R.J. 1997. Not your average density. Structure 5: 1557–1569. [DOI] [PubMed] [Google Scholar]
- Lindahl, L. and Zengel, J.M. 1979. Operon-specific regulation of ribosomal protein synthesis in Escherichia coli. Proc. Natl. Acad. Sci. 76: 6542–6546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattheakis, L. and Nomura, M. 1988. Feedback regulation of the spc operon in Escherichia coli: Translational coupling and mRNA processing. J Bacteriol. 170: 4482–4492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattheakis, L., Vu, L., Sor, F., and Nomura, M. 1989. Retroregulation of the synthesis of ribosomal proteins L14 and L24 by feedback repressor S8 in Escherichia coli. Proc. Natl. Acad. Sci. 86: 448–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mougel, M., Ehresmann, B., and Ehresmann, C. 1986. Binding of Escherichia coli ribosomal protein S8 to 16S rRNA: Kinetic and thermodynamic characterization. Biochemistry 25: 2756–2765. [DOI] [PubMed] [Google Scholar]
- Murshudov, G.N., Vagin, A.A., and Dodson, E.J. 1997. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 53: 240–255. [DOI] [PubMed] [Google Scholar]
- Muto, A., Ehresmann, C., Fellner, P., and Zimmermann, R.A. 1974. RNA–protein interactions in the ribosome, I: Characterization and ribonuclease digestion of 16 S RNA–ribosomal protein complexes. J. Mol. Biol. 86: 411–432. [DOI] [PubMed] [Google Scholar]
- Nevskaya, N., Tishchenko, S., Nikulin, A., Al-Karadaghi, S., Liljas, A., Ehresmann, B., Ehresmann, C., Garber, M., and Nikonov, S. 1998. Crystal structure of ribosomal protein S8 from Thermus thermophilus reveals a high degree of structural conservation of a specific RNA binding site. J. Mol. Biol. 279: 233–244. [DOI] [PubMed] [Google Scholar]
- Nomura, M., Yates, J.L., Dean, D., and Post, L.E. 1980. Feedback regulation of ribosomal protein gene expression in Escherichia coli: Structural homology of ribosomal RNA and ribosomal protein mRNA. Proc. Natl. Acad. Sci. 77: 7084–7088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otwinowski, Z. and Minor, W. 1997. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276: 307–326. [DOI] [PubMed] [Google Scholar]
- Schwarzbauer, J. and Craven, G.R. 1981. Apparent association constants for E. coli ribosomal proteins S4, S7, S8, S15, S17 and S20 binding to 16S RNA. Nucleic Acid Res. 9: 2223–2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studier, F.W., Rosenberg, A.H., Dunn, J.J., and Dubendorff, J.W. 1990. Use of T7 RNA polymerase to direct expression of cloned genes. Methods Enzymol. 185: 60–89. [DOI] [PubMed] [Google Scholar]
- Terwilliger, T.C. 2000. Maximum-likelihood density modification. Acta Crystallogr. D Biol. Crystallogr. 56: 965–972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tishchenko, S.V., Nikulin, A., Fomenkova, N.P., Nevskaya, N., Nikonov, O., Dumas, P., Moine, H., Ehresmann, B., Ehresmann, C., Piendl, W., et al. 2001. Detailed analysis of RNA–protein interactions within ribosomal protein S8–rRNA complex from the archaeon Methanococcus jannaschii. J. Mol. Biol. 311: 311–324. [DOI] [PubMed] [Google Scholar]
- Tong, L. and Rossman, M.G. 1990. The locked rotation function. Acta Crystallogr. A 46: 783–792. [DOI] [PubMed] [Google Scholar]
- Wimberly, B.T., Brodersen, D.E., Clemons Jr., W.M., Morgan-Warren, R.J., Carter, A.P., Vonrhein, C., Hartsch, T., and Ramakrishnan, V. 2000. Structure of the 30S ribosomal subunit. Nature 407: 327–339. [DOI] [PubMed] [Google Scholar]
- Wower, I., Kowaleski, M.P., Sears, L.E., and Zimmermann, R.A. 1992. Mutagenesis of ribosomal protein S8 from Escherichia coli: Defects in regulation of the spc operon. J. Bacteriol. 174: 1213–1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, H., Wower, I., and Zimmermann, R.A. 1993. Mutagenesis of ribosomal protein S8 from Escherichia coli: Expression, stability, and RNA-binding properties of S8 mutants. Biochemistry 32: 1461–1468. [DOI] [PubMed] [Google Scholar]
- Wu, H., Jiang, L., and Zimmermann, R.A. 1994. The binding site for ribosomal protein S8 in 16S rRNA and spc mRNA from Escherichia coli: Minimum structural requirements and the effects of single bulged bases on S8-RNA interaction. Nucleic Acid. Res. 22: 1687–1695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeates, T.O. 1997. Detecting and overcoming crystal twinning. Methods Enzymol. 276: 344–358. [PubMed] [Google Scholar]
- Yusupova, G.Z., Yusupov, M.M., Cate, J.H.D., and Noller, H.F. 2001. The path of messenger RNA through the ribosome. Cell 106: 233–241. [DOI] [PubMed] [Google Scholar]
- Zengel, J.M. and Lindahl, L. 1994. Diverse mechanisms for regulating ribosomal protein synthesis in Escherichia coli. Prog. Nucleic Acid. Res. Mol. Biol. 47: 331–370. [DOI] [PubMed] [Google Scholar]