Abstract
The Tetrahymena group I intron splices 20 to 50 times faster in Tetrahymena than in vitro, implying that the intron rapidly adopts its active conformation in the cell. The importance of cotranscriptional folding and the contribution of the rRNA exons to the stability of the active pre-RNA structure were investigated by comparing the activity of minimal pre-RNAs expressed in Escherichia coli. Pre-RNAs containing exons derived from E. coli 23 S rRNA were three to four times more active than the wild-type Tetrahymena pre-RNA. E. coli transcripts of the chimeric E. coli pre-RNA were two to eight times more active than were T7 transcripts. However, the effect of cotranscriptional folding depends on exon sequences. Unexpectedly, the unspliced pre-RNA decays more slowly than predicted from the rate of splicing. This observation is best explained by partitioning of transcripts into active and inactive pools. We propose that the active pool splices within a few seconds, whereas the inactive pool is degraded without appreciable splicing.
Keywords: self-splicing, ribozyme, RNA folding, T7 RNA polymerase
INTRODUCTION
The Tetrahymena group I intron splices 20- to 50-fold faster in vivo than it self-splices in vitro (Brehm and Cech 1983; Bass and Cech 1984). The basis for its higher intracellular activity is not understood. Because the intron is spliced as rapidly in Escherichia coli as in Tetrahymena when inserted near the homologous position in the 23S rRNA of E. coli, it is unlikely that a species-specific factor such as a maturase is required in vivo (Zhang et al. 1995b). Instead, intracellular conditions may favor formation of the catalytically active form of the pre-rRNA. Folding in vivo may be enhanced by the structure of the flanking rRNA (Woodson and Emerick 1993), favorable ionic conditions (Heilman-Miller et al. 2001; Takamoto et al. 2002), nonspecific RNA-binding proteins (Herschlag et al. 1994; Zhang et al. 1995a; Waldsich et al. 2002), or interactions between the nascent RNA and the polymerase (Pan et al. 1999).
In vitro, the rate of splicing is limited by conformational changes, which are slower than the chemical step of phosphodiester transfer (Herschlag and Cech 1990). Experiments on the td group I intron revealed a strong correlation between the stability of the tertiary structure and intracellular activity (Brion et al. 1999). Similarly, the activity of the hairpin ribozyme in yeast is comparable to that under physiological conditions in vitro (Donahue and Fedor 1997). Thus, intracellular activity provides a direct readout of the folding process.
Under physiological conditions in vitro, roughly 90% of the Tetrahymena pre-RNA misfolds (Pan et al. 1997). Because inactive pre-rRNAs refold very slowly in vitro, apparent splicing activity is reduced (Emerick et al. 1996). When the pre-rRNA is expressed in E. coli, mutations that cause misfolding in vitro are partially or fully rescued, although splicing becomes more cold sensitive (Nikolcheva and Woodson, 1999). This suggests that intracellular conditions either reduce the probability that the pre-RNA misfolds or allow for rapid refolding of inactive RNAs.
The probability that the pre-RNA folds correctly is likely to depend on the conformation of the surrounding rRNA. Interactions within the flanking rRNA alter the balance between active and inactive conformations of the pre-RNA (Woodson and Cech 1991). An alternative stem-loop in the rRNA [P(−1)] inhibits 5′ splice site recognition by competing with formation of the P1 splice site helix (Fig. 1 ▶; Woodson and Cech 1991). This inhibition is balanced by base pairing interactions further upstream that stabilize the active structure of the pre-rRNA (Woodson and Emerick 1993). In E. coli cells, splicing of mutant pre-RNAs was less cold sensitive when the intron was flanked by E. coli rRNA rather than non-rRNA exons, suggesting that they protect the pre-RNA from becoming trapped in misfolded structures (Nikolcheva and Woodson 1999).
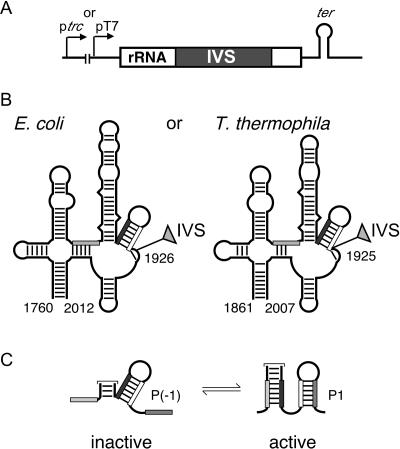
FIGURE 1.
Plasmids for expression of minimal pre-rRNAs in E. coli. (A) Pre-RNA sequences are transcribed from either the E. coli trc promoter or phage T7 promoter. A rrnB or T7 transcription terminator was placed downstream of the cloned insert. Transcripts include 57 or 30 nt of vector sequence upstream of the insert. Ec-Eco, 986 nt; Tt-Eco, 971 nt; Ec-T7, 875 nt; and Tt-T7, 860 nt. (B) Secondary structures of minimal rRNA exons, from E. coli (Ec) and Tetrahymena thermophila (Tt). The intron is naturally found after nt 1925 in Tt LSU rRNA or was artificially inserted after U1926 in the E. coli 23 S rRNA (Zhang et al. 1995b). Numbering is according to E. coli 23 S rRNA (Noller et al. 1981). (C) Alternative secondary structures in the 5′ exon (Woodson and Cech 1991; Woodson and Emerick 1993). P1 is required for 5′ splice site recognition. P(−1) is equivalent to helix 69 in the mature LSU rRNA.
To determine whether the exon sequence matters, we compared the effect of Tetrahymena and E. coli rRNA exons on splicing. Because RNAs are typically express by E. coli RNA polymerase in vivo and in vitro experiments are conducted on T7 transcripts, we also compared the splicing activity of E. coli and T7 transcripts. Slower elongation by E. coli RNA polymerase may enhance RNA activity by giving more opportunity for upstream sequences to fold during transcription. Expression of E. coli 23 S rRNA with T7 RNA polymerase resulted in a lower yield of active 50 S ribosomes (Lewicki et al. 1993). We found that splicing is greatest when the exons are derived from E. coli rRNA and the RNA is transcribed by E. coli RNA polymerase.
RESULTS and DISCUSSION
Comparison of pre-RNAs
Four expression plasmids for pre-RNA were constructed to compare the effects of the transcribing polymerase and the exon sequence on self-splicing (Fig. 1A ▶). Two plasmids were based on pET vectors, for expression of the downstream pre-RNA by T7 RNA polymerase in BL21 cells. The other two contained the E. coli lactose-inducible trc promoter for expression by E. coli RNA polymerase.
Two of the four plasmids contained the sequences of the natural T. thermophila pre-rRNA (Tt). In the other two, the intron was inserted after U1926 of E. coli 23 S rRNA (Ec), and the intron guide sequence was changed to maintain complementarity with the E. coli 5′ exon (Zhang et al. 1995b). The minimal E. coli pre-rRNA was previously found to splice at a rate of 30 min−1 in E. coli cells grown at 42°C (Zhang et al. 1995b), which is comparable to splicing activity in Tetrahymena (Brehm and Cech 1983).
In all cases, the pre-RNAs contained ~250 nucleotides (nt) from domain IV of the large subunit rRNA (Fig. 1B ▶), which correspond to the minimal rRNA exons needed for optimal self-splicing in vitro (Woodson 1992). Because this region of the rRNA is highly conserved, the Tetrahymena and E. coli rRNA exons are expected to adopt similar secondary (and tertiary) structures (Noller et al. 1981; Cannone et al. 2002). However, differences in primary sequence slightly alter the equilibrium between alternative conformations (Fig. 1C ▶).
Splicing in E. coli
The rate of splicing in E. coli was determined from the steady-state ratio of pre-RNA to spliced intron and the decay rate of free intron, as described previously (Brehm and Cech 1983; Zhang et al. 1995b):
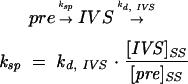 |
(1) |
In cells transformed with the plasmids diagrammed in Figure 1 ▶, induction with IPTG resulted in good expression of plasmid-encoded pre-RNA (Fig. 2A ▶). Levels of the pre-RNA and spliced linear intron (L IVS) were quantified by primer extension and reached a plateau after 90 min.
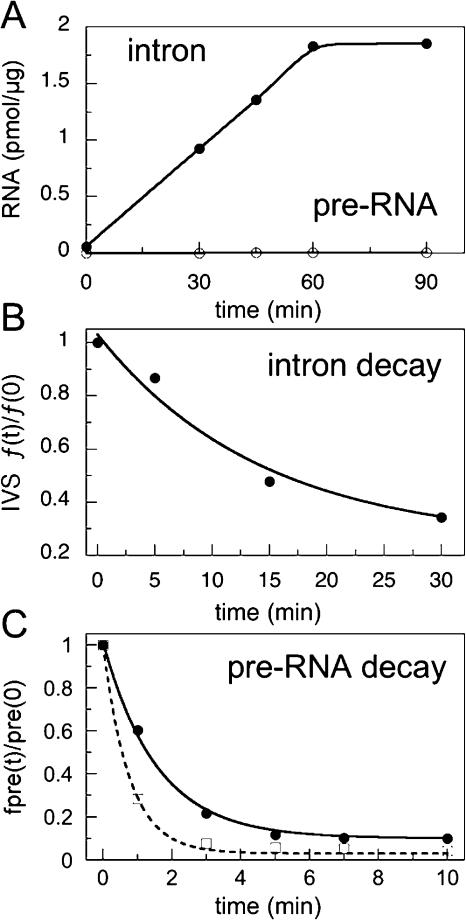
FIGURE 2.
Group 1 splicing in E. coli. (A) Picomoles of intron or pre-RNA per microgram total RNA after induction of plasmid expression (pTN380) with 0.05 mM IPTG at 42°C. RNA was detected by primer extension as previously described (Zhang et al. 1995b). The steady-state ratio of intron to precursor was calculated 90 min after induction (Table 1 ▶). (B) Decay of spliced linear intron RNA at 42°C. Total RNA was isolated from cells harvested 0–30 min after addition of rifampicin. The fraction of spliced intron at each time was normalized to the amount of intron when rifampicin was added, and fit to a first-order rate equation (Table 1 ▶). C. Decay of Tt-Eco pre-RNA at 37°C. Rifampicin was added 60 min after induction with IPTG. Filled circles indicate wild-type (wt) intron core; open squares, C260G core.
To measure the half-life of the spliced intron RNA, transcription by E. coli RNAP was inhibited with rifampicin after a 90-min induction of plasmid-encoded RNA. The amount of intron RNA remaining at various times after addition of rifampicin was measured by primer extension and fit to a first-order rate equation (Fig. 2B ▶). The observed decay rates were 0.08 and 0.18 min−1, respectively, for wild-type (Tt-Eco) and Ec (Ec-Eco) introns at 37°C (Table 1 ▶). Because the primer anneals to the 5′ end of the intron, cyclization products missing the first 15 or 19 nt of the intron (Zaug et al. 1983) were not detected in our assay. Consequently, the decay rates reported here reflect the combined loss of full-length intron via cyclization and nucleolytic degradation.
TABLE 1.
Splicing of the Tetrahymena pre-RNA in E. coli
| Rate constants, 37°C (min−1) | |||||
| Pre-RNA | Splicinga | Intron decayb | Pre-RNA decayb | E. kcal/mol/K | Φc |
| Wild-type core | |||||
| Ec-T7 | 12.0 | — | — | 18 | 0.98 |
| Tt-T7 | 3.0 | — | — | 31 | 0.82 |
| Ec-Eco | 23.0 | 0.08 | 0.26 | 16 | 0.99 |
| Tt-Eco | 9 | 0.18 | 0.64 | NDd | 0.93 |
| C260G core | |||||
| Ec-T7 | 8.0 | — | — | 34 | 0.91 |
| Tt-T7 | 0.40 | — | — | 40 | 0.24 |
| Ec-Eco | 9.0 | 0.24 | 0.75 | 20 | 0.92 |
| Tt-Eco | 0.05 | 0.3 | 1.30 | 14 | 0.04 |
aIn vivo splicing rates were calculated from primer extension assays according to Equation 1. Ec indicates E. coli rRNA exons; Tt, natural Tetrahymena exons; T7, T7 RNAP; and Eco, E. coli RNAP. The steady-state ratios of intron to pre-RNA were from the mean of three trials for the wild-type pre-RNA and four trials for pre-RNA with mutant intron core. These varied 5%–10%, except for Ec-T7 and Ec-Eco which were ±25%. These rates agree within error with previously reported data for splicing in E. coli (Zhang et al. 1995b). Splicing rates at 25°C, 30°C, and 42°C were obtained in the same manner (Fig. 3 ▶) to obtain an activation energy (Ea).
bDecay rates at 37°C (observed) were calculated from primer extension analysis conducted on 0.25 to 0.50 μg of total RNA as in Figure 2 ▶. For the intron, decay rates ranged from 0.02 to 0.3 min−1 (wild type) and 0.07 to 0.5 min−1 (C260G) between 25°C and 42°C. For the wild-type intron, decay rates varied 10%–25% between two and three trials.
cPartition factor Φ at 37°C was calculated from Equation 4.
dND, the activation energy for Tt-Eco transcript was not determined as the splicing rate at 42°C may be influenced by rapid turnover of the pre-RNA at this temperature.
Polymerase and exon effects
Comparison of the splicing activity of the four transcripts diagramed in Figure 1 ▶ showed that both the polymerase used to transcribe the pre-RNA and the rRNA exon sequence affect intracellular processing (Table 1 ▶). At 37°C, E. coli RNAP transcripts spliced two- to threefold faster than T7 transcripts. The pre-RNAs with E. coli rRNA exons (Ec) spliced three- to fourfold faster than pre-RNAs with Tetrahymena rRNA exons (Tt). These effects were cumulative, so that the pre-RNA containing E. coli rRNA exons transcribed by E. coli RNAP (Ec-Eco, kobs = 23 min−1) spliced about eight times faster than the least active pre-RNA (Tt-T7, 3 min−1).
The Ec and Tt pre-RNAs used in these studies had comparable self-splicing activity in vitro, after a brief heat/cool renaturation (data not shown). Hence, differences in intracellular splicing activity likely reflect their ability to fold into the reactive conformation, rather than a change in intrinsic catalytic activity. These results show that the transcribing polymerase or exons have reproducible but subtle effects on pre-RNA folding in vivo.
Temperature dependence of in vivo splicing
To determine whether exon sequence or polymerase choice affect the fraction of misfolded transcripts, the temperature dependence of intracellular splicing activity was compared for the four plasmids in Figure 1 ▶. If the pre-RNA is kinetically trapped in misfolded conformers, the overall folding rate should increase with temperature or denaturant because the inactive RNA refolds more easily (Pan and Sosnick 1997; Pan et al. 1997; Rook et al. 1998). If the native structure is unstable, splicing should decrease with temperature (Brion et al. 1999).
Splicing rates were measured at 25°C, 30°C, 37°C, and 42°C for the Ec and Tt pre-RNAs (Fig. 3A,C ▶). All of the pre-RNAs spliced more rapidly with increasing temperature (Fig. 3C ▶), and the activation energies for T7 and E. coli transcripts of the Ec pre-RNA were similar (Ea = 18 and 16 kcal•mole−1•K−1; Table 1 ▶). Thus, the choice of polymerase made little difference in the energy barrier for splicing of the wild-type RNA.
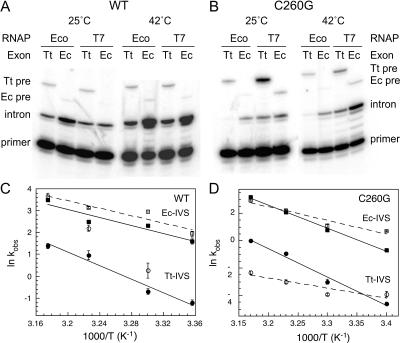
FIGURE 3.
Temperature-dependence of splicing in E. coli. The ratio of intron to pre-RNA at steady state measured by primer extension as in Figure 2 ▶, except that cultures were grown at 25°C, 30°C, 37°C, and 42°C. Splicing rates were calculated as described in Materials and Methods and are listed in Table 1 ▶. (A) Primer extension on pre-RNAs with wild-type intron core. Cells were grown at 25°C and 42°C. Tt indicates Tetrahymena exons; Ec, E. coli exons; T7, phage T7 RNAP; and Eco, E. coli RNAP. (B) Primer extension on pre-RNAs with C260G mutation, as in A. (C) Observed rate constants for splicing (min−1) versus inverse temperature, for wild-type intron core. Squares indicate Ec pre-RNAs; circles, Tt pre-RNAs, open symbols, E. coli RNAP; and closed symbols, T7 RNAP. Arrhenius activation energies are given in Table 1 ▶. Error bars reflect the standard deviation of three trials. (D) Arrhenius plot for C260G pre-RNAs. Symbols as in C. Rapid decay of Tt-Eco pre-RNA may have reduced its apparent splicing activity at 42°C.
Splicing of the natural Tt pre-RNA, however, was more cold sensitive than was splicing of the chimeric Ec pre-RNA. Although the Tt pre-RNAs were less active than Ec pre-RNAs at all temperatures, this gap widened to ~20-fold at 25°C (Fig. 3C ▶). The greater cold sensitivity of the natural Tt pre-RNA also suggested either that a higher fraction of transcripts are partitioned into misfolded structures or that Tt intermediates refold less readily than Ec intermediates.
Splicing of misfolding mutant pre-RNA
To further determine whether the E. coli rRNA exons increase the propensity of the pre-RNA to fold correctly, we mutated C260 to G in the intron core. This mutation disrupts the triple helix at the junction of P4 and P6, and increases misfolding of the Tetrahymena ribozyme (Zarrinkar and Williamson 1996). In E. coli, this mutation caused splicing to become more cold sensitive, particularly in the absence of the flanking rRNA, consistent with increased misfolding in vivo (Nikolcheva and Woodson 1999).
Splicing rates were measured for C260G pre-RNAs between 25°C and 42°C, and activation energies were calculated for each transcript (Fig. 3B,D ▶). As expected, they were less active and more cold sensitive than were their wild-type counterparts, consistent with greater misfolding of the intron core. When comparing the effects of the exons or polymerases, the relative trends were the same as for the analogous wild-type pre-RNAs, except slightly stronger. Surprisingly, the Tt pre-RNAs containing the C260G mutation were 200-fold or 20-fold lower than their wild-type counterparts at 42°C (Fig. 3D ▶). These results suggested that not only are the Tt pre-RNAs more likely to misfold than those with E. coli rRNA exons, but the misfolded RNAs are rapidly turned over in the cell.
Precursor decay in E. coli at 37°C
Although the Tt-Eco:C260G pre-RNA spliced less than the other pre-RNAs, the unspliced pre-RNA did not accumulate, suggesting rapid degradation of the unspliced RNA. To assess this possibility, the half-lives of E. coli RNAP transcripts were compared at 37°C (Fig. 2C ▶). After inhibition of transcription with rifampicin, the pre-RNA disappears at an observed rate that is the sum of the rates for splicing (ksp) and degradation (kd,pre):
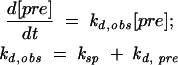 |
(2) |
If these rates are comparable to each other, the splicing rate can be determined from the difference between the observed half-lives of self-splicing and non-self-splicing precursors (Donahue and Fedor 1997; Donahue et al. 2000). Alternatively, the intrinsic decay rate should be equal to the difference between the overall rate of disappearance and the rate of splicing.
Unexpectedly, the observed decay rate was slower than the splicing rate, for all the wild-type and C260G pre-RNAs (Table 1 ▶). Although some pre-RNA may be consumed rapidly after rifampicin is added, the remainder had a longer half-life than expected from Equation 2. This discrepancy can be reconciled by assuming that the measured decay rates represent a population of inactive pre-RNAs. This is not an implausible assumption, as the Tt pre-RNA has been shown to have biphasic splicing kinetics in vitro (Emerick et al. 1996), which is directly related to the partitioning of the RNA into native and misfolded populations (Thirumalai and Woodson 1996).
The Ec pre-RNA had a longer half-life than the Tt pre-RNA (Table 1 ▶), perhaps reflecting a more stable structure (Regnier and Arraiano 2000). The C260G mutants had shorter half-lives than did the wild-type pre-RNAs, consistent with our assumption that loss of pre-RNA primarily represents the decay of misfolded or inactive transcripts. That splicing determines the observed half-life of the pre-RNA is inconsistent with the data, as those pre-RNAs that produced the least spliced product decayed the fastest.
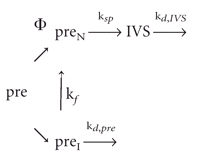
Partitioning into active and inactive populations
The results presented here are inconsistent with a single pool of pre-RNA that splices at a constant rate. Instead, we propose that nascent transcripts are partitioned into active (preN) and inactive (preI) populations (Scheme II). The active RNA splices rapidly, whereas the inactive RNA is either degraded or occasionally refolds into the active form. Although the apparent splicing rates reported in Table 1 ▶ were initially taken to represent the rate constants for splicing itself, in reality it is more probable that they reflect the fraction of transcripts that partition into the active conformation (Φ).
Assuming that most of the active pre-rRNA is converted to product, the ratio of spliced intron (IVS) to pre-RNA at steady state can be expressed as
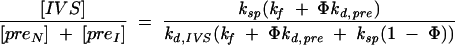 |
(3) |
where Φ is the fraction of active RNA; [preN] and [preI] are the steady-state concentrations of active and misfolded pre-RNA, respectively; and kd,pre represent decay of preI. If all of the pre-RNA folds correctly (Φ = 1), then Equation 3 reduces to Equation 1.
If splicing is rapid compared with refolding of inactive pre-RNA (ksp ≫ kf), then
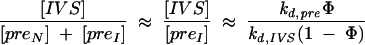 |
(4) |
Under these conditions, the yield of spliced product depends only on the partition factorΦ, because inactive transcripts have no chance to refold. The approximations in Equation 4 lead to estimates of in vivo partitioning that range from Φ = 0.99 for the most active pre-RNAs (Ec-Eco and Ec-T7) to Φ = 0.04 for the least active pre-RNA (Tt-Eco:C260G). Very similar values for Φ are obtained by assuming that the inactive pre-RNAs can refold at rates similar to those obtained from in vitro experiments (kf = 0.14 min−1 at 30°C or ~0.2 min−1 at 37°C; C260G kf = 0.01 min−1 at 30°C; Pan and Woodson 1998). In contrast, assuming that most of the inactive pre-RNA has a chance to refold and splice (kf ≫ kd,pre) overestimates the yield of spliced products, particularly for the C260G mutants. Thus, the data are best explained by assuming that the unspliced pre-RNA represents a pool of inactive transcripts that refold slowly, if at all.
The highest intracellular splicing rates reported here (20–40 min−1) are the same within error as that of the full-length pre-rRNA (Zhang et al. 1995b) and probably represent maximum activity. Unlike the minimal pre-RNAs, the unspliced 23 S rRNA is not degraded rapidly (Nikolcheva and Woodson 1997). Nonetheless, the levels of unspliced pre-23S rRNA are <1%, suggesting very few transcripts become trapped in inactive structures. This can be achieved either by preventing misfolding during transcription or by very rapid refolding of misfolded transcripts by chaperones. Further experiments are needed to distinguish between these possibilities.
Summary
Our results reported show that the intracellular splicing activity of the Tetrahymena group I intron depends on the sequence of the flanking rRNA, and to a lesser extent, the choice of polymerase. Although both the Tetrahymena and E. coli rRNA can form the alternative secondary structures shown in Figure 1C ▶, only in the Tt pre-RNAs is formation of P(−1) directly linked to misfolding of the intron core through an interaction between the internal guide sequence and the 5′ side of P3 (Pan and Woodson 1998). Mutations in the guide sequence of the Ec-IVS abolish this interaction, explaining why the chimeric Ec pre-RNAs misfold less in vivo and are less sensitive to mutations in the intron core.
The choice of polymerase has a small influence on intracellular splicing activity of the group I pre-RNAs, as E. coli transcripts splice two- to threefold faster than do T7 transcripts at 37°C. Such differences could be due to site-specific pausing of the transcription elongation complex, as previously observed for RNase P (Pan et al. 1999). This effect depends on the sequence of the transcript and may reflect different pause sites on the two pre-RNA templates.
In summary, the sequence of upstream RNA, the transcription complex, and the balance between alternative conformations help determine the ultimate fate of a transcript. Importantly, our results are most consistent with rapid partitioning between active and inactive forms, with the inactive pool contributing little to spliced product. Thus, intracellular factors that increase the yield of active pre-RNA presumably act soon after the RNA is transcribed.
MATERIALS AND METHODS
Plasmids
Plasmid pTN112 contains the EC variant of the Tetrahymena IVS inserted after U1926 in E. coli 23S rRNA (Zhang et al. 1995b) and has been described (Nikolcheva and Woodson 1997). Pre-rRNA sequences of pTN112 and pTN112:C260G were amplified by PCR with primers that introduce the BamHI recognition site at both ends of the insert (upstream primer, 5′-GCACGGATCCAGCTG GCTGCAACTGTTTATT; downstream primer, 5′-GCACGGAT CCTGGGGCTCTGAGTCACTT), blunt-end ligated into pTZ19R digested with SmaI. Positive clones were sequenced, and the desired 750-bp fragment was digested with BamHI and inserted into the BamHI site of pET21 (Novagen) and pSE380 (Invitrogen) by standard ligation. The final clones were called pTN21 and pTN380, respectively. The former has an IPTG-inducible T7 promoter and a T7 terminator downstream of the pre-rRNA. The latter has an E. coli trc promoter upstream and an rrnB termination sequence downstream of the insert.
Templates for transcription of a Tetrahymena pre-rRNA fragment, pSW21 and pSW380, were constructed in the same way by amplification of pSW012 (Woodson, 1992) with PCR primers (upstream: 5′-GCAGGGATCCGGTAATCCGACTGTTTAATAA; downstream, 5′-GCACGGATCCTAGTAGATAGGGACAGTG). The resulting eight plasmids contain the Tetrahymena group I intron (wild-type core or C260G) flanked by either E. coli (Ec) rRNA or Tetrahymena thermophila (Tt) rRNA exons. Plasmids contain promoters for either T7 or E. coli (Eco) RNA polymerase (Fig. 1 ▶). Plasmids were transformed into BL21(DE3) E. coli for expression.
E. coli RNA expression at steady state
BL21(DE3) cells transformed with the desired plasmid were grown in 100 mL LB supplemented with 100 mg/mL of ampicillin at 37°C with shaking, until the cell density measured by absorbance at 600 nm was between 0.2 and 0.4. IPTG (0.05 mM final concentration) was added to induce plasmid transcription. Where stated, the culture was shifted to 25°C, 30°C, or 42°C at the time of induction. Aliquots (20 mL) were removed at various times after induction, supplemented with 66.6 μL of 30 mg/mL chloramphenicol and chilled 1 min in an ice-ethanol bath before harvesting by centrifugation. Total RNA was extracted from the cells by the guanidinium isothiocyanate method (Eppendorf-3 Prime, Inc.).
Primer extension
Levels of precursor and intron RNA were determined by primer extension as previously described (Zhang et al. 1995b). Primer IP006 is complementary to the 5′ end of the intron (5′-CCAAAG GTAAATATTGCT); 2 μM 5′-32P-labeled IP006 (1 pmol) was annealed with 250 ng total RNA and extended with AMV reverse transcriptase (USB) in the presence of ddCTP. Analysis of C260G mutants required 500 ng RNA. Products were analyzed by gel electrophoresis and quantified by using a phosphorimager (Molecular Dynamics). The ratio of intron to pre-RNA was obtained from ratio of counts in each extension product. Control reactions containing different amounts of total RNA confirmed that the amount of extended product varies linearly with the amount of RNA template under these conditions. Intracellular splicing rates were calculated from Equation 1.
Intron and precursor decay rates
Transformed cells were grown as above at 25°C, 30°C, 37°C, or 42°C, for 90–120 min after induction of transcription with IPTG to allow expression of plasmid RNA to reach steady state. Transcription was stopped by the addition of 0.8 mg/mL rifampicin, and 20 mL aliquots of culture were removed at various times. Total RNA was extracted and analyzed by primer extension as described above. The fraction of spliced IVS RNA remaining was plotted versus time and fit to a first order rate equation. Precursor decay rates were determined in a similar manner, except cells were harvested at shorter intervals (1–10 and 30 min).
Acknowledgments
We thank Robert Moss for technical assistance. This work was supported by a grant from the National Institutes of Health (GM46686).
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.7880404.
REFERENCES
- Bass, B.L. and Cech, T.R. 1984. Specific interaction between the self-splicing RNA of Tetrahymena and its guanosine substrate: Implications for biological catalysis by RNA. Nature 308: 820–826. [DOI] [PubMed] [Google Scholar]
- Brehm, S.L. and Cech, T.R. 1983. Fate of an intervening sequence ribonucleic acid: excision and cyclization of the Tetrahymena ribosomal ribonucleic acid intervening sequence in vivo. Biochemistry 22: 2390–2397. [DOI] [PubMed] [Google Scholar]
- Brion, P., Schroeder, R., Michel, F., and Westhof, E. 1999. Influence of specific mutations on the thermal stability of the td group I intron in vitro and on its splicing efficiency in vivo: a comparative study. RNA 5: 947–958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannone, J.J., Subramanian, S., Schnare, M.N., Collett, J.R., D’Souza, L.M., Du, Y., Feng, B., Lin, N., Madabusi, L.V., Muller, K.M., et al. 2002. The comparative RNA web (CRW) site: An online database of comparative sequence and structure information for ribosomal, intron, and other RNAs. BMC Bioinformatics 3: 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donahue, C.P. and Fedor, M.J. 1997. Kinetics of hairpin ribozyme cleavage in yeast. RNA 3: 961–973. [PMC free article] [PubMed] [Google Scholar]
- Donahue, C.P., Yadava, R.S., Nesbitt, S.M., and Fedor, M.J. 2000. The kinetic mechanism of the hairpin ribozyme in vivo: Influence of RNA helix stability on intracellular cleavage kinetics. J. Mol. Biol. 295: 693–707. [DOI] [PubMed] [Google Scholar]
- Emerick, V.L., Pan, J., and Woodson, S.A. 1996. Analysis of rate-determining conformational changes during self- splicing of the Tetrahymenaintron. Biochemistry 35: 13469–13477. [DOI] [PubMed] [Google Scholar]
- Heilman-Miller, S.L., Pan, J., Thirumalai, D., Woodson, S.A. 2001. Counterion condensation in folding of the Tetrahymena ribozyme, II: Counterion dependence of folding kinetics. J. Mol. Biol. 309: 57–68. [DOI] [PubMed] [Google Scholar]
- Herschlag, D. and Cech, T.R. 1990. Catalysis of RNA cleavage by the Tetrahymena thermophila ribozyme, 1: Kinetic description of the reaction of an RNA substrate complementary to the active site. Biochemistry 29: 10159–10171. [DOI] [PubMed] [Google Scholar]
- Herschlag, D., Khosla, M., Tsuchihashi, Z., and Karpel, R.L. 1994. An RNA chaperone activity of non-specific RNA binding proteins in hammerhead ribozyme catalysis. EMBO J. 13: 2913–2924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewicki, B.T., Margus, T., Remme, J., and Nierhaus, K.H. 1993. Coupling of rRNA transcription and ribosomal assembly in vivo: Formation of active ribosomal subunits in Escherichia coli requires transcription of rRNA genes by host RNA polymerase which cannot be replaced by bacteriophage T7 RNA polymerase. J. Mol. Biol. 231: 581–593. [DOI] [PubMed] [Google Scholar]
- Nikolcheva, T. and Woodson, S.A. 1997. Association of a group I intron with its splice junction in 50S ribosomes: Implications for intron toxicity. RNA 3: 1016–1027. [PMC free article] [PubMed] [Google Scholar]
- ——— 1999. Facilitation of group I splicing in vivo: misfolding of the Tetrahymena IVS and the role of ribosomal RNA exons. J. Mol. Biol. 292: 557–567. [DOI] [PubMed] [Google Scholar]
- Noller, H.F., Kop, J., Wheaton, V., Brosius, J., Gutell, R.R., Kopylov, A.M., Dohme, F., Herr, W., Stahl, D.A., Gupta, R., et al. 1981. Secondary structure model for 23S ribosomal RNA. Nucleic Acids Res. 9: 6167–6189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan, J. and Woodson, S.A. 1998. Folding intermediates of a self-splicing RNA: mispairing of the catalytic core. J. Mol. Biol. 280: 597–609. [DOI] [PubMed] [Google Scholar]
- Pan, J., Thirumalai, D., and Woodson, S.A. 1997. Folding of RNA involves parallel pathways. J. Mol. Biol. 273: 7–13. [DOI] [PubMed] [Google Scholar]
- Pan, T., Artsimovitch, I., Fang, X.W., Landick, R., Sosnick, T.R. 1999. Folding of a large ribozyme during transcription and the effect of the elongation factor NusA. Proc. Natl. Acad. Sci. 96: 9545–9550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regnier, P. and Arraiano, C.M. 2000. Degradation of mRNA in bacteria: Emergence of ubiquitous features. Bioessays 22: 235–244. [DOI] [PubMed] [Google Scholar]
- Rook, M.S., Treiber, D.K., and Williamson, J.R. 1998. Fast folding mutants of the Tetrahymena group I ribozyme reveal a rugged folding energy landscape. J. Mol. Biol. 281: 609–620. [DOI] [PubMed] [Google Scholar]
- Takamoto, K., He, Q., Morris, S., Chance, M.R., and Brenowitz, M. 2002. Monovalent cations mediate formation of native tertiary structure of the Tetrahymena thermophila ribozyme. Nat. Struct. Biol. 9: 928–933. [DOI] [PubMed] [Google Scholar]
- Thirumalai, D. and Woodson, S.A. 1996. Kinetics of folding of protein and RNA. Acc. Chem. Res. 29: 433–439. [Google Scholar]
- Waldsich, C., Grossberger, R., and Schroeder, R. 2002. RNA chaperone StpA loosens interactions of the tertiary structure in the td group I intron in vivo. Genes Dev. 16: 2300–2312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodson, S.A. 1992. Exon sequences distant from the splice junction are required for efficient self-splicing of the Tetrahymena IVS. Nucleic Acids Res. 20: 4027–4032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodson, S.A. and Cech, T.R. 1991. Alternative secondary structures in the 5′ exon affect both forward and reverse self-splicing of the Tetrahymena intervening sequence RNA. Biochemistry 30: 2042–2050. [DOI] [PubMed] [Google Scholar]
- Woodson, S.A. and Emerick, V.L. 1993. An alternative helix in the 26S rRNA promotes excision and integration of the Tetrahymena intervening sequence. Mol. Cell. Biol. 13: 1137–1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarrinkar, P.P. and Williamson, J.R. 1996. The kinetic folding pathway of the Tetrahymena ribozyme reveals possible similarities between RNA and protein folding. Nat. Struct. Biol. 3: 432–438. [DOI] [PubMed] [Google Scholar]
- Zaug, A.J., Grabowski, P.J., and Cech, T.R. 1983. Autocatalytic cyclization of an excised intervening sequence RNA is a cleavage-ligation reaction. Nature 301: 578–583. [DOI] [PubMed] [Google Scholar]
- Zhang, A., Derbyshire, V., Salvo, J.L., and Belfort, M. 1995a. Escherichia coli protein StpA stimulates self-splicing by promoting RNA assembly in vitro. RNA 1: 783–793. [PMC free article] [PubMed] [Google Scholar]
- Zhang, F., Ramsay, E.S., and Woodson, S.A. 1995b. In vivo facilitation of Tetrahymena group I intron splicing in Escherichia coli pre-ribosomal RNA. RNA 1: 284–292. [PMC free article] [PubMed] [Google Scholar]