Abstract
5′ tRNA editing has been demonstrated to occur in the mitochondria of the distantly related rhizopod amoeba Acanthamoeba castellanii and the chytridiomycete fungus Spizellomyces punctatus. In these organisms, canonical tRNA structures are restored by removing mismatched nucleotides at the first three 5′ positions and replacing them with nucleotides capable of forming Watson–Crick base pairs with their 3′ counterparts. This form of editing seems likely to occur in members of Amoebozoa other than A. castellanii, as well as in members of Heterolobosea. Evidence for 5′ tRNA editing has not been found to date, however, in any other fungus including the deeply branching chytridiomycete Allomyces macrogynus. We predicted that a similar form of tRNA editing would occur in members of the chytridiomycete order Monoblepharidales based on the analysis of complete mitochondrial tRNA complements. This prediction was confirmed by analysis of tRNA sequences using a tRNA circularization/ RT-PCR-based approach. The presence of partially and completely unedited tRNAs in members of the Monoblepharidales suggests the involvement of a 5′-to-3′ exonuclease rather than an endonuclease in removing the three 5′ nucleotides from a tRNA substrate. Surprisingly, analysis of the mtDNA of the chytridiomycete Rhizophydium brooksianum, which branches as a sister group to S. punctatus in molecular phylogenies, did not suggest the presence of editing. This prediction was also confirmed experimentally. The absence of tRNA editing in R. brooksianum raises the possibility that 5′ tRNA editing may have evolved twice independently within Chytridiomycota, once in the lineage leading to S. punctatus and once in the lineage leading to the Monoblepharidales.
Keywords: 5′ tRNA editing, chytridiomycete fungi, tRNA evolution, tRNA processing
INTRODUCTION
RNA editing, the programmed conversion of RNA transcripts from their gene-encoded sequence to an altered sequence, has been described in a wide range of eukaryotes, predominantly in organelles (kinetoplasts, chloroplasts, and mitochondria). Efficient and complete editing is often essential for the survival of an organism, as only converted RNA sequences are able to assume their appropriate cellular function(s) (Brennicke et al. 1999; Gott and Emeson 2000; Simpson et al. 2000). Although many instances have been reported of mRNA molecules being altered by various RNA editing mechanisms (Benne 1996; Simpson et al. 1996), structural RNAs such as ribosomal RNAs (Adler et al. 1991; Schuster et al. 1991; Mahendran et al. 1994; Barth et al. 1999) and tRNAs (Janke and Pääbo 1993; Maréchal-Drouard et al. 1996a; Laforest et al. 1997; Price and Gray 1998; Price and Gray 1999b; Schock et al. 1998) are also subject to alteration by RNA editing processes.
Base modification, substitution, and insertion/deletion editing mechanisms are known to contribute to the maturation of tRNAs in some mitochondrial systems. For example, C-to-U editing corrects base-pair mismatches in the mitochondrial tRNAs of plants (Maier et al. 1996; Maréchal-Drouard et al. 1996b; Fey et al. 2002) and changes the decoding properties of tRNATrp in trypanosome mitochondria (Alfonzo et al. 1999). In the myxomycetes Physarum and Didymium, C and U insertions restore base-pairing in tRNA helical regions as well as create the conserved GUUC sequence in the T stem–loop (Antes et al. 1998). In the mitochondria of several animals, insertions of A and/or C residues at tRNA 3′ ends complete acceptor stems and create discriminator nucleotides (Yokobori and Pääbo 1995, 1997; Tomita et al. 1996; Reichert et al. 1998). Another recently identified form of tRNA editing in the centipede Lithobius forficatus replaces up to 5 nt at tRNA 3′ ends, apparently by a novel mechanism that uses the 5′ end of the acceptor stem as template (Lavrov et al. 2000). Interestingly, a similar type of editing occurs in the mitochondria of the jakobid flagellate Seculamonas ecuadoriensis (Leigh and Lang 2004). Finally, an additional mechanism of tRNA editing in Metazoa modifies the second position of the tRNAAsp anticodon from C to U, thus changing its decoding identity (Janke and Pääbo 1993; Borner et al. 1996).
The first example of tRNA editing was discovered in mitochondria of Acanthamoeba castellanii, an amoeboid protist. This form of editing was found to correct mismatches in the first 3 bp of tRNA acceptor stems by removing the three 5′ nucleotides and replacing them sequentially in a 3 ′-to-5′ direction (contrary to polymerases, which add in a 5′-to-3′ direction) with nucleotides that can form Watson–Crick base pairs (G-C/A-U) with their counterparts in the 3′ half of the acceptor stem (Lonergan and Gray 1993; Price and Gray 1998 Price and Gray 1999a). Sequencing of the mitochondrial DNA (mtDNA) of Spizellomyces punctatus, a chytridiomycete fungus, revealed a reduced set of eight tRNA genes that all contained from one to three non-Watson–Crick base pairs in the three terminal base pairs of the acceptor stem. Direct sequencing with reverse-transcriptase showed examples in five of the eight tRNAs where a predicted mismatch was corrected at the RNA level to give a standard Watson–Crick base pair by substitution of the 5′ nucleotide in the pair (Laforest et al. 1997). The pattern of editing in S. punctatus mitochondrial tRNA genes was remarkably similar to that found in A. castellanii. Because the members of Amoebozoa (the phylum to which A. castellanii has been assigned; Cavalier-Smith 1998) and Chytridiomycota have no specific phylogenetic link, and are in fact very distantly related to each other, it was proposed that this form of editing arose independently in these two lineages.
The discovery of an analogous 5′ tRNA editing system in S. punctatus was unexpected, as other examined fungal mitochondrial tRNAs did not appear to require editing, including that of the chytridiomycete Allomyces macrogynus (Paquin and Lang 1996). In this study, we identify and verify additional cases of 5′ tRNA editing in chytridiomycete fungi. These data shed light onto the emergence and evolution of this type of editing, as well as providing insights into the biochemistry of the activities involved.
RESULTS AND DISCUSSION
Prediction of editing by analysis of tRNA acceptor stem base-pairings
The majority of tRNAs conform to a standard secondary structure consisting of three short stem–loops and a terminal acceptor stem. The nucleotides in tRNA molecules are numbered in a system that begins (in most cases) with the most 5′ nucleotide in a mature tRNA (position 1) and ends with the discriminator nucleotide (position 73). The nucleotide at position 1 forms a base pair with the nucleotide at position 72, thereby forming the terminal base pair (1–72) of the seven acceptor stem base pairs (1–72 to 7–66).
Not all acceptor stem base pairs involve standard Watson–Crick (WC; G-C/C-G/A-U/U-A) base pairs. Non-WC base pairs can be divided into “wobble” pairs (G-U/U-G pairs) and “mismatch” pairs (non-WC/nonwobble). Wobble base pairs are well known to be able to substitute for WC pairs in certain contexts (Masquida and Westhof 2000) and, as shown in Table 1, wobble base pairs are tolerated to a relatively high percentage in the acceptor stems of all the organisms compared. For example, 8.6% of acceptor stem base pairs in the mitochondrial tRNAs of Schizosaccharomyces pombe are wobble base pairs. In contrast, mismatches are tolerated only to a very low percentage; for example, in the archaeal tRNA acceptor stems analyzed, no mismatch pairs were identified.
TABLE 1.
WC and non-WC base pairs in tRNA acceptor stems
| Acceptor stem base pairs (%) | |||||||||
| 1–72 to 7–66 | 1–72 to 3–70 | ||||||||
| Organism | # of tRNAs | WC | Non-WC | G-U U-G | mm | WC | Non-WC | G-U U-G | mm |
| Eukaryaa | 2025 | 89.5 | 10.5 | 9.3 | 1.1 | 92.8 | 7.2 | 6.9 | 0.2 |
| Archaeaa | 581 | 97.6 | 2.4 | 2.4 | 0.0 | 97.4 | 2.6 | 2.6 | 0.0 |
| Eubacteriaa | 1598 | 94.7 | 5.3 | 4.6 | 0.7 | 94.0 | 6.0 | 4.8 | 1.2 |
| S. pombe | 25 | 89.1 | 10.9 | 8.6 | 2.3 | 88.0 | 12.0 | 10.7 | 1.3 |
| A. macrogynus | 25 | 91.4 | 8.6 | 7.4 | 1.1 | 88.0 | 12.0 | 10.7 | 1.3 |
| R. brooksianum | 7 | 93.9 | 6.1 | 6.1 | 0.0 | 90.5 | 9.5 | 9.5 | 0.0 |
| N. gruberi | 21 | 88.4 | 11.6 | 3.4 | 8.2 | 76.2 | 23.8 | 4.8 | 19.0 |
| D. discoideum | 18 | 78.6 | 21.4 | 11.1 | 10.3 | 63.0 | 37.0 | 13.0 | 24.1 |
| A. castellanii | 15 | 76.2 | 23.8 | 3.8 | 20.0 | 48.9 | 51.1 | 4.4 | 46.7 |
| S. punctatus | 8 | 67.9 | 32.1 | 3.6 | 28.6 | 29.2 | 70.8 | 4.2 | 66.7 |
| Monoblepharella15 | 9 | 74.6 | 25.4 | 9.5 | 15.9 | 40.7 | 59.3 | 22.2 | 37.0 |
| Harpochytrium94 | 8 | 76.8 | 23.2 | 1.8 | 21.4 | 45.8 | 54.2 | 4.2 | 50.0 |
| Harpochytrium105 | 8 | 71.4 | 28.6 | 1.8 | 26.8 | 33.3 | 66.7 | 4.2 | 62.5 |
| H. curvatum | 7 | 83.7 | 16.3 | 8.2 | 8.2 | 76.2 | 23.8 | 9.5 | 14.3 |
Bold type indicates organisms with confirmed/predicted editing and highlights the percentage of WC base-pairing in their mitochondrial tRNA acceptor stems. (WC) Watson–Crick; (mm) mismatch. GenBank accession numbers for mitochondrial data: S. pombe, NC001326; A. macrogynus, NC001715; R. brooksianum, NC0030503; N. gruberi, NC002573; D. discoideum, AB000109; A. castellanii, U12386; S. punctatus, NC003052; Monoblepharella15, AY1820007; Harpochytrium94, AY182005; Harpochytrium105, AY1820006, H. curvatum, NC003048.
aData from the analysis of Marck and Grosjean (2002).
Editing has been confirmed experimentally at the first three 5′ nucleotides of mitochondrial tRNAs in A. castellanii and S. punctatus by identifying differences between genomic and cDNA sequences. Analysis of the acceptor stem base pairs inferred from the mtDNA sequences of these two species reveals a lower percentage of WC base pairs at all seven acceptor stem positions compared to organisms lacking editing, and this trend is particularly pronounced in the terminal 3 bp of this stem (Table 1). When non-WC pairs are separated into wobble and mismatch pairs, mismatches represent a large majority of the increase in non-WC base-pairing in these acceptor stems. These trends are also evident in the mtDNA-encoded tRNAs of the amoebozoan Dictyostelium discoideum and the heterolobosean amoeba Naegleria gruberi (Table 1), two organisms in which 5′ tRNA editing is likely to occur (Ogawa et al. 2000; M.W. Gray, unpubl. observation). Only 1/102 mismatches in the acceptor stems of organisms with predicted/confirmed editing are found at positions 4–69 to 7–66, supporting the strong selection against mismatches in nonedited positions.
No evidence for mitochondrial tRNA editing has been found in members of the other three fungal divisions (Ascomycota, Basidiomycota, and Zygomycota; Bullerwell et al. 2003b). Similarly, the mitochondrial tRNAs of Allomyces macrogynus (a member of the deeply diverging chytridiomycete order Blastocladiales) lack features in their acceptor stems that would suggest the presence of editing (Table 1). In light of these data, it was uncertain whether chytridiomycetes other than S. punctatus would require tRNA editing. In contrast to the situation in A. macrogynus, non-WC base pairs are abundant at the first three acceptor stem base pairs of the mtDNA-encoded tRNAs in Monoblepharella15, Harpochytrium94, and Harpochytrium105 (Bullerwell et al. 2003a,b; Table 1). Three non-WC base pairs at these positions are also present in Hyaloraphidium curvatum, a fourth examined member of Monoblepharidales (Forget et al. 2002). Based on these data, we sought to obtain experimental confirmation of the presence of tRNA editing in chytridiomycete fungi other than S. punctatus.
Further exploration of 5′ tRNA editing in S. punctatus (order Spizellomycetales)
To further examine 5′ tRNA editing in S. punctatus, a mitochondrial RNA fraction enriched for tRNA was circularized with T4 RNA ligase, an RT-PCR strategy was designed to amplify the acceptor stem region (including the ligation site) of three mtDNA-encoded tRNAs, and cDNA sequences were determined (see Materials and Methods). The data presented here for S. punctatus (Table 2) has confirmed and expanded the findings of the earlier study (Laforest et al. 1997): Nucleotide substitution occurs on the 5′ side of the acceptor stem, resulting in the replacement of nucleotides involved in non-WC base pairs at positions 1–3 with nucleotides that can form WC pairs with their counterparts in the 3′ half of the stem. The activity does not correct non-WC pairs outside of the first 3 bp, as evidenced by the retention of a U6-G67 wobble pair in tRNATyr (Laforest et al. 1997). Thus, as expected, this study confirms that tRNA editing in S. punctatus resembles very closely the situation in A. castellanii.
TABLE 2.
cDNA sequences of circularized tRNAs
| Speciesa | tRNA | Originb | Sequencec | # of cDNAs | Commentsd |
| S.punc | Lys | mtDNA | 3′-GUGAAUU-----AUCCUCAC-5′ | — | — |
| cDNA | 3′-GUGAGGA--ACCAUCCUCAC-5′ | 8 | E | ||
| fMet | mtDNA | 3′-GGCCUUC-----UUCAGGCC-5′ | — | — | |
| cDNA | 3′-GGCCUGA--ACCUUCAGGCC-5′ | 5 | E | ||
| Pro | mtDNA | 3′-GCGGAAU-----AGUCCCGC-5′ | — | — | |
| cDNA | 3′-GCGGGAC--ACCAGUCCCGC-5′ | 15 | E | ||
| R.brook | Leu | mtDNA | 3′-AUCCCCG-----AUGGGGAU-5′ | — | — |
| cDNA | 3′-AUCCCCG--ACCAUGGGGAU-5′ | 16 | NE | ||
| cDNA | 3′-AUCCCCGauACCAUGGGGAU-5′ | 1 | NE | ||
| Mono15 | Glu | mtDNA | 3′-UCCAAGG-----GCUCUGGA-5′ | — | — |
| cDNA | 3′-UCCAGAG--ACCGCUCUGGA-5′ | 23 | E | ||
| cDNA | 3′-UCCAAGG--ACCGCUCUGGA-5′ | 4 | NE | ||
| cDNA | 3′-UCCAAGGg-ACCGCUCUGGA-5′ | 2 | NE | ||
| cDNA | 3′-UCCAAGG---CCGCUCUGGA-5′ | 4 | NE | ||
| fMet | mtDNA | 3′-AAGAUAG-----AACGUCUU-5′ | — | — | |
| cDNA | 3′-AAGACGU--ACCAACGUCUU-5′ | 21 | E | ||
| cDNA | 3′-AAGAUGU--ACCAACGUCUU-5′ | 15 | PE | ||
| Pro | mtDNA | 3′-AAGGAGU-----AGUCCCUU-5′ | — | — | |
| cDNA | 3′-AAGGGAC--ACCAGUCCCUU-5′ | 10 | E | ||
| Gly | mtDNA | 3′-UGGAUAU-----AGGAUCCA-5′ | — | — | |
| cDNA | 3′-UGGAUCC--ACCAGGAUCCA-5′ | 8 | E | ||
| Harp94 | Glu | mtDNA | 3′-UCCAAUU-----GCUCUGGA-5′ | — | — |
| cDNA | 3′-UCCAGAG--ACCGCUCUGGA-5′ | 3 | E | ||
| fMet | mtDNA | 3′-AAGAUGU-----AACGUCUU-5′ | — | — | |
| cDNA | 3′-AAGAUGU--ACCAACGUCUU-5′ | 2 | NE | ||
| Pro | mtDNA | 3′-AAGAAAA-----AGUCUCUU-5′ | — | — | |
| cDNA | 3′-AAGAGAC--ACCAGUCUCUU-5′ | 7 | E | ||
| cDNA | 3′-AAGAGACug---AGUCUCUU-5′ | 1 | E | ||
| cDNA | 3′-AAGAGAC-----AGUCUCUU-5′ | 1 | E | ||
| cDNA | 3′-AAGAAAA--ACCAGUCUCUU-5′ | 1 | NE | ||
| Harp105 | Glu | mtDNA | 3′-UCCAACA-----GCUCUGGA-5′ | — | — |
| cDNA | 3′-UCCAGAG--ACCGCUCUGGA-5′ | 13 | E | ||
| cDNA | 3′-UCCAGAGa-ACCGCUCUGGA-5′ | 1 | E | ||
| cDNA | 3′-UCCAGAGuu--CGCUCUGGA-5′ | 1 | E | ||
| cDNA | 3′-UCCAAAG--ACCGCUCUGGA-5′ | 1 | PE | ||
| fMet | mtDNA | 3′-AAGAUAA-----AACGUCUU-5′ | — | — | |
| cDNA | 3′-AAGAUGU--ACCAACGUCUU-5′ | 15 | PE | ||
| Pro | mtDNA | 3′-AAGGAUA-----AGUCCCUU-5′ | — | — | |
| cDNA | 3′-AAGGGAC--ACCAGUCUCUU-5′ | 5 | E |
aS.punc, S. punctatus; R.brook, R. brooksianum; Mono15, Monoblepharella15; Harp94, Harpochytrium94; Harp105, Harpochytrium105.
bSequence inferred from either the mtDNA gene sequence or cDNA sequences.
cIsolated tRNAs were circularized and amplified by RT-PCR (see Materials and Methods). Sequences are given from 3′ to 5′ starting at position 7 on the 5′ side of the acceptor stem and ending at position 66 on the 3′ side of the acceptor stem and including other nucleotides present at the ligation junction (e.g., the CCA tail; other nucleotides of uncertain origin are indicated in lowercase).
dE, edited; NE, not edited; PE, partially edited
Confirmation of 5′ tRNA editing in the order Monoblepharidales
To confirm that editing was in fact occurring in mono-blepharidalean mitochondrial tRNAs, cDNA sequences were obtained for the acceptor stem region of several tRNAs from Monoblepharella15, Harpochytrium94, and Harpochytrium105 using the procedure described above for S. punctatus (Table 2; for predicted tRNA structures in these species, see http://megasun.bch.umontreal.ca/papers/tRNAstructures). The pattern of editing was found to be as in S. punctatus and A. castellanii, in that WC base pairs are created by replacement of the first three 5′ nucleotides in the acceptor stem. However, in contrast to the situation in those species, some 5′ positions that were predicted to be edited based on genomic data are left completely or partially unedited (Table 2; discussed below).
Absence of tRNA editing in Rhizophydium brooksianum (order Chytridiales)
A member of the order Chytridiales, R. brooksianum (formerly Rhizophydium136; Longcore 2004), was found to have only two non-WC base pairs, both G-U, in the first 3 bp of the acceptor stems of its mtDNA-encoded tRNAs (Table 2). Therefore it did not appear to be a likely candidate for 5′ tRNA editing. This finding was unexpected, due to the phylogenetic position of R. brooksianum within Chytridiomycota (Fig. 1; see below). To confirm the absence of this form of editing in R. brooksianum, cDNA data was acquired for tRNALeu, which contains a G1-U72 base pair. All 17 clones examined retained this non-WC pairing, confirming the absence of 5′-tRNA editing in this organism.
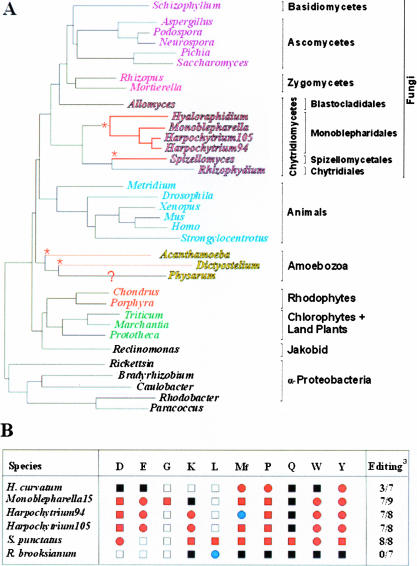
FIGURE 1.
Phylogenetic distribution of 5′ tRNA editing. (A) Schematic tree based on the branching order of published phylogenetic trees (Forget et al. 2002; Bullerwell et al. 2003a). The branching order of the animal, fungal, and amoebozoan lineages is consistent with phylogenies based on nucleus-encoded protein sequence data (Baldauf et al. 2000). Red asterisks mark the presumed, independent origins of 5′ tRNA editing. (B) Editing status of tRNAs in chytridiomycete mtDNAs. atRNAs where editing is predicted or confirmed/total. (Open box) tRNA gene not present; ((Black filled box) tRNA gene present, editing not predicted; (red filled box) tRNA gene present, editing confirmed; (red filled circle) tRNA gene present, editing predicted but not confirmed by sequencing; (blue filled circle) tRNA gene present, confirmed not editing.
tRNA processing and mechanism of tRNA editing in chytridiomycete fungi
tRNA processing/editing intermediates are observed in the cDNA sequence data obtained from monoblepharidalean mitochondrial tRNAs (Fig. 2; Table 2). Completely unedited cDNAs were obtained for 10/33 clones of Monoblepharella15 tRNAGlu, 1/10 clones of Harpochytrium94 tRNAPro and 2/2 clones of Harpochytrium94 tRNAfMet. Interestingly, all of the clones obtained with complete absence of editing encode a CCA tail (note that one Monoblepharella15 tRNAGlu sequence has only CC). This indicates that editing is not required for addition of the CCA tail by nucleotidyl transferase, even in the one unedited clone of Harpochytrium94 tRNAPro, which retains two mismatches in the first 3 bp of its acceptor stem. Similarly, the CCA tail does not appear to be necessary for editing, as 2/10 clones of Harpochytrium94 tRNAPro are completely edited, yet lack the CCA sequence. These results suggest that these two processes, CCA addition and editing, function independently of each other, at least in Harpochytrium94.
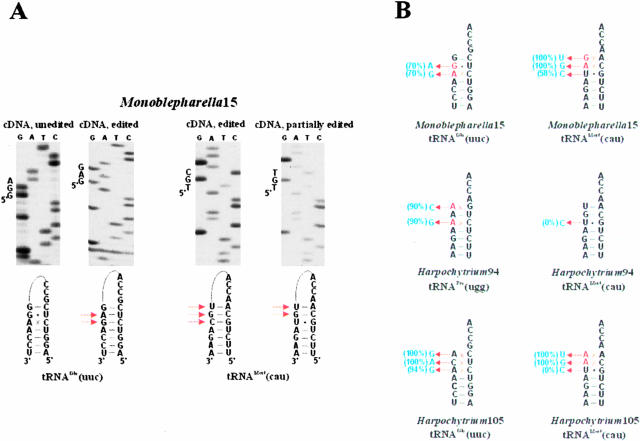
FIGURE 2.
Partially and completely unedited monoblepharidalean mitochondrial tRNAs. (A) cDNA sequencing gels of unedited and partially edited tRNAs from Monoblepharella15. Sequence of the first three 5′ nucleotides of tRNA acceptor stems is indicated to the left of the sequencing gels, and the inferred structure of the circularized tRNA is indicated below each gel. Positions where a nucleotide change has occurred relative to the genomic sequence are indicated by arrows. (B) tRNA acceptor stems for all examples where partial or incomplete editing is observed. Predicted nucleotide substitutions are indicated by arrows, and the percentage of cDNA clones in which the predicted change occurs is indicated in brackets.
Partial editing is observed in Monoblepharella15 tRNAfMet, where the G1xA72 and A2xC71 mismatches are edited in all 36 cases, but U3-G70 is altered to C3-G70 in only 21/36 cDNA clones. Similar situations are found in Harpochytrium105 tRNAGlu, where the A1xC72 and C2xU71 mismatches are corrected in all 16 clones whereas the A3xC70 pair is retained in 1/16 clones, and Harpochytrium105 tRNAfMet, where the A1xA72 and A2xC71 mismatches are repaired in all 15 analyzed cDNA clones, whereas the U3-G70 pair is not corrected in any case (Fig. 2). These results support the involvement of an exonuclease as opposed to an endonuclease in the removal of the 5′ nucleotides from a substrate tRNA prior to nucleotide incorporation: If an endo-nuclease were responsible, the first 3 nt should either be found to be completely edited or completely unedited in all cases. Further, partially edited positions are found to be at position 3 in all examples, supporting sequential 5′-to-3′ exonuclease activity from position 1 through position 3.
All cDNAs analyzed in S. punctatus and A. castellanii were found to have WC base pairs at the first three positions of the acceptor stem, suggesting that the editing activities are very efficient in these systems. The lack of intermediates in these two species is unfortunate as it offers no insight into, for example, whether editing occurs before or after addition of the CCA tail to tRNA 3′ ends by nucleotidyl transferase, and whether the nuclease component is an endo- or exonuclease. In contrast, the tRNA processing/editing intermediates observed in the mono-blepharidalean mitochondrial tRNAs give us insights into the mechanism of editing in this lineage as well as the relationship to tRNA processing (Fig. 3).
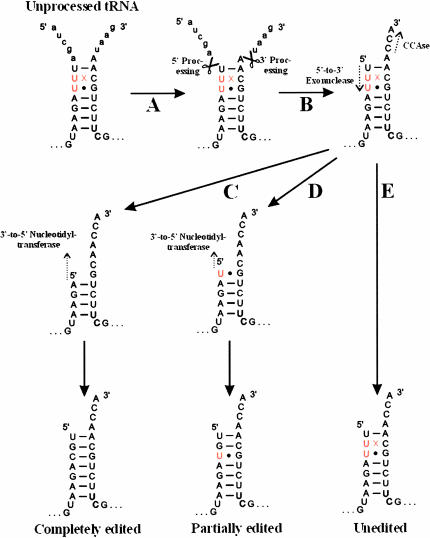
FIGURE 3.
Model of tRNA processing and editing in monoblepharidalean mitochondria. A hypothetical unprocessed tRNA acceptor stem is shown. (A) 5′ and 3′ extensions are first removed by nuclease activities (the order of events is not known). (B) CCA tails are added by nucleotidyl transferase (CCAse) and a 5′-to-3′ exonuclease activity removes nucleotides from tRNA 5′ ends. The CCAse and exonuclease activities apparently function independently of one another. A presumed 3′-to-5′ nucleotidyltransferase (as proposed for A. castellanii based on in vitro results; Price and Gray 1999b) then fills in nucleotides removed by the 5′-to-3′ exonuclease. (C) If all three 5′ nucleotides have been removed by the exonuclease, three nucleotides are added that can form WC base pairs, resulting in a completely edited tRNA. (D) If only one or two 5′ nucleotides have been removed, a partially edited tRNA results. (E) If no nucleotides have been removed, an unedited tRNA results.
Independent origins of 5′ tRNA editing within Chytridiomycota?
The data presented in this study indicate that the form of 5′ tRNA editing that occurs in S. punctatus and mono-blepharidalean fungi does not occur in R. brooksianum. As R. brooksianum branches as a sister lineage to S. punctatus to the exclusion of the Mono-blepharidales in molecular phylogenies based on mitochondrial protein sequences (Fig. 1), the most parsimonious description of the evolution of tRNA editing in Chytridiomycota would appear to be that tRNA editing emerged once at the base of the chytridiomycete lineage subsequent to the divergence of A. macrogynus, and was then lost in the branch leading to R. brooksianum.
The principal argument at variance with this interpretation is that all of the 5′ nucleotides involved in mismatches in the first three acceptor stem base pairs would have to revert to nucleotides that could form WC or wobble pairs with their 3′ partners. In other words, multiple back mutations would have to occur to render the editing activity superfluous, a seemingly unlikely prospect. In addition, because 5′ tRNA editing likely evolved independently in the distantly related groups Amoebozoa and Chytridiomycota (Price and Gray 1998), the idea that this type of editing could also have emerged independently on two occasions in chytridiomycete fungi is less surprising. In any case, further exploration will be necessary to firmly establish how many times 5′ tRNA editing has emerged in the fungi and throughout Eukarya.
CONCLUSIONS
In this study, we present data that help to better understand the emergence, evolution, and mechanism of 5′ tRNA editing in chytridiomycete fungi. To supplement these data, purification and analysis of the enzyme activities involved will be necessary to determine whether the A. castellanii, S. punctatus, and monoblepharidalean forms of 5′ tRNA editing have common origins and/or common biochemical mechanisms. The activity responsible for editing has in fact been partially purified from A. castellanii, and an in vitro assay for this system has been developed (Price and Gray 1999b). An equivalent system is currently being established in S. punctatus (C.E. Bullerwell and M.W. Gray, unpubl.). Identification of the component(s) of these editing complexes will be a definitive method for addressing mechanistic and evolutionary issues.
MATERIALS AND METHODS
Fungal strains and culture conditions
R. brooksianum, Harpochytrium94, and Harpochytrium105 were kindly provided by J.E. Longcore (University of Maine), and Monoblepharella15 by M.R.N. Mollicone (University of Maine). R. brooksianum, the two Harpochytrium species and Monoblepharella15 were grown in a liquid medium containing 0.25% tryptone, 0.125% yeast extract, and 3% glucose, and S. punctatus in a liquid medium containing 0.5% yeast extract, and 3% glycerol (pH 5.8). All cell cultures were performed at room temperature with shaking (100 rpm), with the exception of Monoblepharella15, which was grown without shaking. Further details on these chytridiomycete species are available at http://megasun.bch.umontreal.ca/People/lang/FMGP/FMGP.html.
Purification of mitochondrial tRNA
Fungal cells were disrupted by shaking with glass beads (Lang et al. 1977), with the exception of Monoblepharella15 cells that were crushed in a mortar in the presence of glass beads. A crude mitochondrial fraction was isolated by differential centrifugation and lysed in guanidinium buffer (100 mM Tris-HCl at pH 8.0, 5 mM EDTA, 8 M Guanidine-HCl). After removal of cell debris by centrifugation, nucleic acids were ethanol precipitated, redissolved in H2O containing 0.1% SDS, and precipitated with 2 M LiCl for 1 h on ice. After centrifugation at 14,000g at 4°C, the pellet was dissolved in H2O, and the LiCl precipitation was repeated. The supernatant of the second LiCl precipitation, enriched in mitochondrial tRNAs, was precipitated twice with ethanol, redissolved in H2O, and stored frozen at −80°C.
tRNA circularization and RT-PCR
Oligonucleotides used in this study:
Ligation of mitochondrial tRNAs was performed according to Yokobori and Pääbo (1995), with minor modifications. Mitochondrial RNA (10 μg) was ligated in the presence of 50 mM HEPES (pH 8.3), 10 mM MgCl2, 3.5 mM DTT, 2 μg/mL BSA, 1 mM ATP, 20 U RNase inhibitor (Promega), 10% DMSO (Sigma-Aldrich), and 8 U of T4 RNA ligase (Gibco-BRL) in a final volume of 25 μL, at 37°C for 2 h. After phenol/chloroform extraction, RNA was precipitated with ethanol and redissolved in 10 μL H2O. For each of the RT-PCR experiments, an aliquot of 2 μg of ligated RNA, plus 1 μL of the appropriate first-strand primer (1 pmole/μL), were brought with TE (10 mM Tris at pH 8.0, 1 mM EDTA) to a final volume of 23 μL. The solution was heated for 2 min at 90°C, left at room temperature for 15 min, and then placed on ice for 15 min. cDNA was synthesized using 12 U of AMV Reverse Transcriptase (Roche Diagnostics), in the presence of 50 mM Tris-HCl (pH 8.5), 8 mM MgCl2, 30 mM KCl, 2 mM DTT, and 1 mM each dNTP in a final volume of 40 μL, for 45 min at 45°C.
Aliquots of the resulting cDNAs (0.5 μL) were used for PCR amplification, in the presence of 0.5 μM of appropriate first- and second-strand primers, 1 U Taq DNA polymerase (Roche Diagnostics), and 0.2 mM each dNTPs, in the reaction buffer supplied by the company. cDNAs were then amplified by PCR (40 cycles), in a Perkin-Elmer-Cetus 9600 system. Control PCR experiments without first-strand cDNA synthesis were performed for each RT-PCR to demonstrate that the resulting DNA fragments were amplified from cDNAs and not from genomic DNA. The amplified DNAs were separated on 2% agarose gels, and fragments corresponding to the predicted sizes (approximately 75 bp) were isolated from the gel, cloned, and sequenced.
Table t3.
| Species | tRNA | Strand | DNA sequence |
| S. punctatus | tRNALys | 1 | 5′-TCCGTTGCTCTAGCCATTGAGCT-3′ |
| 2 | 5′-CTTTTAATCCGTGGGTTGCAGGT-3′ | ||
| tRNAPro | 1 | 5′-ACCAGTATTCTAACCATTGAACT-3′ | |
| 2 | 5′-TTTGGGAACCAGCGATACAG-3′ | ||
| tRNAfMet | 1 | 5′-CCTAGGACGCTACCATTACAAT-3′ | |
| 2 | 5′-CTCATAACCTGGTAGTGTAGG-3′ | ||
| R. brooksianum | tRNALeu | 1 | 5′-ACCGATGAATCTACCAATTCTTCT-3′ |
| 2 | 5′-CTCTAAAATCGAATTTTGTTGGTT-3′ | ||
| Monoblepharidalesa | tRNAGlu | 1 | 5′-TCCAGAGTTCTAACCATTAAACT-3′ |
| 2 | 5′-TTTTCGTTCCAGTAATAGGGGT-3′ | ||
| Monoblepharella15 | tRNAGlu | 1 | 5′-TCCAGGGTTCTACCATTAAACTA-3′ |
| Monoblepharidalesa | tRNAfMet | 1 | 5′-CCAGCGAGTTACCCTTACTCC-3′ |
| 2 | 5′-TCTCATCATCCGGAAATGGAGG-3′ | ||
| Monoblepharidalesa | tRNAPro | 1 | 5′-CTTTCGTGCTACCAATTACACTA-3′ |
| 2 | 5′-TTTTGGGTACTTTTAGCTTGGG-3′ | ||
| Monoblepharella15 | tRNAGly | 1 | 5′-ACCAAAGTTCTACCGTTAAACTA-3′ |
| 2 | 5′-CTTCCAAACCAAGAATGCAGG-3′ |
aUsed for Monoblepharella15, and both Harpochytrium species.
Acknowledgments
We thank G. Burger and D. Lavrov (Université de Montréal) for helpful comments on the manuscript, J.E. Longcore, M.R.N. Mollicone (both of the University of Maine), and K. O’Donnell (National Center for Agricultural Utilization Research, Peoria, IL) for supplying of fungal strains. We also acknowledge the contributions of H. Hamel in DNA sequencing and the excellent technical assistance of Z. Wang. Salary and interaction support from the Canadian Institutes of Health Research (MOP 42475; B.F.L.), the Canadian Institute for Advanced Research (B.F.L.), and supply of laboratory equipment and informatics infrastructure by Genome Quebec/Canada is gratefully acknowledged.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.7330504.
REFERENCES
- Adler, B.K., Harris, M.E., Bertrand, K.I., and Hajduk, S.L. 1991. Modification of Trypanosoma brucei mitochondrial rRNA by posttran-scriptional 3′ polyuridine tail formation. Mol. Cell. Biol. 11: 5878–5884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alfonzo, J.D., Blanc, V., Estévez, A.M., Rubio, M.A.T., and Simpson, L. 1999. C to U editing of the anticodon of imported mitochondrial tRNATrp allows decoding of the UGA stop codon in Leishmania tarentolae. EMBO J. 18: 7056–7062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antes, T., Costandy, H., Mahendran, R., Spottswood, M., and Miller, D. 1998. Insertional editing of mitochondrial tRNAs of Physarum polycephalum and Didymium nigripes. Mol. Cell. Biol. 18: 7521–7527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldauf, S.L., Roger, A.J., Wenk-Siefert, I., and Doolittle, W.F. 2000. A kingdom-level phylogeny of eukaryotes based on combined protein data. Science 290: 972–977. [DOI] [PubMed] [Google Scholar]
- Barth, C., Greferath, U., Kotsifas, M., and Fisher, P.R. 1999. Polycistronic transcription and editing of the mitochondrial small subunit (SSU) ribosomal RNA in Dictyostelium discoideum. Curr. Genet. 36: 55–61. [DOI] [PubMed] [Google Scholar]
- Benne, R. 1996. RNA editing: How a message is changed. Curr. Opin. Genet. Dev. 6: 221–231. [DOI] [PubMed] [Google Scholar]
- Borner, G.V., Morl, M., Janke, A., and Pääbo, S. 1996. RNA editing changes the identity of a mitochondrial tRNA in marsupials. EMBO J. 15: 5949–5957. [PMC free article] [PubMed] [Google Scholar]
- Brennicke, A., Marchfelder, A., and Binder, S. 1999. RNA editing. FEMS Microbiol. Rev. 23: 297–316. [DOI] [PubMed] [Google Scholar]
- Bullerwell, C.E., Forget, L., and Lang, B.F. 2003a. Evolution of mono-blepharidalean fungi based on complete mitochondrial genome sequences. Nucleic Acids Res. 31: 1614–1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bullerwell, C.E., Leigh, J., Seif, E., Longcore, J.E., and Lang, B.F. 2003b. Evolution of the fungi and their mitochondrial genomes. In Applied mycology and biotechnology (eds. D.K. Arora and G.G. Khachatourians), pp. 133–159. Elsevier Science, Amsterdam.
- Cavalier-Smith, T. 1998. A revised six-kingdom system of life. Biol. Rev. Camb. Philos. Soc. 73: 203–266. [DOI] [PubMed] [Google Scholar]
- Fey, J., Weil, J.H., Tomita, K., Cosset, A., Dietrich, A., Small, I., and Marechal-Drouard, L. 2002. Role of editing in plant mitochondrial transfer RNAs. Gene 286: 21–24. [DOI] [PubMed] [Google Scholar]
- Forget, L., Ustinova, J., Wang, Z., Huss, V.A., and Lang, B.F. 2002. Hyaloraphidium curvatum: A linear mitochondrial genome, tRNA editing, and an evolutionary link to lower fungi. Mol. Biol. Evol. 19: 310–319. [DOI] [PubMed] [Google Scholar]
- Gott, J.M. and Emeson, R.B. 2000. Functions and mechanisms of RNA editing. Annu. Rev. Genet. 34: 499–531. [DOI] [PubMed] [Google Scholar]
- Janke, A. and Pääbo, S. 1993. Editing of a tRNA anticodon in marsupial mitochondria changes its codon recognition. Nucleic Acids Res. 21: 1523–1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laforest, M.J., Roewer, I., and Lang, B.F. 1997. Mitochondrial tRNAs in the lower fungus Spizellomyces punctatus: tRNA editing and UAG ‘stop’ codons recognized as leucine. Nucleic Acids Res. 25: 626–632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang, B., Burger, G., Doxiadis, I., Thomas, D.Y., Bandlow, W., and Kaudewitz, F. 1977. A simple method for the large-scale preparation of mitochondria from microorganisms. Anal. Biochem. 77: 110–121. [DOI] [PubMed] [Google Scholar]
- Lavrov, D.V., Brown, W.M., and Boore, J.L. 2000. A novel type of RNA editing occurs in the mitochondrial tRNAs of the centipede Lithobius forficatus. Proc. Natl. Acad. Sci. 97: 13738–13742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leigh, J. and Lang, B.F. 2004. Mitochondrial 3′ tRNA editing in the jakobid Seculamonas ecuadoriensis: A novel mechanism and implications for tRNA processing. RNA 10: 615–621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lonergan, K.M. and Gray, M.W. 1993. Editing of transfer RNAs in Acanthamoeba castellanii mitochondria. Science 259: 812–816. [DOI] [PubMed] [Google Scholar]
- Longcore, J.E. 2004. Rhizophydium brooksianum sp. nov., a mulitpored chytrid from soil. Mycologia 96: 162–171. [PubMed] [Google Scholar]
- Mahendran, R., Spottswood, M.S., Ghate, A., Ling, M.L., Jeng, K., and Miller, D.L. 1994. Editing of the mitochondrial small subunit rRNA in Physarum polycephalum. EMBO J. 13: 232–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maier, R.M., Zeltz, P., Kossel, H., Bonnard, G., Gualberto, J.M., and Grienenberger, J.M. 1996. RNA editing in plant mitochondria and chloroplasts. Plant Mol. Biol. 32: 343–365. [DOI] [PubMed] [Google Scholar]
- Marck, C. and Grosjean, H. 2002. tRNomics: Analysis of tRNA genes from 50 genomes of Eukarya, Archaea, and Bacteria reveals anti-codon-sparing strategies and domain-specific features. RNA 8: 1189–1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maréchal-Drouard, L., Cosset, A., Remacle, C., Ramamonjisoa, D., and Dietrich, A. 1996a. A single editing event is a prerequisite for efficient processing of potato mitochondrial phenylalanine tRNA. Mol. Cell. Biol. 16: 3504–3510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maréchal-Drouard, L., Kumar, R., Remacle, C., and Small, I. 1996b. RNA editing of larch mitochondrial tRNA(His) precursors is a prerequisite for processing. Nucleic Acids Res. 24: 3229–3234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masquida, B. and Westhof, E. 2000. On the wobble G.U and related pairs. RNA 6: 9–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa, S., Yoshino, R., Angata, K., Iwamoto, M., Pi, M., Kuroe, K., Matsuo, K., Morio, T., Urushihara, H., Yanagisawa, K., et al. 2000. The mitochondrial DNA of Dictyostelium discoideum: Complete sequence, gene content and genome organization. Mol. Gen. Genet. 263: 514–519. [DOI] [PubMed] [Google Scholar]
- Paquin, B. and Lang, B.F. 1996. The mitochondrial DNA of Allomyces macrogynus: The complete genomic sequence from an ancestral fungus. J. Mol. Biol. 255: 688–701. [DOI] [PubMed] [Google Scholar]
- Price, D.H. and Gray, M.W. 1998. Editing of tRNA. In Modification and editing of RNA (eds. H. Grosjean and R. Benne), pp. 289–305. ASM Press, Washington, DC.
- ———. 1999a. Confirmation of predicted edits and demonstration of unpredicted edits in Acanthamoeba castellanii mitochondrial tRNAs. Curr. Genet. 35: 23–29. [DOI] [PubMed] [Google Scholar]
- ———. 1999b. A novel nucleotide incorporation activity implicated in the editing of mitochondrial transfer RNAs in Acanthamoeba castellanii. RNA 5: 302–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reichert, A., Rothbauer, U., and Morl, M. 1998. Processing and editing of overlapping tRNAs in human mitochondria. J. Biol. Chem. 273: 31977–31984. [DOI] [PubMed] [Google Scholar]
- Schock, I., Maréchal-Drouard, L., Marchfelder, A., and Binder, S. 1998. Processing of plant mitochondrial tRNAGly and tRNASer-(GCU) is independent of RNA editing. Mol. Gen. Genet. 257: 554–560. [DOI] [PubMed] [Google Scholar]
- Schuster, W., Ternes, R., Knoop, V., Hiesel, R., Wissinger, B., and Brennicke, A. 1991. Distribution of RNA editing sites in Oenothera mitochondrial mRNAs and rRNAs. Curr. Genet. 20: 397–404. [DOI] [PubMed] [Google Scholar]
- Simpson, L., Frech, G.C., and Maslov, D.A. 1996. RNA editing in trypanosomatid mitochondria. Methods Enzymol. 264: 99–121. [DOI] [PubMed] [Google Scholar]
- Simpson, L., Thiemann, O.H., Savill, N.J., Alfonzo, J.D., and Maslov, D.A. 2000. Evolution of RNA editing in trypanosome mitochondria. Proc. Natl. Acad. Sci. 97: 6986–6993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomita, K., Ueda, T., and Watanabe, K. 1996. RNA editing in the acceptor stem of squid mitochondrial tRNA(Tyr). Nucleic Acids Res. 24: 4987–4991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokobori, S. and Pääbo, S. 1995. Transfer RNA editing in land snail mitochondria. Proc. Natl. Acad. Sci. 92: 10432–10435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ———. 1997. Polyadenylation creates the discriminator nucleotide of chicken mitochondrial tRNA(Tyr). J. Mol. Biol. 265: 95–99. [DOI] [PubMed] [Google Scholar]