Abstract
MicroRNAs (miRNAs) are ~21-nt-long RNAs involved in regulating development, differentiation, and other processes in eukaryotes. In metazoa, nearly all miRNAs control gene expression by imperfectly base-pairing with the 3′slated region (3′f target mRNAs and repressing protein synthesis by an unknown mechanism. It is also unknown whether miRNA–mRNA duplexes containing mismatches and bulges provide specific features that are recognized by factors mediating the repression. miRNAs form part of ribonucleoprotein complexes, miRNPs, that contain Argonaute (Ago) and other proteins. Here we demonstrate that effects of miRNAs on translation can be mimicked in human HeLa cells by the miRNA-independent tethering of Ago proteins to the 3′ a reporter mRNA. Inhibition of protein synthesis occurred without a change in the reporter mRNA level and was dependent on the number, but not the position, of the hairpins tethering hAgo2 to the 3′hese findings indicate that a primary function of miRNAs is to guide their associated proteins to the mRNA.
Keywords: Argonaute proteins, miRNA, microRNA, RNAi, RNA–protein interactions, translation
INTRODUCTION
MicroRNAs (miRNAs) are a large family of ~21-nt-long regulatory RNAs expressed in metazoan animals and plants. In animals, nearly all miRNAs investigated to date appear to regulate gene expression by imperfectly base-pairing to the 3′slated region (3′-UTR) of target mRNAs and inhibiting protein synthesis by an unknown mechanism (for review, see Bartel 2004). Although an example of a miRNA affecting mRNA translation in Arabidopsis has been reported (Chen 2004), most plant miRNAs show nearly precise complementarity to target mRNAs, and trigger mRNA degradation via a mechanism similar to that operating during RNA interference (RNAi), which involves ~21-nt small interfering RNAs (siRNAs) (Bartel 2004).
The first miRNAs, lin-4 and let-7, were discovered in Caenorhabditis elegans, as RNAs that regulate expression of mRNAs, which control the timing of larval development (Lee et al. 1993; Wightman et al. 1993; Reinhart et al. 2000; Slack et al. 2000; Bartel 2004). More recently, hundreds of new miRNAs have been identified through cloning, and genetic and bioinformatic methods (Bartel 2004). Biological processes regulated by miRNAs in metazoa include developmental timing, cell differentiation and proliferation, neuronal asymmetry, apoptosis, and fat and possibly amino acid metabolism. Existence of numerous tissue-and developmental stage-specific miRNAs, and the evolutionary conservation of many miRNAs, argue for numerous additional, yet unidentified, functions of miRNAs (Bartel 2004).
Initial studies aimed at understanding how miRNAs repress gene expression were carried out in C. elegans using lin-4 miRNA and its target lin-14 mRNA. They indicated that lin-4 interacts with multiple, partially complementary sequences at the mRNA 3′ down-regulate LIN-14 protein accumulation. The down-regulation was not accompanied by changes in mRNA level or its association with polysomes, suggesting that protein synthesis is repressed at steps downstream of translation initiation (Lee et al. 1993; Wightman et al. 1993; Ha et al. 1996; Olsen and Ambros 1999). Subsequent studies with other natural and artificial miRNAs in C. elegans, Drosophila, and mammals also indicated that these interact with the mRNA 3′-UTR to form imperfect bulged duplexes, and inhibit protein synthesis without decreasing mRNA levels (Zeng et al. 2002; Brennecke and Cohen 2003; Doench et al. 2003; Moss and Tang 2003; Zeng et al. 2003; Doench and Sharp 2004; Kiriakidou et al. 2004; Nelson et al. 2004; Vella et al. 2004).
Although miRNA and RNAi pathways have different outcomes, they are closely related mechanistically. Both miRNAs and siRNAs are part of ribonucleoprotein (RNP) particles that have overlapping protein composition (for review, see Denli and Hannon 2003; Bartel 2004). The best characterized proteins associated with these RNPs are members of the PAZ and Piwi domain (PPD) family (for review, see Carmell et al. 2002). Members of the Argonaute (Ago) subgroup of the PPD proteins were initially identified as components of the RNA-induced silencing complex (RISC) enzyme, which catalyzes the siRNA-directed cleavage of target mRNAs (Hammond et al. 2001; Martinez et al. 2002), but subsequently were also shown to be part of miRNPs (Caudy et al. 2002; Ishizuka et al. 2002; Mourelatos et al. 2002). Additional proteins cofractionating with both siRNAs and miRNAs include members of the fragile X mental retardation protein family, Tudor-SN and VIG (Caudy et al. 2002, 2003; Ishizuka et al. 2002; Jin et al. 2004). However, proteins were also identified, as exemplified by different PPD proteins in C. elegans (Tabara et al. 1999; Grishok et al. 2001) and Drosophila (Okamura et al. 2004), or different Dicers in Drosophila (Lee et al. 2004), which are either exclusively or preferentially required for RNAi but not miRNA function, and vice versa.
RISC and miRNP complexes are also related functionally. The mammalian let-7 and other miRNPs can function as RISC nucleases, able to cleave RNAs that bear sequences perfectly complementary to miRNAs (Hutvagner and Zamore 2002; Zeng et al. 2003; Meister et al. 2004). Likewise, siRNAs can repress protein synthesis, much like the endogenous miRNAs, when confronted with mRNA targets containing partially complementary sites in their 3′-UTRs (Doench et al. 2003; Zeng et al. 2003; Doench and Sharp 2004). It appears that it is the degree of mRNA complementarity to miRNA or siRNA that primarily determines whether the mRNA will undergo cleavage or translational repression.
It is not known whether miRNA–mRNA duplexes require specific features to be recognized by factors mediating the translational repression. In C. elegans, particular secondary structures containing bulged C residues appear to be essential for the inhibition of lin-14 mRNA by lin-4 (Ha et al. 1996). Moreover, in the lin-41 mRNA 3′-UTR, only two of the six predicted let-7 complementary sites are functional, together with the 27-nt spacer separating them (Vella et al. 2004). In mammalian cells, however, reporter mRNAs containing sequences capable of forming imperfect duplexes of rather arbitrary sequence are repressed by natural or artificial miRNAs (Doench et al. 2003; Zeng et al. 2003; Doench and Sharp 2004), arguing against specific structural requirements.
To investigate whether repression of translation in fact requires the miRNA–mRNA interaction, or whether miRNAs merely function as guides, positioning repressive factors on the 3′-UTR, we directly tethered protein components of miRNPs to the mRNA. We found the outcome of tethering Ago proteins to the 3′-UTR of mRNA in HeLa cells to be similar to that of the repression mediated by miRNAs. These findings demonstrate that a primary function of miRNAs is to guide their associated proteins to the mRNA.
RESULTS AND DISCUSSION
Tethering of hAgo2 to reporter mRNAs inhibits protein synthesis
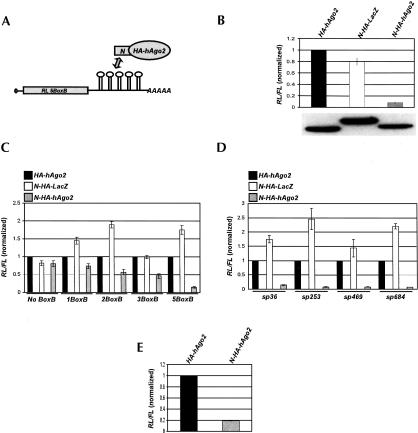
We investigated whether the inhibitory effect of miRNAs on gene expression can be mimicked in transfected human HeLa cells by tethering a human hAgo2 protein, a member of the PPD family and an established component of RISC and miRNPs (Martinez et al. 2002; Mourelatos et al. 2002), to the mRNA reporter. The tethering was achieved through coexpression of the Renilla reniformis luciferase (RL) mRNA containing five B-box hairpins (Gehring et al. 2003) in its 3′-UTR (the reporter referred to as RL-5BoxB), and the N-HA-hAgo2 protein, which is a fusion of the HA-tagged hAgo2 with a 22-amino-acid-long N peptide specifically recognizing the B box hairpin (Fig. 1A ▶; Gehring et al. 2003). RL activity was measured 48 h posttransfection by the dual luciferase assay, with the Photinus pyralis (firefly) luciferase (FL) activity, expressed from a cotransfected plasmid, serving as transfection control.
FIGURE 1.
Tethered hAgo2 down-regulates protein synthesis. (A) Scheme of the basic RL mRNA reporter, containing five 19-nt BoxB hairpins, interacting with N-HA-hAgo2. (B) RL activity detected in extracts from HeLa cells expressing the indicated fusion proteins. Cells were cotransfected with constructs expressing the RL-5BoxB reporter, FL, and indicated fusion proteins. Histograms in panels B through D represent normalized mean values (±SD) of RL/FL activities from a minimum of three experiments. RL activity values seen in the presence of HA-hAgo2 were set as 1. Expression levels of fusion proteins, as determined by Western analysis using anti-HA mAb, are shown below the histograms. The aliquot of the N-HA-LacZ-expressing extract applied to the gel was 10 times smaller than aliquots of other extracts. Generally, the N-HA-LacZ protein is expressed at an ~10-fold higher level than N-HA-hAgo2 or HA-hAgo2. Increased RL activity in extracts expressing N-HA-LacZ is likely due to the effect of the protein on the stability of mRNA reporters containing BoxB sequences (see also Fig. 5 ▶). (C) Activity of reporter RL mRNAs containing different numbers of BoxB hairpins. Relative activities of different reporters in cells coexpressing HA-hAgo2 were as follows: No BoxB, 1.0; 1BoxB, 1.0; 2BoxB, 0.7; 3BoxB, 1.4; and 5BoxB, 1.2. (D) Activity of RL-5BoxB mRNAs with hairpins spaced (sp) 36, 253, 469, or 684 nt downstream of the translation termination codon. Relative activities of different reporters in cells coexpressing HA-hAgo2 were as follows: sp36, 1.0; sp253, 1.6; sp469, 1.7; and sp684, 1.0. (E) Activity of the FL-5BoxB reporter mRNA in extracts from cells expressing the indicated fusion proteins. Histograms represent normalized mean values of FL/RL activities from three experiments. FL activity seen in the presence of HA-hAgo2 was set to 1.
The results shown in Figure 1B ▶ indicated that RL activity is inhibited by N-HA-hAgo2 but not HA-hAgo2 or N-HA-LacZ control proteins expressed at comparable or higher levels, as established by Western blotting using the anti-HA antibody. The extent of inhibition was proportional to the number of B-box hairpins in the 3′-UTR (Fig. 1C ▶), consistent with observations that the presence of multiple miRNA target sites in the 3′-UTR enhances repression (Doench et al. 2003; Bartel 2004; Doench and Sharp 2004). The inhibition was independent of the position of the hairpins (Fig. 1D ▶), also in agreement with a variable placement of miRNA target sites in the 3′-UTR of natural mRNAs (Bartel 2004). The finding that increasing the distance between the termination codon and the hairpins to >600 nt does not eliminate inhibition argues against the repression being due to sterical interference with translation termination. In addition to the RL mRNA, a reporter encoding the firefly luciferase and containing five B-box hairpins in its 3′-UTRs (referred to as FL-5BoxB) was tested in the tethering assay. Expression of the FL-5BoxB reporter mRNA was inhibited by the coexpression of N-HA-hAgo2 but not the HA-hAgo2 control protein (Fig. 1E ▶). Moreover, inhibition occurred irrespective of whether expression of the hairpin-containing reporter mRNA was driven by thymidine kinase (TK) (Fig. 1B–D ▶) or cytomegalovirus (CMV) (Fig. 1E ▶; and see following) promoters.
Together, these results indicate that tethering of hAgo2 to the mRNA 3′-UTR results in inhibition of protein synthesis in transfected HeLa cells.
Cellular localization and properties of the RL-5BoxB reporter and N-HA-hAgo2
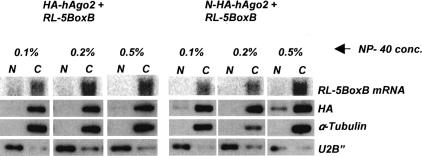
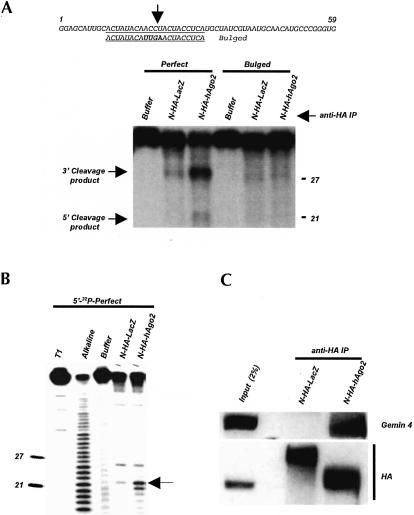
Additional control experiments were performed to demonstrate that the effects described earlier are unlikely due to mRNA reporter mislocalization or an aberrant function of the N-HA-Ago2 protein. Cell fractionation experiments indicated that both HA-hAgo2 and N-HA-hAgo2 proteins, and also the RL-5BoxB mRNA, are present in the cytoplasmic and not the nuclear compartment (Fig. 2 ▶). To acquire information about functionality of the N-HA-hAgo2 protein, we tested its competence to assemble into an enzymatically active RISC-like miRNP, and to associate with Gemin 4, an established component of miRNPs (Mourelatos et al. 2002). We found that N-HA-Ago2-containing immunoprecipitates, selected by the anti-HA antibody, catalyzed in vitro cleavage of an RNA substrate containing a perfect but not bulged let-7 miRNA target sequence (Fig. 3A ▶), as previously described for the endogenous let-7 miRNP (Hutvagner and Zamore 2002). Mapping of the cleavage site, performed with the 5′-terminally labeled substrate, revealed that the cleavage occurred at an expected position, across from miRNA nucleotides 10 and 11, measured from the miRNA 5′ end (Fig. 3B ▶; Martinez and Tuschl 2004). Immunoprecipitation experiments revealed that N-HA-hAgo2 associates with endogenous Gemin 4 in the transfected cells (Fig. 3C ▶), further indicating that the tagged protein is functional.
FIGURE 2.
Cytoplasmic localization of tagged hAgo2 proteins and the RL-5BoxB reporter mRNA. HeLa cells were cotransfected with pRL-5BoxB and plasmids expressing either N-HA-hAgo2 or HA-hAgo2. Cells were lysed in a buffer containing the different indicated concentrations of NP-40 and fractionated into nuclear (N) and cytoplasmic (C) fractions. Equivalent amounts of the fractions were analyzed for RL-5BoxB mRNA by Northern blotting and for the indicated proteins by Western blotting. The U2B″ protein and α-tubulin served as markers for nuclear and cytoplasmic proteins, respectively.
FIGURE 3.
N-HA-hAgo2 protein is active in the let-7-directed RNA cleavage and interacts with endogenous Gemin 4. (A) Processing of perfectly complementary and bulged substrates uniformly labeled with [α-32P]-UTP. The sequence of the RNA substrate is shown at the top, with the sequence complementary to let-7 RNA underlined. The corresponding sequence of the mutant RNA substrate, forming a bulged duplex on base-pairing to let-7 RNA, is shown below, with mutated nucleotides indicated in bold. (Lanes N-HA-LacZ and N-HA-hAgo2) Anti-HA immunoprecipitates (IPs) of extracts expressing N-HA-LacZ and N-HA-hAgo2 proteins, respectively. (Lane “Buffer”) Incubations performed in the absence of anti-HA IPs. Positions of the 5′ and 3′ cleavage products and 27- and 21-nt RNA markers are indicated. Note that the 3′ cleavage product contains 2.5 times more label than does the 5′ product. (B) Mapping of the cleavage site in the 5′-terminally labeled fully complementary RNA. (Lanes T1 and Alkaline) RNase T1 and alkaline ladders prepared with the same substrate RNA. Note that the ladder oligoribonucleotides contain a 3′-terminal phosphate, which is absent in the let-7-mediated 5′-cleavage product. Position of the cleavage between C and U residues is indicated by an arrow. Low processing activity seen in N-HA-LacZ control lanes in panels A and B is due to unspecific retention of endogenous let-7 miRNP on the HA-antibody-containing beads (data not shown). (C) Interaction of N-HA-Ago2 protein with endogenous Gemin 4. Anti-HA IPs from extracts expressing N-HA-LacZ and N-HA-hAgo2 fusion proteins were analyzed by Western for Gemin 4 and HA-tagged proteins. Input fraction represents 2% of the N-HA-hAgo2 extract used for IP.
In summary, these experiments indicate that both the RL-5BoxB reporter mRNA and the N-HA-hAgo2 fusion protein have cytoplasmic localization and that N-HA-hAgo2 is able to interact with established components of miRNPs. Hence, the tethering effects described earlier are unlikely to be due to the mRNA reporter mislocalization or an aberrant function of the N-HA-Ago2 protein.
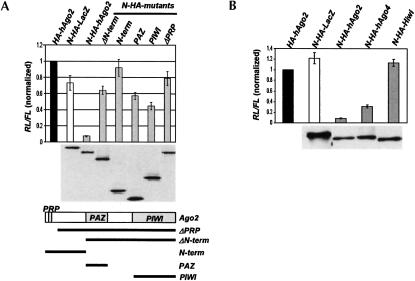
Tethering of hAgo2 mutants and the Hiwi protein is not inhibitory
We have tested different hAgo2 mutants and also other members of the PPD family of proteins for their ability to inhibit protein synthesis in the tethering assay. Analysis of the hAgo2 mutants revealed that integrity of the protein is important. None of the tested domains of hAgo2 (PRP, PAZ, and Piwi), or combinations thereof, was able to effectively inhibit RL activity at concentrations comparable to that of the complete hAgo2 (Fig. 4A ▶). The most obvious interpretation of these findings is that none of the hAgo2 fragments represents an effector domain that mediates the inhibitory effect of the miRNP. Rather, integrity of hAgo2 may be required for assembly of the functional complex.
FIGURE 4.
Tethered hAgo2 and hAgo4, but not hAgo2 mutants and Hiwi, induce the repression. (A) RL activity in extracts from HeLa cells cotransfected with plasmids expressing RL-5BoxB reporter and N-HA-tagged mutant hAgo2 fusions. Western analysis of fusion protein expression is shown below the histograms. Schematic representation of hAgo2 and its deletion mutants is shown in lower panel. (B) Tethering of hAgo4 but not Hiwi represses translation. For other details, see legend to Figure 1 ▶.
Eight different PPD proteins, four belonging to the Ago group and four to the Hiwi/Piwi group, are expressed in humans, and hAgo1 and hAgo2 were shown to form part of RISC or miRNP complexes (Martinez et al. 2002; Mourelatos et al. 2002). Like hAgo2, two other tested members of the Ago subgroup of PPD proteins, hAgo4 (Fig. 4B ▶) and hAgo3 (data not shown), inhibited RL activity when tethered to the mRNA. Hiwi, a member of a second group of PPD proteins, however, was inactive (Fig. 4B ▶). Hiwi and other members of its group do not contain the PRP motif, an ~60-amino-acid-long N-terminal domain present in the Ago PPD proteins (Doi et al. 2003). Because deletion of the PRP region completely abolished the inhibitory effect of hAgo2 (Fig. 4A ▶), we tested activity of hybrid Hiwi proteins having the N-terminal 68 or 275 amino acids replaced by the corresponding regions of hAgo2. Neither of the hybrid proteins inhibited activity of the RL-5BoxB reporter (data not shown). Hiwi is exclusively expressed in spermatocytes and hematopoietic stem cells (for review, see Carmell et al. 2002). Possibly, other proteins expressed in these cells are required for its function as a mediator of the miRNA function. Alternatively, although Hiwi shares some properties with hAgo2 (Carmell et al. 2002; Doi et al. 2003; Tahbaz et al. 2004), it may not be a part of the miRNA pathway.
The experiments presented earlier, demonstrating that full-length hAgo proteins, but not hAgo2 mutants, the Hiwi protein, or LacZ, repress protein expression when tethered to the mRNA, further argue for the specificity of the observed inhibitory effects.
Inhibition by tethered hAgo proteins occurs without changes in reporter mRNA levels
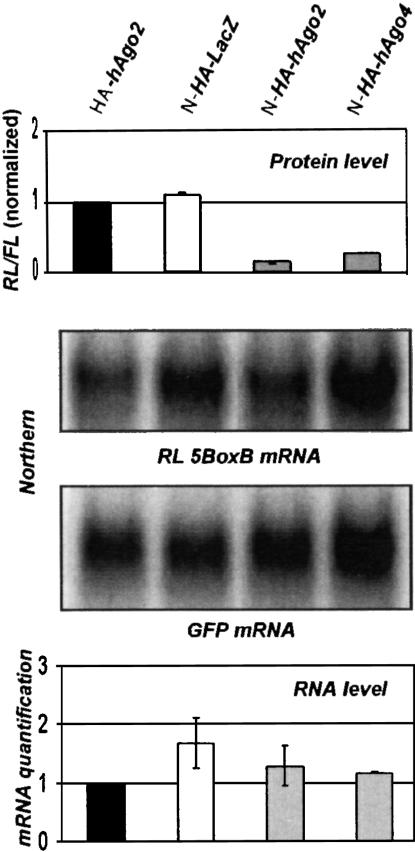
Previous work has shown that inhibition of protein synthesis by miRNAs in C. elegans and mammalian cells occurs without a significant decrease in target mRNA levels (see Introduction). To obtain additional evidence that protein tethering inhibits translation by a mechanism similar to that of miRNAs, we compared the levels of RL-5BoxB mRNA isolated from HeLa cells in which RL expression was inhibited either by hAgo2 or hAgo4 tethering. To facilitate Northern analysis, we recloned the RL-5BoxB reporter into the expression plasmid containing the CMV promoter. Similar to TK-promoter-directed expression (see Figs. 1B–D ▶, 4 ▶), activity of the RL reporter driven by a CMV promoter was strongly inhibited by the coexpression of N-HA-hAgo2 and N-HA-hAgo4 but not by HA-hAgo2 or N-HA-lacZ control proteins (Fig. 5 ▶, upper panel). Northern analysis of the RL-5BoxB mRNA indicated that for both investigated proteins, N-HA-hAgo2 and N-HA-hAgo4, repression occurred without any change in reporter mRNA level (Fig. 5 ▶, middle panels). This is further confirmed by the quantification of the RL-5BoxB mRNA Northern data from three independent experiments, normalized to GFP mRNA coexpressed in transfected cells (Fig. 5 ▶, lower panel).
FIGURE 5.
Repression by N-HA-hAgo2 and N-HA-hAgo4 occurs without changes in reporter mRNA level. Northern analysis (middle panels) was performed with total RNA isolated from transfected cells, using probes specific for RL-5BoxB mRNA and green fluorescence protein (GFP) mRNA, expressed from the cotransfected plasmid. The RL activity in extracts from the same transfected cells is shown in the upper panel. PhosphorImaging quantification of the RL-5BoxB mRNA, normalized to GFP mRNA, is shown in the lower panel; values are means (±SD) from three independent experiments. The RL-5BoxB mRNA level in cells cotransfected with HA-hAgo2 is set to 1.
We conclude that inhibition of protein synthesis by the tethering of hAgo2 and hAgo4 to the 3′-UTR is not accompanied by degradation of the reporter mRNA. This offers further support to the notion that tethering-induced inhibition occurs by a mechanism similar to that mediated by miRNAs.
CONCLUSIONS
The findings presented in this work strongly argue that the tethering of hAgo proteins to the 3′-UTR closely mimics the repression seen with miRNAs. The extent of the inhibition was directly proportional to the number of tethering hairpins in the 3′-UTR, consistent with observations that mRNAs regulated by miRNAs often contain several target sequences, and that multiplication of the miRNA binding sites in a model mRNA enhances repression (Doench et al. 2003; Bartel 2004). Inhibition was independent of the distance between the termination codon and the hairpins, in agreement with a variable placement of miRNA target sites in the 3′-UTR of natural mRNAs (Bartel 2004). In addition, tethering-induced inhibition was observed with two different reporter mRNAs, transcribed from two different promoters. Most important, repression by both hAgo2 and hAgo4 tethering occurred without a significant effect on the mRNA level. These data, taken together with the expected cellular localization and other properties of the RL-5BoxB reporter and the tethered hAgo2 protein, show unequivocally that a primary function of miRNAs is to guide their associated proteins to the mRNA. Perfect base-pairing, devoid of G:U wobbles, of the miRNA 5′-region was recently reported to be critical for efficient mRNA repression in mammalian cells (Doench and Sharp 2004). Our data would suggest that this interaction is important for the miRNP targeting and not for the inhibition reaction per se. The described tethering assay will facilitate studies of other miRNP components and the identification of proteins, or their subdomains, which mediate translational repression.
We found that tethering of hAgo2, hAgo3, and hAgo4, but not Hiwi, represses protein synthesis. In contrast to hAgo proteins, the expression of which is ubiquitous, Hiwi is exclusively expressed in spermatocytes and hematopoietic stem cells (for review, see Carmell et al. 2002). Possibly, other proteins expressed in these cells but not in HeLa cells are required for its function. Alternatively, although Hiwi shares many properties with hAgo proteins (e.g., it has a similar, although not identical, domain structure [Doi et al. 2003], and, like Ago proteins, it interacts with Dicer [Tahbaz et al. 2004]), it may not be involved in the miRNA function. We also found that none of the tested domain deletion mutants of hAgo2 effectively inhibited protein synthesis when tethered to the 3′-UTR. This suggests that none of the expressed hAgo2 regions is acting as an effector domain mediating the inhibitory effect of miRNPs. More plausibly, the integrity of hAgo2 is important for assembly of the functional complex. It will be interesting to tether other known miRNP components, and individual domains thereof, to reporter mRNAs and to measure their effect on protein synthesis.
MATERIALS AND METHODS
Construction of plasmids
Plasmids expressing tagged proteins
Annealed DNA oligonucleotides encoding the influenza hemagglutinin (HA) epitope were inserted into the vector pCI-λN (Gehring et al. 2003) to get pCI-λNHA or into pCIneo (Promega) to get the pCI-HA expression vector. For expression constructs encoding either N-HA or HA fusions, protein-coding regions were inserted at EcoRI–NotI sites downstream of the tags in the earlier-mentioned vectors. The open reading frame (ORF) sequences were PCR-amplified from plasmids originating from the following sources: hAgo2 (Tahbaz et al. 2004), hAgo4 (GenBank Accession number AB046787; obtained from the Kazusa DNA Research Institute), and Hiwi (BC028581; obtained from IMAGE Consortium). hAgo2 deletion mutants were generated by PCR amplifying required regions and inserting them into the pCI-λNHA vector. Different hAgo2 mutants encompass the following amino acid residues: ΔN-term (230–861), N-term (1–235), PAZ (230–385), Piwi (515–861), and ΔPRP (63–861); numbering is as described by Tahbaz et al. (2004). The LacZ ORF was also inserted into the pCI-λNHA vector for expression as an N-HA-tagged protein. All PCR products were verified for correctness by sequencing.
Luciferase reporter constructs
Reporters encoding R. reniformis (phRL-TK; expressing the “humanized” protein) and P. pyralis (pGL3 Promoter) enzymes were purchased from Promega. To obtain the pRL-5BoxB construct, we PCR amplified BoxB sequences from the plasmid β-globin/5boxB (Gehring et al. 2003) and inserted it into the XbaI site downstream of the stop codon of phRL-TK, resulting in a spacer of 36 nt between the stop codon and BoxB sequence cluster. An ~200-bp inert DNA fragment was sequentially inserted into the XbaI site to generate constructs with 253-, 469-, and 684-bp spacers. Constructs with 1, 2, and 3 BoxB sequences were prepared by sequential insertion of annealed oligonucleotides containing a single BoxB sequence into the XbaI site in the phRL-TK vector. For expression from a CMV promoter, cassettes containing the luciferase coding region and downstream 5BoxB sequences were inserted into the pCIneo vector. Plasmid pCMV-FL-5BoxB was generated by inserting a 5BoxB PCR product 36 nt downstream of the firefly luciferase ORF.
HeLa cell transfections, luciferase assays, cell fractionations, and Western analyses
HeLa cells grown in DMEM supplemented with 10% FBS were transiently transfected with Lipofectamine Plus (Invitrogen). Tethering experiments were performed by cotransfecting cells in triplicates in six-well plates with 500 ng of plasmids expressing N-HA or HA fusion proteins, 100 ng of pRL-5boxB or pCMV-FL-5BoxB, and 100 ng of either pGL3 Promoter or phRL-TK reference vector. When indicated, 75 ng of a GFP expression vector was also included. Luciferase assays were performed 48 h posttransfection using the Dual-Luciferase Reporter Assay kit (Promega) as per the manufacturer’s instructions. Normalized mean values and standard deviations (SD) for all experiments were calculated from at least three independent experiments. RL or FL activity values seen in the presence of HA-hAgo2 were set as 1.
For cell fractionation experiments, HeLa cells were cotransfected with pRL-5BoxB and plasmids expressing either N-HA-hAgo2 or HA-hAgo2. Cells were lysed with a buffer (5 mM Tris-Cl at pH 7.5, 1.5 mM KCl, 2.5 mM MgCl2) containing different indicated concentrations of NP-40. Following a 10-min incubation on ice, extracts were separated into nuclear (N) and cytoplasmic (C) fractions by centrifugation at 3000g for 8 min. The nuclear pellet was washed twice with lysis buffer. Equivalent amounts of the fractions were analyzed for RL-5BoxB mRNA by Northern blotting and, for the indicated proteins, by Western blotting. The U2B″ protein and α-tubulin served as markers for nuclear and cytoplasmic proteins, respectively.
For Western analysis, the following primary antibodies were used: anti-HA mAb (at 1:1000 dilution, BabCo; for visualization of N-HA-and HA-tagged proteins), α-tubulin (1:1000; Neomarkers), anti-U2B″ (1:100; ICN Biochemicals), and anti-Gemin 4 (mAb 17D10 [Mourelatos et al. 2002], at 1:500 dilution; antibody kindly supplied by Dr. G. Dreyfuss).
RNA cleavage assays and Northern analysis
For cleavage assays, HeLa cells were cotransfected with plasmids expressing either N-HA-LacZ or N-HA-hAgo2, and the siRNA-like double-stranded form of let-7 miRNA (20 nM). One strand of this siRNA corresponds to the human let-7a RNA (sequence UGAGGUAGUAGGUUGUAUAGUU), whereas the other is complementary to it (sequence CUAUACAACCUACUACCU CAUU; oligonucleotides were obtained from Qiagen). Whole-cell lysates, prepared as described earlier from HeLa cells 48 h post-transfection, were incubated for 2 h at 4°C with beads coated with anti-HA mAb (Roche), and subsequently washed five times with 10 mM Tris-Cl (pH 8), containing 300 mM NaCl and 0.1% NP-40. Cleavage assays were performed as described (Martinez et al. 2002). The reactions contained equivalent amounts of immunoprecipitates and radiolabeled substrates at 0.5 nM concentration, and were incubated for 90 min at 30°C. In vitro transcriptions were carried out with T7 polymerase (Ambion) from annealed oligonucleotides to prepare either unlabeled RNAs or RNAs uniformly labeled with [α-32P]-UTP. Cold transcripts were 5′-terminally labeled in the presence of [α-32P]-ATP and T4 polynucleotide kinase (New England Biolabs). The products of cleavage reactions were analyzed by a denaturing 15% polyacrylamide gel electrophoresis.
For Northern analysis, total RNA was isolated from transfected cells 48 h posttransfection with the Absolutely RNA RT-PCR Miniprep Kit (Stratagene). Northern blot analysis was performed with 10 μg of total RNA according to standard protocols, using probes specific for RL or GFP mRNAs. Radioactive signals were quantified by a Phosphorimager. The RL mRNA expression levels were calculated after correction for transfection efficiency, measured by GFP mRNA levels.
Acknowledgments
We thank N. Gehring and M. Hentze for plasmids for the tethering assay and G. Dreyfuss and T. Hobman for anti-Gemin 4 antibodies and hAgo2 cDNA, respectively. We also thank L. Saleh for help with cleavage assays, J. Filkowski for corrections of the manuscript, and G. Meister and members of the group for discussions. Friedrich Miescher Institute for Biomedical Research is a part of the Novartis Research Foundation.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.7131604.
REFERENCES
- Bartel, D.P. 2004. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 116: 281–297. [DOI] [PubMed] [Google Scholar]
- Brennecke, J. and Cohen, S.M. 2003. Towards a complete description of the microRNA complement of animal genomes. Genome Biol. 4: 228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmell, M.A., Xuan, Z., Zhang, M.Q., and Hannon, G.J. 2002. The Argonaute family: Tentacles that reach into RNAi, developmental control, stem cell maintenance, and tumorigenesis. Genes & Dev. 16: 2733–2742. [DOI] [PubMed] [Google Scholar]
- Caudy, A.A., Myers, M., Hannon, G.J., and Hammond, S.M. 2002. Fragile X-related protein and VIG associate with the RNA interference machinery. Genes & Dev. 16: 2491–2496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caudy, A.A., Ketting, R.F., Hammond, S.M., Denli, A.M., Bathoorn, A.M., Tops, B.B., Silva, J.M., Myers, M.M., Hannon, G.J., and Plasterk, R.H. 2003. A micrococcal nuclease homologue in RNAi effector complexes. Nature 425: 411–414. [DOI] [PubMed] [Google Scholar]
- Chen, X. 2004. A microRNA as a translational repressor of APETALA2 in Arabidopsis flower development. Science 303: 2022–2025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denli, A.M. and Hannon, G.J. 2003. RNAi: An ever-growing puzzle. Trends Biochem. Sci. 28: 196–201. [DOI] [PubMed] [Google Scholar]
- Doench, J.G. and Sharp, P.A. 2004. Specificity of microRNA target selection in translational repression. Genes & Dev. 18: 504–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doench, J.G., Petersen, C.P., and Sharp, P.A. 2003. siRNAs can function as miRNAs. Genes & Dev. 17: 438–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doi, N., Zenno, S., Ueda, R., Ohki-Hamazaki, H., Ui-Tei, K., and Saigo, K. 2003. Short-interfering-RNA-mediated gene silencing in mammalian cells requires Dicer and eIF2C translation initiation factors. Curr. Biol. 13: 41–46. [DOI] [PubMed] [Google Scholar]
- Gehring, N.H., Neu-Yilik, G., Schell, T., Hentze, M.W., and Kulozik, A.E. 2003. Y14 and hUpf3b form an NMD-activating complex. Mol. Cell 11: 939–949. [DOI] [PubMed] [Google Scholar]
- Grishok, A., Pasquinelli, A.E., Conte, D., Li, N., Parrish, S., Ha, I., Baillie, D.L., Fire, A., Ruvkun, G., and Mello, C.C. 2001. Genes and mechanisms related to RNA interference regulate expression of the small temporal RNAs that control C. elegans developmental timing. Cell 106: 23–34. [DOI] [PubMed] [Google Scholar]
- Ha, I., Wightman, B., and Ruvkun, G. 1996. A bulged lin-4/lin-14 RNA duplex is sufficient for Caenorhabditis elegans lin-14 temporal gradient formation. Genes & Dev. 10: 3041–3050. [DOI] [PubMed] [Google Scholar]
- Hammond, S.M., Caudy, A.A., and Hannon, G.J. 2001. Post-transcriptional gene silencing by double-stranded RNA. Nat. Rev. Genet. 2: 110–119. [DOI] [PubMed] [Google Scholar]
- Hutvagner, G. and Zamore, P.D. 2002. A microRNA in a multiple-turnover RNAi enzyme complex. Science 297: 2056–2060. [DOI] [PubMed] [Google Scholar]
- Ishizuka, A., Siomi, M.C., and Siomi, H. 2002. A Drosophila fragile X protein interacts with components of RNAi and ribosomal proteins. Genes & Dev. 16: 2497–2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin, P., Zarnescu, D.C., Ceman, S., Nakamoto, M., Mowrey, J., Jongens, T.A., Nelson, D.L., Moses, K., and Warren, S.T. 2004. Biochemical and genetic interaction between the fragile X mental retardation protein and the microRNA pathway. Nat. Neurosci. 7: 113–117. [DOI] [PubMed] [Google Scholar]
- Kiriakidou, M., Nelson, P.T., Kouranov, A., Fitziev, P., Bouyioukos, C., Mourelatos, Z., and Hatzigeorgiou, A. 2004. A combined computational-experimental approach predicts human microRNA targets. Genes & Dev. 18: 1165–1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, R.C., Feinbaum, R.L., and Ambros, V. 1993. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 75: 843–854. [DOI] [PubMed] [Google Scholar]
- Lee, Y.S., Nakahara, K., Pham, J.W., Kim, K., He, Z., Sontheimer, E.J., and Carthew, R.W. 2004. Distinct roles for Drosophila Dicer-1 and Dicer-2 in the siRNA/miRNA silencing pathways. Cell 117: 69–81. [DOI] [PubMed] [Google Scholar]
- Martinez, J. and Tuschl, T. 2004. RISC is a 5′ phosphomonoester-producing RNA endonuclease. Genes & Dev. 18: 975–980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez, J., Patkaniowska, A., Urlaub, H., Luhrmann, R., and Tuschl, T. 2002. Single-stranded antisense siRNAs guide target RNA cleavage in RNAi. Cell 110: 563–574. [DOI] [PubMed] [Google Scholar]
- Meister, G., Landthaler, M., Dorsett, Y., and Tuschl, T. 2004. Sequence-specific inhibition of microRNA- and siRNA-induced RNA silencing. RNA 10: 544–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moss, E.G. and Tang, L. 2003. Conservation of the heterochronic regulator Lin-28, its developmental expression and microRNA complementary sites. Dev. Biol. 258: 432–442. [DOI] [PubMed] [Google Scholar]
- Mourelatos, Z., Dostie, J., Paushkin, S., Sharma, A., Charroux, B., Abel, L., Rappsilber, J., Mann, M., and Dreyfuss, G. 2002. miRNPs: A novel class of ribonucleoproteins containing numerous micro-RNAs. Genes & Dev. 16: 720–728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson, P.T., Hatzigeorgiou, A.G., and Mourelatos, Z. 2004. miRNP: mRNA association in polyribosomes in a human neuronal cell line. RNA 10: 387–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamura, K., Ishizuka, A., Siomi, H., and Siomi, M.C. 2004. Distinct roles for Argonaute proteins in small RNA-directed RNA cleavage pathways. Genes & Dev. 18: 1655–1666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen, P.H. and Ambros, V. 1999. The lin-4 regulatory RNA controls developmental timing in Caenorhabditis elegans by blocking LIN-14 protein synthesis after the initiation of translation. Dev. Biol. 216: 671–680. [DOI] [PubMed] [Google Scholar]
- Reinhart, B.J., Slack, F.J., Basson, M., Pasquinelli, A.E., Bettinger, J.C., Rougvie, A.E., Horvitz, H.R., and Ruvkun, G. 2000. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature 403: 901–906. [DOI] [PubMed] [Google Scholar]
- Slack, F.J., Basson, M., Liu, Z., Ambros, V., Horvitz, H.R., and Ruvkun, G. 2000. The lin-41 RBCC gene acts in the C. elegans heterochronic pathway between the let-7 regulatory RNA and the LIN-29 transcription factor. Mol. Cell 5: 659–669. [DOI] [PubMed] [Google Scholar]
- Tabara, H., Sarkissian, M., Kelly, W.G., Fleenor, J., Grishok, A., Timmons, L., Fire, A., and Mello, C.C. 1999. The rde-1 gene, RNA interference, and transposon silencing in C. elegans. Cell 99: 123–132. [DOI] [PubMed] [Google Scholar]
- Tahbaz, N., Kolb, F.A., Zhang, H., Jaronczyk, K., Filipowicz, W., and Hobman, T.C. 2004. Characterization of the interactions between mammalian PAZ PIWI domain proteins and Dicer. EMBO Rep. 5: 189–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vella, M.C., Choi, E.Y., Lin, S.Y., Reinert, K., and Slack, F.J. 2004. The C. elegans microRNA let-7 binds to imperfect let-7 complementary sites from the lin-41 3′UTR. Genes & Dev. 18: 132–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wightman, B., Ha, I., and Ruvkun, G. 1993. Posttranscriptional regulation of the heterochronic gene lin-14 by lin- 4 mediates temporal pattern formation in C. elegans. Cell 75: 855–862. [DOI] [PubMed] [Google Scholar]
- Zeng, Y., Wagner, E.J., and Cullen, B.R. 2002. Both natural and designed micro RNAs can inhibit the expression of cognate mRNAs when expressed in human cells. Mol. Cell 9: 1327–1333. [DOI] [PubMed] [Google Scholar]
- Zeng, Y., Yi, R., and Cullen, B.R. 2003. MicroRNAs and small interfering RNAs can inhibit mRNA expression by similar mechanisms. Proc. Natl. Acad. Sci. 100: 9779–9784. [DOI] [PMC free article] [PubMed] [Google Scholar]