Abstract
Ribonuclease P (RNase P) is the endonuclease responsible for the removal of 5′ leader sequences from tRNA precursors. The crystal structure of an archaeal RNase P protein, Ph1771p (residues 36–127) from hyperthermophilic archaeon Pyrococcus horikoshii OT3 was determined at 2.0 Å resolution by X-ray crystallography. The structure is composed of four helices (α1–α4) and a six-stranded antiparallel β-sheet (β1–β6) with a protruding β-strand (β7) at the C-terminal region. The strand β7 forms an antiparallel β-sheet by interacting with strand β4 in a symmetry-related molecule, suggesting that strands β4 and β7 could be involved in protein-protein interactions with other RNase P proteins. Structural comparison showed that the β-barrel structure of Ph1771p has a topological resemblance to those of Staphylococcus aureus translational regulator Hfq and Haloarcula marismortui ribosomal protein L21E, suggesting that these RNA binding proteins have a common ancestor and then diverged to specifically bind to their cognate RNAs. The structure analysis as well as structural comparison suggested two possible RNA binding sites in Ph1771p, one being a concave surface formed by terminal α-helices (α1–α4) and β-strand β6, where positively charged residues are clustered. A second possible RNA binding site is at a loop region connecting strands β2 and β3, where conserved hydrophilic residues are exposed to the solvent and interact specifically with sulfate ion. These two potential sites for RNA binding are located in close proximity. The crystal structure of Ph1771p provides insight into the structure and function relationships of archaeal and eukaryotic RNase P.
Keywords: archaea, Pyrococcus horikoshii, RNA binding protein, RNase P, X-ray crystallography
INTRODUCTION
Ribonuclease P (RNase P) is responsible for generating the mature 5′-end of tRNAs by a single endonucleolytic cleavage of their precursors (Guerrier-Takada et al. 1983; Pace and Brown 1995; Frank and Pace 1998; Jarrous 2002). It is an essential, ubiquitous enzyme present in all three phylo-genetic kingdoms. Although the functionality of this endoribonuclease remains almost the same from bacteria to humans, the chemical composition and enzymatic property of this enzyme differ in various organisms. Eubacterial RNase P consists of a single RNA subunit of ~400 nucleotides and a single protein subunit of ~120 amino acids, and in the presence of a high concentration of Mg2+, the RNA subunit itself can hydrolyze tRNA precursors in vitro; hence RNase P RNA is a ribozyme (Guerrier-Takada et al. 1983). In contrast, the eukaryotic RNase P, such as RNase P from humans, is composed of at least 10 protein subunits designated Rpp14, Rpp20, Rpp21, Rpp25, Rpp29, Rpp30, Rpp38, Rpp40, hPop1, and hPop5, associated with a single RNA subunit, H1 RNA, and in the absence of protein subunits, the RNA subunit does not alone exhibit enzyme activity in vitro (Bartkiewicz et al. 1989; Yuan and Altman 1995; Xiao et al. 2001). As for archaeal RNase P, some RNase P holo-enzymes have been isolated from the thermoacidophilic crenarchaeote Sulfolobus acidocaldorius (Darr et al. 1990), the extremely halophilic euroyarchaeote Haloferax volcanii (Nieuwlandt et al. 1991), and two methanogenic archaea, Methanococcus jannaschii and Methanothermobacter thermoautorophicus (Andrews et al. 2001), and characterized in terms of susceptibility to micrococcal nuclease treatment and the buoyant density in Cs2SO4. These results suggested the presence of a few protein components associated with their RNase P RNAs. Moreover, Pannucci et al. (1999) tested a spectrum of archaeal RNase P RNAs for enzyme activity and found that some archaeal RNase P RNAs have no enzyme activity in cleavage reactions of pre-tRNA, though some of the RNase P RNA do have catalytic activity under unusual high salt conditions.
We earlier found in reconstitution experiments that RNase P RNA and four proteins (Ph1481p, Ph1601p, Ph1771p, and Ph1877p) are essential for the RNase P activity of the hyperthermophilic archaeon Pyrococcus horikoshii OT3 (Kouzuma et al. 2003). Subsequently, the crystal structure of Ph1877p was determined at 1.8 Å resolution, and its mutagenesis study identified Arg90, Arg107, Lys123, Arg176, and Lys196 as essential amino acids for the RNase P activity (Takagi et al. 2004). In the present study, we extended the structural study to the P. horikoshii RNase P protein Ph1771p. The protein Ph1771p is composed of 127 amino acid residues, and its amino acid sequence shows a significant homology to those of Rpp29 and Pop4p from human and Saccharomyces cerevisiae, respectively. Mann et al. (2003) recently reported that the reconstitution of human RNase P activity was achieved by the use of in vitro-transcribed H1 RNA and just two recombinant proteins of Rpp21 and Rpp29 (Ph1601p and Ph1771p homologs of P. horikoshii, respectively), despite the existence of at least 10 protein subunits in human RNase P holoenzyme (Mann et al. 2003). Moreover, it was revealed in yeast two- and three-hybrid systems that human Rpp29 and yeast Pop4p bind to each cognate RNA subunit (Jiang et al. 2001; Houser-Scott et al. 2002) as well as to several other RNase P protein subunits (Jiang and Altman 2001; Houser-Scott et al. 2002). Similar results of protein-protein interactions were reported in an archaeal homolog from Methanothermobacter thermoautotrophicus (Hall and Brown 2004). These results indicate that human Rpp29 and its homologous proteins have important roles in catalytic reactions and could be located in close proximity to the active center of archaeal and eukaryotic RNase P holoenzyme.
To provide structural insight into the function of an archaeal homolog of Rpp29, we report here the X-ray crystal structure of an archaeal RNase P protein of Ph1771p from P. horikoshii determined at 2.0 Å resolution.
RESULTS AND DISCUSSION
Protein preparation and characterization
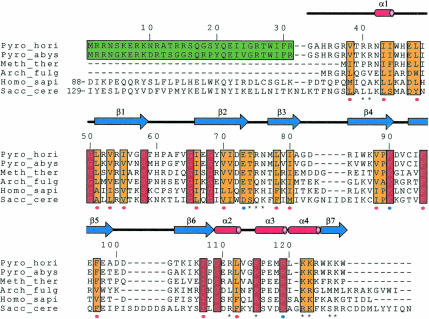
Although we initially attempted to crystallize full-length protein, these trials failed: no crystal of full-length Ph1771p grew under any conditions used. This is probably due to heterogeneity of the purified protein, as the full-length Ph1771p was produced as an inclusion body and then purified under denaturing conditions (Kouzuma et al. 2003). Thus, it should be required that the protein be produced as a soluble fraction in Escherichia coli cells to purify under nondenaturing conditions. For this purpose, we attempted to construct the expression system for the mutant form Ph1771p. Ph1771p has a single cysteine residue (Cys93) that forms an artificial intermolecular disulfide bond between molecules. Therefore, production of Ph1771p as an inclusion body in E. coli cells may be due in part to the formation of an aggregate through the intermolecular disulfide bond between molecules. Furthermore, sequence alignment of Ph1771p with its homologous proteins from archaea revealed that Ph1771p has a long extended segment (residues 1–31) at its N-terminal region (Fig. 1). It is thus suggested that the presence of the N-terminal extension of Ph1771p may have a negative effect on crystallization of this protein. To obtain the protein suitable for crystallography study, the N-terminal extension composed of 31 amino acids was truncated and a single cysteine residue was mutated to Ser as described in Materials and Methods. As anticipated, the mutant form referred to as mPh1771p was produced as a soluble fraction and subsequently purified to apparent homogeneity in a series of chromatography studies. The direct sequencing of the purified protein gave a single N-terminal amino acid sequence, Gly-Ala-His-Arg-Gly-Arg-, indicating that methionine at the N-terminus, a cloning artifact derived from the restriction site of expression vector, was processed. To confirm the biological activity of mPh1771p in the RNase P holoenzyme, it was characterized by reconstitution of the RNase P enzymatic activity. The reconstituted RNase P containing mPh1771p retained 15% enzymatic activity compared to wild-type RNase P activity (data not shown), suggesting that mPh1771p has a functional conformation despite truncation of the N-terminal extension and the mutation of a single cysteine residue. However, this result also suggests the important roles of the N-terminal extension for the catalytic activity, as described below.
FIGURE 1.
Sequence alignment of Ph1771p and its homologous proteins from archaea and eukaryote: archaeal proteins from Pyrococcus horikoshii (Pyro_hori), Pyrococcus abyssi (Pyro_abys), Methanobacterium thermoautotrophicum (Meth_ther), Archaeoglobus fulgidus (Arch_fulg), human Rpp29 (Homo_sapi), and yeast Pop4p (Sacc_cere) are compared. Secondary structure elements of Ph1771p determined in the present work are also shown. Amino acid residue numbering is based on that for Ph1771p. The N-terminal extension observed in Pyrococcus sp. is boxed in green. The residues highlighted in the red box represent complete conservation, and those in the yellow box conservative mutation. (*) The amino acids that are likely to be involved in the interaction with RNase P RNA. Filled circles in red and blue represent residues involved in structural stability by forming a hydrophobic core and intramolecular salt bridges, respectively. The single cysteine residue, Cys93, in Ph1771p was mutated to Ser in the present study.
Structure description
The crystal structure of mPh1771p was determined by the MAD method and has been refined at 2.0 Å resolution to an Rwork of 20.9% and Rfree of 25.7% with good geometry (Table 1). The final model of the asymmetric unit consists of 183 protein residues, 144 ordered water molecules, and two sulfate ions. Two molecules of mPh1771p occupy the crystallographic asymmetric unit (termed molecule A and molecule B). The N-terminal four residues (residues 32–35) in molecule A and the N-terminal five residues (residues 32–36) in molecule B are disordered, whereas a clear electron density is observed for residual amino acids of both molecules. A ribbon representation of the peptide backbone of the molecule is shown in Figure 2. Although the two molecules in the asymmetric unit adopt a very similar conformation, there is some difference between them. Superimposition of these two molecules reveals that the difference occurs at a loop region (residues 59–65) connecting strands β1 and β2, thereby indicating flexibility of this region. The overall structures except for residues 59–65 of these two molecules could be superimposed with an rms deviation of 0.375 Å (data not shown).
TABLE 1.
Data collection and refinement statistics
| Data collection | ||
| Peak | Remote | |
| Beam line | SPring-8 BL38B1 | |
| Space group | P21 | |
| Unit cell parameters | a = 24.1 Å, b = 56.3 Å, c = 71.8 Å, 3 = 96.8° | |
| Wavelength (Å) | 0.97972 | 0.90 |
| Resolution (Å)a | 50-2.0 (2.07-2.0) | 50-2.0 (2.07-2.0) |
| Number of reflections | ||
| Observed | 92,735 | 45,932 |
| Unique | 25,204 | 24,391 |
| Rsymab | 0.096 (0.240) | 0.082 (0.199) |
| I/σ(I)a | 14.6 (8.8) | 9.4 (6.1) |
| Completeness (%)a | 99.7 (99.1) | 99.7 (99.4) |
| Refinement statistics | ||
| Resolution (Å) | 30.1-2.0 | |
| Number of reflections | ||
| Working set | 22,292 | |
| Test set | 2,296 | |
| Completeness (%) | 97.1 | |
| Rwork (%)c | 20.9 | |
| Rfree (%)d | 25.7 | |
| Number of atoms | ||
| Protein | 1,516 | |
| Sulfate ion | 10 | |
| Water | 144 | |
| Average B-factor (Å2) | ||
| Protein | 17.5 | |
| Sulfate ion | 40.7 | |
| Water | 27.3 | |
| rms deviation | ||
| Bond lengths (Å) | 0.005 | |
| Bond angles (°) | 1.30 | |
| Ramachandran analysis | Molecule A | Molecule B |
| Most favored (%) | 94.9 | 96.1 |
| Allowed (%) | 5.1 | 2.6 |
| Generously allowed (%) | 0 | 1.3 |
| Disallowed (%) | 0 | 0 |
aValues in parentheses are for the highest resolution shell. bRsym = ∑(I − 〈I〉)/∑〈I〉, where I is the intensity measurement for a given reflection and 〈I〉 is the average intensity for multiple measurements of this reflection. cRwork = ∑|Fobs − Fcal|/∑Fobs, where Fobs and Fcal are observed and calculated structure factor amplitudes. dRfree value was calculated for Rwork, using only an unrefined subset of reflection data (10%).
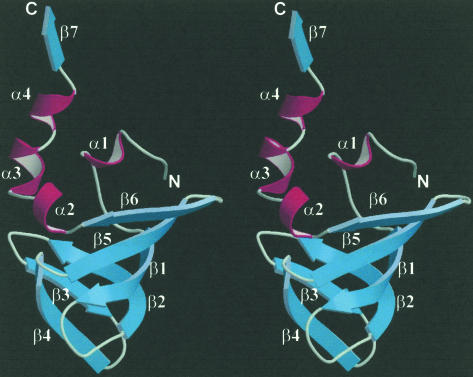
FIGURE 2.
Ribbon representation of the RNase P protein Ph1771p (residues 35–127) from P. horikoshii. The helices are in magenta, β-strands in light blue, and the loops in gray. The image was produced with the programs MOLSCRIPT (Kraulis 1991) and Raster 3D (Merrit and Murphy 1994).
As shown in Figure 2, mPh1771p is folded into a central globular domain with a protruding well structured C-terminal region, with the approximate dimensions 25 Å × 30 Å × 45 Å. The central globular domain of this molecule consists of an N-terminal 310-helix (α1, residues 42–44), followed by six β-strands (β1, residues 51–58; β2, residues 66–73; β3, residues 77–81; β4, residues 84–90; β5, residues 93–99; and β6, residues 103–108), and further followed by three helices [α2, residues 109–112 (310); α3, residues 115–119; and α4, residues 120–123 (310)]. The protruding C-terminal region (residue 124–127) is folded into a β-strand (β7), which forms an antiparallel intersubunit β-sheet by interacting with strand β4 in a symmetry-related molecule (see below). A striking feature of this molecule is its formation of six stranded (β1–β6) antiparallel highly twisted β-sheets. The formation of such a high degree of β-strand curvature is apparently related to several strictly conserved glycines, particularly Gly68, which is located in the middle of strand β2 and is critical to the maintenance of the highly distorted structure.
The central globular domain forms a hydrophobic core with residues Val38, Ile43, Leu48, Leu51, Val53, Ile55, Ile66, Val70, Leu78, Ile80, Val88, Phe95, Phe97, Ile107, and Leu112. This result, together with the fact that most of these hydrophobic residues are highly conserved in homologous proteins (Fig. 1), strongly suggests that these residues play important roles not only in structural stability but also in maintenance of the specific β-barrel structure.
Two strands of the molecule, β2 and β4 and helix α3, are connected by a hydrogen-bond network that includes the highly conserved residues Glu73, Lys90, and Arg119. The side chain of Glu73 forms a salt bridge with the side chain of Lys90, which is in turn hydrogen-bonded to the main chain carbonyl oxygen atoms of Thr74, Leu112, and Gly114. This network is further stabilized by a salt bridge between the side chains of Glu73 and Arg119, which further donate hydrogen bonds to the main chain carbonyl oxygen of Ile44 and His46. It is of interest to note that this hydrogen-bond network (salt bridge) is formed on the interior of the molecule. Therefore, it seems apparent that these intramolecular hydrogen-bonding networks, including highly conserved charged residues, are involved in structural stability and/or molecular folding.
Similarity to other structures
The three-dimensional structures of three eubacterial RNase P proteins from Bacillus subtilis (Stams et al. 1998), Staphylococcus aureus (Spitzfaden et al. 2000), and Thermotoga maritima (Kazantsev et al. 2003) were determined by X-ray crystallography and NMR. The structure of Ph1771p from the hyperthermophilic archaeon P. horikoshii has neither marked sequence homology nor a structure similar to those of eubacterial RNase P proteins.
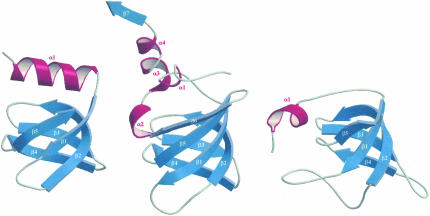
A DALI server (Holm and Sander 1993) and VAST search (Gibrat et al. 1996) found that the structure of Ph1771p resembles the translational regulator Hfq from Staphylococcus aureus (Schumacher et al. 2002) and ribosomal protein L21E from Haloarcula marismortui (Ban et al. 2000). The bacterial Hfq protein is known to be a structural homolog of the eukaryotic Sm proteins (Kambach et al. 1999), the core components of spliceosomal small nuclear ribonucleo-protein particles. The Sm protein family contains an N-terminal α-helix followed by a twisted five-stranded β-sheet known as an Sm-like fold in the SCOP database (Fig. 3; Murzin et al. 1995). The most conserved features of the Sm-like fold are in β2 and β5 strands: (1) the β2 strand is long and strongly coiled, which is promoted by a conserved Gly at the middle of the strand, and (2) the last strand, β5, forms an antiparallel β-sheet with the first strand, β1. The first five β-strands of Ph1771p are topologically similar to those of Hfq and L21E with rms deviations of 1.29 Å and 2.38 Å for 41 and 40 corresponding Cα atoms, respectively. There are, however, slight differences among these; that is, Ph1771p has an additional strand, β6, which forms an antiparallel β-sheet with strand β5, and an extended C-terminal region containing three helices (α2–α4) and strand β7. Furthermore, the location and orientation of helix α1 of Ph1771p are slightly different from those of the two other proteins. Nevertheless, the β-barrel structures of Ph1771p and Sm family proteins exhibit essentially identical core conformations, as described above, and in addition, Gly68 in Ph1771p, located in the middle of strand β2, is highly conserved among the Sm-like folds to maintain the highly distorted β-barrel structure, though Ph1771p has insignificant sequence identity (<12%) with these two RNA binding proteins. It is thus likely that Ph1771p and RNA binding proteins belonging to the Sm family have evolved from a common ancestor to bind to each cognate RNA molecule.
FIGURE 3.
Structure comparison of Ph1771p with other proteins of a similar fold. (Left) Translational regulator Hfq (residues 6–65) from Staphylococcus aureus; (center) archaeal RNase P protein Ph1771p from P. horikoshii (residues 35–127); and (right) ribosomal protein L21E (residues 24–95) from Haloarcula marismortui. The first five β-strands (β1–β5) superimpose well.
Among RNA binding motifs with a β-barrel, OB-fold (oligonucleotide/oligosaccharide binding-fold; Murzin 1993) and PAZ (Piwi/Argonaute/Zwille) domain (Lingel et al. 2003) are known to have a five- or six-stranded β-barrel structure. The OB-fold comprises a five-stranded antiparallel β-barrel. The most conserved features of the OB-fold are in strands β1 and β4 of the barrel: β1 is in the middle of the β-sheet and is long and strongly coiled, and β4 is connected to β5 by a short loop, forming an antiparallel β-sheet. In contrast, the Sm-like fold found in the Sm family proteins is described as a β-sandwich comprising two sets of three-stranded antiparallel β-sheets and is characteristic in that the last strand is interrupted by a turn of 310-helix. As for the PAZ domain, it consists of a five-stranded β-barrel capped at one end by two α-helices and wrapped on one side by an additional long β-hairpin and a short α-helix; the central β-barrel has some resemblance to that of Ph1771p. However, the β-hairpin structure and α-helix are inserted between β2 and β3 in the PAZ domain; the additional domain plays a role in RNA binding (Lingel et al. 2003). In this context, the β-barrel structure of Ph1771p seems to be more closely similar to the Sm-like fold than to the OB-fold and PAZ domain.
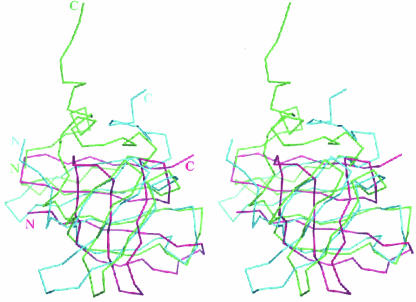
During the preparation of the present report, the solution structures of archaeal homologous proteins of Ph1771p from Archaeoglobus fulgidus (referred to as aRpp29; Sidote and Hoffman 2003) and M. thermoautotrophicus (referred to as Mth11/MthRpp29; Boomershine et al. 2003) were reported. As shown in Figure 1, the P. horikoshii protein Ph1771p shares 51% and 36% identical residues with the two archaeal homologous proteins Mth11/MthRpp29 and aRpp29, respectively, although it has an N-terminal extension with about 30 amino acids. Figure 4 provides a comparison of the molecular structures of Ph1771p and two archaeal homologous proteins. Although N- and C-terminal regions of both aRpp29 and Mth11/MthRpp29 are not defined in the structures, it is still evident that the core region (residues 50–110) of Ph1771p has a folding topology similar to the corresponding regions in the two archaeal molecules. Particularly, the Ph1771p structure can be superimposed with that of Mth11/MthRpp29, giving the rms deviation value of 2.79 Å for 60 corresponding Cα atoms. There are, however, some differences in the corresponding structural elements between Ph1771p and aRpp29 in terms of their relative locations and orientations, as indicated by the larger rms deviation value (4.08 Å for 60 Cα atoms). The obvious differences are at the loop regions connecting strands β1 and β2, β2 and β3, β3 and β4, and β5 and β6. In addition, the orientations of the three strands β3, β4, and β6 differ slightly. These structural differences including their N- and C-terminal regions attached to the central β-barrel probably reflect a structural diversity of RNase Ps in P. horikoshii and A. fulgidus. Alternatively, the structural differences observed could be partly attributed to a methodological difference between X-ray crystallography and NMR spectroscopy.
FIGURE 4.
Stereoview of the superposition of the molecular structures of Ph1771p from P. horikoshii and homologous proteins aRpp29 and Mth11/Mth Rpp29 from A. fulgidus (Sidote and Hoffman 2003) and M. thermoautotrophicus (Boomershine et al. 2003), respectively. The α-carbon tracings of Ph1771p, aRpp29, and Mth/Mth Rpp29 are shown in green, violet, and blue, respectively.
Potential sites for RNA binding
In our foregoing study, gel retardation analysis demonstrated that Ph1771p is implicated in interaction with RNase P RNA (Kouzuma et al. 2003). Similar observations were also reported in eukaryotic RNase P: the homologous proteins, Rpp29 and Pop4p, from human and Saccharomyces cerevisiae nuclear RNase P, respectively, were found to interact with each RNase P RNA subunit in yeast three-hybrid systems (Jiang et al. 2001; Houser-Scott et al. 2002). Although the role of the protein subunits in the RNase P reaction is not entirely clear, the fact that they confer high activity on the RNA suggests that RNase P proteins may function as an RNA chaperon and stabilize the holoenzyme against electrostatic distortion. As described above, Ph1771p belongs to the Sm protein family, members of which control many aspects of RNA processing in eukaryotic and eubacterial cells. The crystal structures showed that the Sm family proteins form homo- or hetero-oligomeric complexes to bind to single-stranded RNA molecules (Kambach et al. 1999; Collins et al. 2001; Mura et al. 2001; Schumacher et al. 2002). On the basis of structural analysis as well as structural comparison with the Sm protein family, we propose two likely RNA binding regions in Ph1771p.
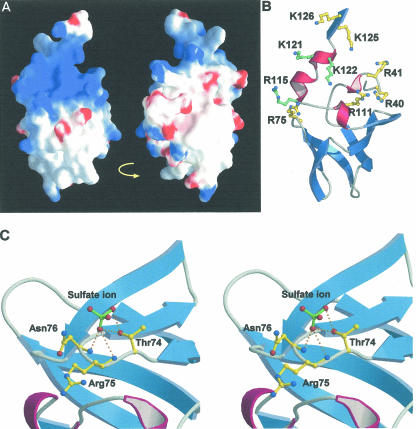
The first RNA binding region is the concave surface, where basic amino acids are clustered (Fig. 5A,B). The electrostatic potential on the molecular surface of the Ph1771p, depicted in Figure 5A, reveals that the basic patch occurred around the concave surface formed at the upper side of the molecule. Positively charged residues Arg40, Arg41, Arg75, Arg111, Arg115, Lys121, Lys122, Lys125, and Lys126 are clustered at this concave surface (Fig. 5B). Among them, Arg115, Lys121, and Lys122 are strictly conserved as positively charged residues (either lysine or arginine) not only in archeal but also in eukaryotic homologs (Fig. 1). It is thus suggested that the concave surface at the upper part of the protein may present a potential site for interaction with RNase P RNA. Indeed, our recent mutational analysis showed that double mutations of Lys121 and Lys122 significantly decreased the enzymatic activity of the reconstituted RNase P (K. Kimura and M. Kimura, unpubl.). In addition, Boomershine et al. (2003) found by NMR spectroscopic analysis of Mth11/Mth Rpp that the presence of the RNase P RNA caused shift perturbations, and the largest measurable shift perturbations were induced in amides of Arg25, Phe63, and Thr65, and in residues at the N- and C-terminal segments. Interestingly, Phe97 in Ph1771p, which corresponds to Phe63 in Mth11/Mth Rpp29, is located in close proximity to the concave surface. It is likely that the concave surface formed by the terminal helices and β-strand β6 may serve as one RNA binding site in Ph1771p.
FIGURE 5.
Possible RNA binding site in Ph1771p. (A) Surface representation of the electrostatic potential of Ph1771p, as calculated by GRASP (Nicholls et al. 1991). Blue is electropositive and red is electronegative. (Left) Front view of Ph1771p: The orientation of this image is the same as in (B). (Right) Back view of Ph1771p: View is rotated by 180° from left panel. (B) A cluster of positively charged residues located at the concave surface of Ph1771p. Basic amino acids that are candidates for the interaction with RNase P RNA are shown in a ball and stick representation. Positively charged residues that are strictly conserved in homologous proteins are drawn in green, the others in yellow. (C) Stereoview of a ball and stick representation of the second potential site for RNA binding located at the conserved loop connecting strands β2 and β3. Sulfur atom is in green. Hydrogen bonds are shown as dotted lines. The side chains of Thr74, Arg75, and Asn76 are exposed to the solvent. The side chain of Thr74 and main chain atoms of Arg75 and Asn76 are responsible for specific binding of sulfate anion. These observations may reflect the biological significance that the phosphate moiety of RNase P RNA specifically binds to this region of Ph1771p.
The second site for RNA binding is located at the loop region connecting strands β2 and β3, in close proximity to the basic patch (Fig. 5C). As described above, Ph1771p most closely resembles the fold of the translational regulator Hfq, which forms a homohexameric ring for RNA binding. Schumacher et al. (2002) revealed the Hfq-RNA interactions, where a single-stranded RNA molecule specifically binds in a circular conformation along with a central cleft of a homohexameric ring. The side chains of Lys41 and Tyr42 in Hfq interact with the base moieties of oligoribonucleotide by hydrogen bonding and a stacking interaction, respectively. On the basis of structural alignment of Ph1771p and Hfq proteins, the amino acids of Lys41 and Tyr42 in Hfq correspond to Arg75 (also one of the amino acids composing the basic patch) and Asn76 in Ph1771p, respectively. A comparison of primary structures of Ph1771p homologs revealed that amino acids Arg75 and Asn76, located at the loop region connecting strands β2 and β3, are well conserved as hydrophilic residues (Fig. 1). In the crystal structure of mPh1771p, the amino acids of Arg75 and Asn76 are exposed to the solvent (Fig. 5C), suggesting that the side chains of these residues could be involved in intermolecular contacts with RNase P RNA.
Intriguingly, each of the two sulfate anions specifically bound to the loop region connecting strands β2 and β3 in a similar manner in each molecule of Ph1771p observed in the crystallographic asymmetric unit (Fig. 5C). Therefore, it could be assumed that the sulfate anion bound to this site may have a specific biological significance. The oxygen atom of sulfate anion accepts hydrogen bonds from the side chain hydroxyl group of Thr74 and main chain amide nitrogen atoms of Arg75 and Asn76. The structure of the Hfq-RNA complex showed that in addition to interactions of the side chains of Lys41 and Tyr42 with the RNA, the main chain atom of Lys41 (corresponding to Arg75 in Ph1771p) forms a hydrogen bond to the base moiety, and that of Tyr42 (corresponding to Asn76 in Ph1771p) participates in the interaction with the phosphate backbone of the RNA molecule (Schumacher et al. 2002). These results, together with the fact that Thr74 in Ph1771p is also highly conserved in the homologous proteins (Fig. 1), indicate that the loop region connecting strands β2 and β3 are likely to contact the phosphate moiety of the RNA molecule, as is the case in the specific sulfate anion bound to it.
Possible interactions with other proteins
In contrast to the protein component of the bacterial RNase P, the archaeal and eukaryotic RNase P are composed of several protein subunits associated with a single RNA subunit. Thus, protein subunits in the archaeal and eukaryotic RNase P have crucial roles not only in the interaction with RNase P RNA but also in the protein-protein interactions for assembly of the RNase P molecule. The protein-protein interactions between archaeal RNase P protein subunits from Methanothermobacter thermoautotrophicus were recently studied by means of a yeast two-hybrid system (Hall and Brown 2004). As a result, a strong interaction between Mth11p (M. thermoautotrophicus homolog of Ph1771p) and Mth1618p (M. thermoautotrophicus homolog of Ph1601p) was observed. It was further revealed that Mth11p weakly interacts with Mth687p and Mth688p (M. thermoautotrophicus homologs of Ph1481p and Ph1877p, respectively) and with Mth11p by itself. Similar results were obtained using eukaryotic RNase P (Jiang and Altman 2001; Houser-Scott et al. 2002). It is thus tempting to speculate that archaeal and eukaryotic homologs of Ph1771p may function as a scaffold in the assembly of protein components on the RNA molecule.
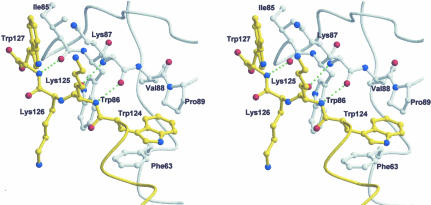
In the crystal structure of mPh1771p, strand β7 (residues 124–127) located at the C-terminal forms an antiparallel intersubunit β-sheet by interacting with strand β4 in a symmetry-related molecule (Fig. 6). At the same time, its own strand β4 hydrogen bonds with strand β7 of a second symmetry-related molecule at the opposite interface. The interface of the adjacent molecules was formed by hydrophobic interaction. Two hydrophobic residues, Trp124 and Trp127, and the aliphatic portion of Lys125 and Lys126 in the C-terminal region are capped by the hydrophobic surface formed by Phe63, Ile85, Trp86, Lys87, Val88, and Pro89 from a symmetry-related molecule. Although we cannot exclude the possibility that the interaction of β7 in one molecule with β4 in the symmetry-related molecule observed is a crystallographic artifact, the weak interaction between Mth11p (Ph1771p homolog) and itself could reflect the protein-protein interaction between one molecule and an adjacent symmetry-related molecule, as observed in this study.
FIGURE 6.
Intermolecular β-sheet pairing in Ph1771p. A close-up stereoview of the interface of two Ph1771p molecules including hydrophobic cluster. One molecule is yellow and the other white. Interacting residues are shown as a ball and stick representation. Hydrogen bonding formed between the two β-strands are represented as dotted lines.
It is of interest to note that Sm family proteins, exhibiting a structural resemblance to Ph1771p (described above), oligomerize to form a homo- or hetero-oligomeric complex through the β strand pairing and hydrophobic interactions (Kambach et al. 1999; Collins et al. 2001; Mura et al. 2001; Schumacher et al. 2002). As for the β strand pairing, strand β4 in one subunit, together with strand β5 in an adjacent subunit, forms an intersubunit antiparallel β-sheet, and then in turn its own strand β5 forms an antiparallel β-sheet by interacting with strand β4 at the opposite interface. Although unlike Sm family proteins Ph1771p molecules do not form a ring structure, Ph1771p interacts with itself in a similar manner to the protein-protein interaction in Sm family proteins; that is, β-sheet pairing (antiparallel β-sheet) occurred at strand β4 and β7 in Ph1771p, but for strand β4 and β5 in Sm family proteins. In both cases, the hydrophobic interactions formed between each molecule are of critical importance for stability on the interface of the molecules. These results, together with the finding that the Ph1771p homolog is involved in interactions with other protein subunits, suggest that strands β4 and β7 in the Ph1771p homolog may be responsible for intermolecular interactions with other RNase P proteins, particularly the Ph1601p homolog. Further studies such as mutational and crystallographic analyses will be needed to confirm this speculation.
Summary
In the present study, the structure of the mutant mPh1771p, which lacks the N-terminal 31 amino acids and contains a C93S mutation, was determined in an X-ray crystallographic study. The N-terminal deletion was chosen by a sequence comparison with other homologous proteins. Although the protein mPh1771p was found to be resistant to protease digestions (data not shown), we have no experimental data indicating a core structure of Ph1771p. We found that the RNase P reconstituted with mPh1771p had reduced enzymatic activity (15%) compared to that of the reconstituted RNase P with the wild-type Ph1771p. This finding indicated that the N-terminal extension in Ph1771p is involved in the catalytic activity of RNase P. As the N-terminal extension of Ph1771p is highly positively charged because of its high content of basic amino acids (32%), it could play an important role in interactions with either RNA molecules such as RNase P RNA and pre-tRNA or the other protein subunits. Alternatively, the N-terminal residues may participate in a proper folding of Ph1771p; the truncation may cause an incorrect folding of mPh1771p, thereby resulting in the reduced enzymatic activity of the reconstituted RNase P. Therefore, a structural analysis of the N-terminal extension of Ph1771p is a prerequisite for a full understanding of the structure-function relationship of Ph1771p. For this purpose, we intend to crystallize the overall structure of Ph1771p liganded with the interacting partners; this may provide insight into enzymatic functions of archaeal and eukaryotic RNase P.
MATERIALS AND METHODS
Protein preparation
In our earlier study, full-length Ph1771p was produced as an inclusion body (Kouzuma et al. 2003). Although we initially attempted to crystallize the full-length Ph1771p, which was purified under denaturing conditions, it has yet to be crystallized under any conditions we used. Therefore, we constructed an expression system for the mutant form Ph1771p (referred to as mPh1771p), in which 31 amino acids at the N-terminal region were truncated and a single cysteine residue, Cys93, was further replaced by Ser (Fig. 1). The gene encoding mPh1771p was generated as follows: Cys93 was mutated to Ser by site-directed mutagenesis on the template DNA coding for the full-length Ph1771p in the pGEM T-easy vector using QuikChange Site-Directed Mutagenesis Kits (Strata-gene), done as suggested by the manufacturer. Subsequently, the gene encoding mPh1771p was PCR-amplified from the template DNA containing the C93S mutation described above. Oligo-nucleotide primers were designed to include artificial sites for NdeI and HindIII to allow the PCR product to be placed under control of the T7 phage promoter on the expression plasmid pET-22b (Studier et al. 1990). Selenomethionyl recombinant protein was produced in E. coli strain B834(DE3), then purified from a total cell extract by a series of column chromatography, as follows: after sonication of E. coli cells in 50 mM Tris-HCl buffer, pH 7.0, the supernatant was first put on a RESOURCE S column (16 × 30 mm) previously equilibrated with the same buffer. The protein was eluted with a linear gradient of 0–1.0 M NaCl, in the same buffer, using a gradient volume of 60 mL. The fractions being rich in the protein, as evaluated by SDS-PAGE, were pooled and dialyzed against 50 mM sodium acetate buffer, pH 4.0 containing 0.5 M NaCl. The recombinant protein was again purified on a RESOURCE S column (16 × 30 mm) that had been equilibrated with the same buffer. The elution process followed a linear gradient of 0.5–1.5 M NaCl, using a gradient volume of 60 mL. The purified protein was extensively dialyzed against 50 mM sodium acetate buffer, pH 4.0, containing 0.4 M NaCl.
Crystallization and data collection
The purified protein was concentrated to 6–8 mg/mL in 50 mM sodium acetate buffer, pH 4.0, containing 0.4 M NaCl. Crystallization conditions were screened using the sparse-matrix sampling method by sitting drop vapor diffusion at 20°C. Each drop was formed by mixing equal volumes (0.8 μL:0.8 μL) of protein and reservoir solution. Small crystals of mPh1771p were obtained under condition 4 (1.0 M ammonium sulfate in 100 mM Tris-HCl, pH 8.5) of Crystal Screen Lite (Hampton Research). To obtain a large single crystal suitable for X-ray analysis, the crystallization method was changed to the hanging-drop vapor diffusion method by mixing 1 μL of protein solution and 1 μL of reservoir solution. Plate-like crystals were grown within a few days to a size of up to 0.2 × 0.2 × 0.02 mm3.
For data collection, crystals of mPh1771p were transferred into a cryoprotectant solution containing 15% glycerol in reservoir solution, then mounted in a nylon loop and flash-frozen in a nitrogen stream at 100 K. MAD data of selenomethionyl recombinant protein was collected from a single crystal at cryogenic temperature (100 K) at the BL38B1 station of Spring-8, Japan. A crystal of mPh1771p diffracts to 2.0 Å and belongs to space group P21 with cell dimensions of a = 24.1 Å, b = 56.3 Å, c = 71.8 Å, β = 96.8°. The crystal contains two molecules in the asymmetric unit, with a solvent content of 41.1% and a VM value of 2.17 Å3/Da, which is in the range of most common protein crystals. Two data sets were collected on and around the selenium K absorption edge. All data were integrated and scaled with HKL2000 (Otwinowski and Minor 1997). The processing statistics are given in Table 1.
Structure determination and refinement
The structure of mPh1771p was determined using the MAD data with the program package SOLVE/RESOLVE (Terwilliger and Berendzen 1999). Four sites of selenium atom were determined from the difference Patterson map. The density modification was applied to electron density derived from the MAD phases. The model of mPh1771p was built automatically using RESOLVE and ARP/wARP (Perrakis et al. 1999), and manually modified using program O (Jones et al. 1991). Because the N-terminal regions in both molecules were unclear in the electron density map, these segments were excluded from model building. The molecular model was subsequently improved with several rounds of rigid-body, simulated annealing, and individual temperature factor refinements with CNS (Brunger et al. 1998) and manual rebuilding in O. At the current stage of refinement, the model has an Rwork of 20.9% for 90% of the data between 30.1 and 2.0 Å resolution, including residues 36–127 (molecule A) and 37–127 (molecule B) for crystallographically independent molecules, 144 water molecules, and two sulfate anions, for a total of 1670 atoms. The Rfree for the remaining 10% of the data within this resolution range is 25.7%. The rms deviations from standard values of bond length and bond angles are 0.005 Å and 1.30°, respectively. The stereochemistry of the models was verified using PROCHECK (Laskowski et al. 1993). Refinement statistics are summarized in Table 1.
Protein Data Bank entry
The atomic coordinates and structure factors have been deposited in the Protein Data Bank under the accession code 1V76.
Acknowledgments
We thank Dr. Kazuya Hasegawa of JASRI for assistance with data collection using synchrotron radiation at BL38B1 of Spring-8. We are grateful to M. Ohara (Fukuoka) for helpful comments on the manuscript. This work was supported in part by a grant from the National Project on Protein Structural and Functional Analyses, and by a Grant-in-Aid for Scientific Research from the Ministry of Education, Science, Technology, Sports and Culture of Japan, and further by Research Fellowships of the Japan Society for the Promotion of Science for Young Scientists.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.rnajournal.org/cgi/doi/10.1261/rna.7560904.
REFERENCES
- Andrews, A.J., Hall, T.A., and Brown, J.W. 2001. Characterization of RNase P holoenzymes from Methanococcus jannaschii and Methanothermobacter thermoautotrophicus. Biol. Chem. 382: 1171–1177. [DOI] [PubMed] [Google Scholar]
- Ban, N., Nissen, P., Hansen, J., Moore, P.B., and Steitz, T.A. 2000. The complete atomic structure of the large ribosomal subunit at 2.4 Å resolution. Science 289: 905–920. [DOI] [PubMed] [Google Scholar]
- Bartkiewicz, M., Gold, H., and Altman, S. 1989. Identification and characterization of an RNA molecule that copurifies with RNase P activity from HeLa cells. Genes & Dev. 3: 488–499. [DOI] [PubMed] [Google Scholar]
- Boomershine, W.P., McElroy, C.A., Tsai, H-Y., Wilson, R.C., Gopalan, V., and Foster, M.P. 2003. Structure of Mth11/Mth Rpp29, an essential protein subunit of archaeal and eukaryotic RNase P. Proc. Natl. Acad. Sci. 100: 15398–15403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunger, A.T., Adams, P.D., Clore, G.M., DeLano, W.L., Gros, P., Grosse-Kunstleve, R.W., Jiang, J.S., Kuszewski, J., Nilges, M., Pannu, N.S., et al. 1998. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. D54: 905–921. [DOI] [PubMed] [Google Scholar]
- Collins, B.M., Harrop, S.J., Kornfeld, G.D., Dawes, I.W., Curmi, P.M.G., and Mabbutt, B.C. 2001. Crystal structure of a heptameric Sm-like protein complex from archaea: Implications for the structure and evolution of snRNPs. J. Mol. Biol. 309: 915–923. [DOI] [PubMed] [Google Scholar]
- Darr, S.C., Pace, B., and Pace, N.R. 1990. Characterization of ribonuclease P from the archaebacterium Sulfolobus solfataricus. J. Biol. Chem. 265: 12927–12932. [PubMed] [Google Scholar]
- Frank, D.N. and Pace, N.R. 1998. Ribonuclease P: Unity and diversity in a tRNA processing ribozyme. Annu. Rev. Biochem. 67: 153–180. [DOI] [PubMed] [Google Scholar]
- Gibrat, J.F., Madej, T., and Bryant, S.H. 1996. Surprising similarity in structure comparison. Curr. Opin. Struct. Biol. 6: 377–385. [DOI] [PubMed] [Google Scholar]
- Guerrier-Takada, C., Gardiner, K., Marsh, T., Pace, N., and Altman, S. 1983. The RNA moiety of ribonuclease P is the catalytic subunit of the enzyme. Cell 35: 849–857. [DOI] [PubMed] [Google Scholar]
- Hall, T.A. and Brown, J.W. 2004. Interactions between RNase P protein subunits in Archaea. Archaea 1: (in press). [DOI] [PMC free article] [PubMed]
- Holm, L. and Sander, C. 1993. Protein structure comparison by alignment of distance matrices. J. Mol. Biol. 233: 123–138. [DOI] [PubMed] [Google Scholar]
- Houser-Scott, F., Xiao, S., Millikin, C.E., Zengel, J.M., Lindahl, L., and Engelke, D.R. 2002. Interactions among the protein and RNA subunits of Saccharomyces cerevisiae nuclear RNase P. Proc. Natl. Acad. Sci. 99: 2684–2689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarrous, N. 2002. Human ribonuclease P: Subunits, function, and intranuclear localization. RNA 8: 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang, T. and Altman, S. 2001. Protein-protein interactions with subunits of human nuclear RNase P. Proc. Natl. Acad. Sci. 98: 920–925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang, T., Guerrier-Takada, C., and Altman, S. 2001. Protein-RNA interactions in the subunits of human nuclear RNase P. RNA 7: 937–941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, T.A., Zou, J.Y., Cowan, S.W., and Kjeldgaad, M. 1991. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A47: 110–119. [DOI] [PubMed] [Google Scholar]
- Kambach, C., Walke, S., Young, R., Avis, J.M., de la Fortelle, E., Raker, V.A., Luhrmann, R., Li, J., and Nagai, K. 1999. Crystal structures of two Sm protein complexes and their implications for the assembly of the spliceosomal snRNPs. Cell 96: 375–387. [DOI] [PubMed] [Google Scholar]
- Kazantsev, A.V., Krivenko, A.A., Harrington, D.J., Carter, R.J., Holbrook, S.R., Adams, P.D., and Pace, N.R. 2003. High-resolution structure of RNase P protein from Thermotoga maritima. Proc. Natl. Acad. Sci. 100: 7497–7502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouzuma, Y., Mizoguchi, M., Takagi, H., Fukuhara, H., Tsukamoto, M., Numata, T., and Kimura, M. 2003. Reconstitution of archaeal ribonuclease P from RNA and four protein components. Biochem. Biophys. Res. Commun. 306: 666–673. [DOI] [PubMed] [Google Scholar]
- Kraulis, P.J. 1991. MOLSCRIPT: A program to produce both detailed and schematic plots of protein structures. J. Appl. Crystallogr. 24: 946–950. [Google Scholar]
- Laskowski, R.A., MacArthur, M.W., Moss, D.S., and Thornton, J.M. 1993. PROCHECK: A program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 26: 283–291. [Google Scholar]
- Lingel, A., Simon, B., Izaurralde, E., and Sattler, M. 2003. Structure and nucleic-acid binding of the Drosophila Argonaute 2 PAZ domain. Nature 426: 465–469. [DOI] [PubMed] [Google Scholar]
- Mann, H., Ben-Asouli, Y., Schein, A., Moussa, S., and Jarrous, N. 2003. Eukaryotic RNase P: Role of RNA and protein subunits of a primordial catalytic ribonucleoprotein in RNA-based catalysis. Mol. Cell 12: 925–935. [DOI] [PubMed] [Google Scholar]
- Merrit, E.A. and Murphy, M.E.P. 1994. Raster3D version 2.0, a program for photorealistic molecular graphics. Acta Crystallogr. D50: 869–873. [DOI] [PubMed] [Google Scholar]
- Mura, C., Cascio, D., Sawaya, M.R., and Eisenberg, D.S. 2001. The crystal structure of a heptameric archaeal Sm protein: Implications for the eukaryotic snRNP core. Proc. Natl. Acad. Sci. 98: 5532–5537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murzin, A.G. 1993. OB (oligonucleotide/oligosaccharide binding)-fold: Common structural and functional solution for non-homologous sequences. EMBO J. 12: 861–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murzin, A.G., Brenner, S.E., Hubbard, T., and Chothia, C. 1995. SCOP: A structural classification of protein database for the investigation of sequences and structures. J. Mol. Biol. 247: 536–540. [DOI] [PubMed] [Google Scholar]
- Nicholls, A., Sharp, K., and Honig, B. 1991. Protein folding and association: Insights from the interfacial and thermodynamic properties of hydrocarbons. Proteins Struct. Funct. Genet. 11: 281–296. [DOI] [PubMed] [Google Scholar]
- Nieuwlandt, D.T., Haas, E.S., and Daniels, C.J. 1991. The RNA component of RNase P from the archaebacterium Haloferax volcanii. J. Biol. Chem. 266: 5689–5695. [PubMed] [Google Scholar]
- Otwinowski, Z. and Minor, W. 1997. Processing of x-ray diffraction data collected in oscillation mode. Methods Enzymol. 276: 307–326. [DOI] [PubMed] [Google Scholar]
- Pace, N.R. and Brown, J.W. 1995. Evolutionary perspective on the structure and function of ribonuclease P, a ribozyme. J. Bacteriol. 177: 1919–1928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pannucci, J.A., Haas, E.S., Hall, T.A., Harris, J.K., and Brown, J.W. 1999. RNase P RNAs from some Archaea are catalytically active. Proc. Natl. Acad. Sci. 96: 7803–7808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrakis, A., Morris, R., and Lamzin, V.S. 1999. Automated protein model building combined with iterative structure refinement. Nat. Struct. Biol. 6: 458–463. [DOI] [PubMed] [Google Scholar]
- Schumacher, M.A., Pearson, R.F., Moller, T., Valentin-Hansen, P., and Brennan, R.G. 2002. Structures of the pleiotropic translational regulator Hfq and an Hfq-RNA complex: A bacterial Sm-like protein. EMBO J. 21: 3546–3556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidote, D.J. and Hoffman, D.W. 2003. NMR structure of an archaeal homologue of ribonuclease P protein Rpp29. Biochemistry 42: 13541–13550. [DOI] [PubMed] [Google Scholar]
- Spitzfaden, C., Nicholson, N., Jones, J.J., Guth, S., Lehr, R., Prescott, C.D., Hegg, L.A., and Eggleston, D.S. 2000. The structure of ribonuclease P protein from Staphylococcus aureus reveals a unique binding site for single-stranded RNA. J. Mol. Biol. 295: 105–115. [DOI] [PubMed] [Google Scholar]
- Stams, T., Niranjanakumari, S., Fierke, C.A., and Christianson, D.W. 1998. Ribonuclease P protein structure: Evolutionary origins in the translational apparatus. Science 280: 752–755. [DOI] [PubMed] [Google Scholar]
- Studier, F.W., Rosenberg, A.H., Dunn, J.J., and Dubendorff, J.W. 1990. Use of T7 RNA polymerase to direct expression of cloned genes. Methods Enzymol. 185: 60–89. [DOI] [PubMed] [Google Scholar]
- Takagi, H., Watanabe, M., Kakuta, Y., Kamachi, R., Numata, T., Tanaka, I., and Kimura, M. 2004. Crystal structure of the ribonuclease P protein Ph1877p from hyperthermophilic archaeon Pyrococcus horikoshii OT3. Biochem. Biophys. Res. Commun. 319: 787–794. [DOI] [PubMed] [Google Scholar]
- Terwilliger, T.C. and Berendzen, J. 1999. Automated MAD and MIR structure solution. Acta Crystallogr. D55: 849–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao, S., Houser-Scott, F., and Engelke, D.R. 2001. Eukaryotic ribonuclease P: Increased complexity to cope with the nuclear pre-tRNA pathway. J. Cell Physiol. 187: 11–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan, Y. and Altman, S. 1995. Substrate recognition by human RNase P: Identification of small, model substrates for the enzyme. EMBO J. 14: 159–168. [DOI] [PMC free article] [PubMed] [Google Scholar]