Abstract
To investigate the role of 3′ end formation in yeast mRNA export, we replaced the mRNA cleavage and polyadenylation signal with a self-cleaving hammerhead ribozyme element. The resulting RNA is unadenylated and accumulates near its site of synthesis. Nonetheless, a significant fraction of this RNA reaches the cytoplasm. Nuclear accumulation was relieved by insertion of a stretch of DNA-encoded adenosine residues immediately upstream of the ribozyme element (a synthetic A tail). This indicates that a 3′ stretch of adenosines can promote export, independently of cleavage and polyadenylation. We further show that a synthetic A tail-containing RNA is unaffected in 3′ end formation mutant strains, in which a normally cleaved and polyadenylated RNA accumulates within nuclei. Our results support a model in which a polyA tail contributes to efficient mRNA progression away from the gene, most likely through the action of the yeast polyA-tail binding protein Pab1p.
Keywords: RNA export, 3′ end formation, polyA, Pab1, yeast
INTRODUCTION
Eukaryotic mRNA transcripts are modified within the nucleus to generate mRNA that is functional for translation. Nuclear mRNA processing includes capping, in which a 5′ 7-methylguanosine cap is added; splicing, in which noncoding intervening sequences are removed; and 3′ end formation, in which a stretch of 3′ adenosines (the polyA tail) is added. These processing events are interconnected, both with each other and with transcription, and contribute to the stepwise, ordered assembly of mRNA into messenger ribonucleoprotein particles (mRNPs) (for review, see Howe 2002). Mature mRNPs are then exported from the nucleus to the cytoplasm for bulk translation.
3′ end formation is carried out by the cleavage and polyadenylation machinery, which site-specifically cleaves RNA and adds a polyA tail of 70–90 adenosines in yeast and 200–250 adenosines in higher eukaryotes (for reviews, see Zhao et al. 1999; Proudfoot and O’Sullivan 2002). The polyA tail is an important determinant of mRNA metabolism and function, largely through the action of cellular polyA tail-binding proteins (PABPs) (for review, see Mangus et al. 2003). While higher eukaryotes have multiple PABPs, yeast (Saccharomyces cerevisiae) has only one, Pab1p. PABPs have a minimal binding site of 11–12 adenosines, and as a consequence a single polyA tail is bound by multiple PABP molecules (Baer and Kornberg 1983; Sachs et al. 1987; Deo et al. 1999). PABPs have been implicated most prominently in mRNA translation and stability. They physically bind the translational initiation factor eIF4G, which in turn binds the cap binding protein eIF4E to effectively circularize the 5′ and 3′ ends of mRNA (Tarun and Sachs 1996; Wells et al. 1998). This “closed loop” conformation may promote translational competence by enabling the recycling of ribosomes from the 3′ to the 5′ end of the mRNA. Additionally, a major pathway of mRNA turnover is triggered by polyA tail shortening to ~10 residues (Decker and Parker 1993; Muhlrad et al. 1995), which leads to loop disassembly and accessibility of exoribonucleases to the RNA termini. However, these PABP-mediated effects persist under conditions where loop formation cannot occur, indicating other contributions of PABPs to translation and stability (Kessler and Sachs 1998; Wilusz et al. 2001).
While these roles are mediated by PABPs that localize predominantly to the cytoplasm (PABCs), higher eukaryotes have a nuclear PABP (PABN1) that has been implicated in 3′ end formation and polyA tail length control (Wahle 1991; Bienroth et al. 1993; Keller et al. 2000; Kerwitz et al. 2003). Microscopic analysis has further shown that PABN1 associates with nuclear mRNA early and traffics with RNA to the cytoplasm (Bear et al. 2003). Although yeast lack a bona fide PABN1 homolog, similar roles have been attributed to the predominantly but not exclusively cytoplasmic Pab1p (Sachs et al. 1986; Amrani et al. 1997a; Minvielle-Sebastia et al. 1997; Brown and Sachs 1998).
mRNAs within the nucleus are decorated with proteins, some of which serve to target RNA to nuclear pore complexes (NPCs) for export to the cytoplasm (for reviews, see Stutz and Rosbash 1998; Vinciguerra and Stutz 2004). The requirements for mRNA progression from its site of synthesis to the NPC and through the NPC to the cytoplasm are not well-understood. Foci of potentially nonnascent RNA are sometimes observed near higher eukaryotic genes, indicating that RNAs can dwell near their site of synthesis prior to progression toward the pore (Custodio et al. 1999). Once in the nucleoplasm, however, mRNPs appear to move by diffusion through interchromatin space (for review, see Daneholt 1999; Politz et al. 1999; Shav-Tal et al. 2004). mRNAs therefore acquire components during and/or shortly after synthesis that ultimately allow for their interaction with the NPC and subsequent export.
Several studies have demonstrated a connection between mRNA 3′ end formation and export. Yeast transcripts become nuclear accumulated in 3′ end formation mutant strains (Brodsky and Silver 2000; Hilleren et al. 2001), and proper export of RNAs generated in vivo by bacteriophage T7 RNA polymerase is dependent on cleavage and polyadenylation (Dower and Rosbash 2002). In another study, a mutagenesis screen counterselecting against the expression of a heat-shock-GFP fusion gene yielded mutations in cleavage factors and polyA polymerase that were temperature sensitive for export (Hammell et al. 2002). Such a connection may also exist in higher eukaryotes, as RNAs whose 3′ ends are directed by a self-cleaving ribozyme element fractionate to the nuclear compartment in cell culture experiments (Eckner et al. 1991; Huang and Carmichael 1996). As these studies relied primarily on biochemical assays, however, the results could also be influenced by cytoplasmic instability in the absence of a polyA tail.
The evidence linking 3′ end formation to export is therefore more persuasive in yeast, due also to the availability of temperature-sensitive strains. However, the use of elevated temperatures restricts analysis to nonphysiological conditions under which indirect effects may occur. Temperature-sensitive strains are a further concern since there is evidence that the requirements for mRNA export at elevated temperatures may differ from those for normal mRNA export (Saavedra et al. 1997; Krebber et al. 1999; Henry et al. 2003; Takemura et al. 2004).
To test directly for a role of 3′ end formation in yeast export under normal conditions, we analyzed the effect of replacing the RNA cleavage and polyadenylation signals with a self-cleaving hammerhead ribozyme element. These RNAs are predominantly unadenylated and display a partial defect in export, visualized by fluorescent in situ hybridization (FISH) as RNA accumulation near sites of transcription. This defect is rescued by the insertion of a stretch of 3′ DNA-encoded adenosine residues immediately upstream of the ribozyme (a synthetic A tail), implicating the absence of the polyA tail rather than the lack of cleavage and polyadenylation in RNA accumulation. Unlike a normally cleaved and polyadenylated RNA, a synthetic A tail-containing RNA escapes nuclear accumulation in strains temperature sensitive for 3′ end formation. Unadenylated ribozyme-terminated RNA is also deficient in Pab1p binding, and this defect is rescued by a synthetic A tail. Taken together, our results indicate that the polyA tail, most likely through Pab1p binding, promotes the efficient movement of RNA away from its site of synthesis.
RESULTS
A synthetic A tail relieves transcription-site accumulation of ribozyme-terminated RNA
To investigate the role of 3′ end formation in mRNA export, we generated TDH3 promoter-driven GFP constructs containing either 500 base pairs (bp) of TDH3 3′ flanking region (which includes the TDH3 cleavage and polyadenylation, or pA, signals) or a self-cleaving hammerhead ribozyme element (RZ; GFP pA and GFP RZ, respectively). The ribozyme element in GFP RZ is immediately downstream of the GFP stop codon and is predicted to generate an RNA with a 6-nucleotide (nt) extension and a 2′,3′-cyclic phosphate end (Samarsky et al. 1999). We also generated a construct in which a stretch of 75 DNA-encoded adenosine residues has been inserted immediately upstream of the ribozyme element in GFP RZ (a synthetic A tail; GFP A75 RZ). A schematic of these constructs is shown in Figure 1A.
FIGURE 1.
Ribozyme-terminated RNA is predominantly unadenylated. (A) Schematic of the TDH3 promoter-driven GFP constructs: (Txn) transcription start site, (pA) TDH3 polyadenylation signal, (RZ) hammerhead ribozyme element, (A75 RZ) 75 DNA encoded adenosine residues. The start and stop codons of GFP are indicated by ATG and TAA, respectively. All constructs were introduced into yeast on 2μ high-copy plasmids. (B) Primer extension analysis of total (T) RNA, and of flowthrough (FT) and bound (B) RNAs to oligo dT. RNA analysis for total and flowthrough are of 1/10th the amount analyzed for the bound fraction. Primer extension for U2 snRNA is included as an internal control. Construct names have shortened here and in subsequent figures for simplicity. (C) Northern blot analysis for GFP 3′ end fragments. Total RNA from cells was treated with RNase H and an oligonucleotide to internally cleave GFP RNA, in the absence (−) and presence (+) of oligo dT to remove polyA stretches. GFP pA migrates more slowly due to the TDH3 3′ UTR in this RNA.
GFP pA, GFP RZ, and GFP A75 RZ were introduced into yeast on 2μ high-copy plasmids, and the RNAs were assayed for binding to oligo dT (Fig. 1B). By primer extension analysis, steady-state RNA levels of GFP RZ are lower than those of GFP pA and GFP A75 RZ, which are comparable (lanes 1–3). This is consistent with a contribution of the polyA tail to mRNA stability. Both GFP pA and GFP A75 RZ associate with oligo dT (lanes 7 and 9), despite the fact that a substantial fraction of these RNAs are in the flowthrough under these conditions (lanes 4–6). In contrast, GFP RZ is not appreciably bound, indicating that this RNA is predominantly unadenylated (lane 8). The very faint GFP RZ band visible in the bound fraction may be largely due to background binding, as a comparably faint band is seen with the unadenylated U2 snRNA.
We next performed Northern blot analysis to further characterize the 3′ ends of these RNAs (Fig. 1C). Total RNA from transformed strains was treated with an oligonucleotide and RNase H to internally cleave GFP RNA and increase resolution, and the RNase H incubations were carried out in the presence or absence of oligo dT to remove polyA stretches. Northern blot analysis for GFP 3′ end fragments shows that the ribozyme element is active and that GFP RZ is largely if not completely unadenylated. The presence of 3′ adenosines in GFP pA and GFP A75 RZ is indicated by the mobility shift of these RNAs upon RNase H/oligo dT treatment.
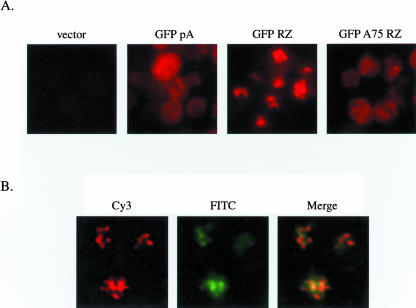
To determine the steady-state localization of these RNAs, we performed FISH analysis with probes spanning the GFP open reading frame (Fig. 2A). GFP pA localizes throughout the cell, as expected for a cleaved and polyadenylated RNA that is exported normally. FISH for GFP RZ reveals a granular signal that is nuclear, i.e., it is coincident with DAPI staining (Fig. 2A; data not shown). In contrast, nuclear accumulation is not visible with GFP A75 RZ, which localizes throughout the cell similarly to GFP pA.
FIGURE 2.
Synthetic A tail rescue of transcription site-associated RNA accumulation. (A) FISH analysis for GFP RNA in cells transformed with empty vector, GFP pA, GFP RZ, or GFP A75 RZ. (B) Combined FISH for GFP RZ (Cy3 channel) and indirect immunofluorescence for lacI-GFP (FITC channel). LacI-GFP expressing cells were transformed with a 2μ high-copy plasmid harboring both the GFP RZ reporter and 256 tandem repeats of lacO sequence. A merged image is shown to the right. Images in B have been additionally software enhanced for clarity.
Previous reports have shown that RNAs accumulate at or near sites of transcription when export is blocked (Jensen et al. 2001; Thomsen et al. 2003). This suggested that the granular RNA accumulation of GFP RZ reflected the multiple sites of transcription present with the 2μ high-copy reporter, which is expected to occur in 10–30 copies/cell. To test this directly, the GFP RZ reporter was inserted into a 2μ high-copy plasmid harboring 256 tandem repeats of lac operator (lacO) sequence. The localization of this plasmid was determined by α-GFP indirect immunofluorescence (IF) in a yeast strain expressing a GFP fusion of lac repressor protein (lacI-GFP), which binds to the lacO sequences. The results of combined FISH and α-GFP IF indicate that GFP RZ accumulation is nearly, although not precisely, coincident with lacI-GFP localization (Fig. 2B). Note that GFP RZ yields little to no protein (Fig. 3C) and that lacI-GFP mRNA levels are low, allowing for this analysis. Taken together, the results demonstrate that unadenylated ribozyme-terminated RNA accumulates near its site of transcription, and that this accumulation is rescued by addition of a synthetic A tail.
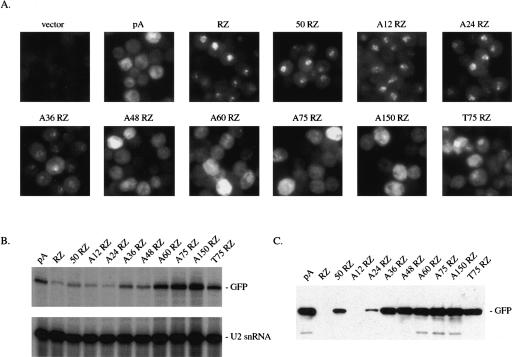
FIGURE 3.
Rescue of nuclear accumulation by a synthetic A tail of 48 adenosines. (A) FISH for GFP RNA in cells transformed with constructs containing various synthetic tails. In addition to GFP pA, GFP RZ and GFP A75 RZ are constructs in which, immediately upstream of the ribozyme, is either 50 bp of TDH3 3′ UTR sequence (GFP 50 RZ), DNA encoded stretches of 12, 24, 36, 48, 60, or 150 adenosines (GFP A12 RZ, GFP A24 RZ, etc.), or 75 DNA encoded uridine residues (GFP T75 RZ). (B) Primer extension for GFP RNA and U2 snRNA in strains transformed with these synthetic tail constructs. (C) α-GFP Western blot for GFP protein generated from these constructs.
A stretch of 48 adenosines is sufficient to rescue nuclear accumulation
To investigate the synthetic A tail length requirements for rescue, we generated a series of constructs containing synthetic tails of varying size (A12 RZ, A24 RZ, A36 RZ, A48 RZ, and A60 RZ, where A12 RZ has 12 adenosines, A24 RZ has 24 adenosines, etc.). Two additional constructs were included to test the specificity for adenosines: GFP T75 RZ contains 75 DNA encoded thymidine residues (to generate a synthetic uridine tail) and GFP 50 RZ contains 50 nt of TDH3 3′ UTR sequence between the GFP stop codon and the ribozyme. These additional sequences did not result in spurious polyadenylation of ribozyme-terminated ends (data not shown). We were unsuccessful in generating extended synthetic guanidine and cytidine constructs, as these sequences collapsed in the PCR-based cloning strategy. We also generated a construct with a synthetic A tail of 150 adenosines (GFP A150 RZ), as a correlation between nuclear accumulation and RNA hyperadenylation has been reported (Jensen et al. 2001; Hector et al. 2002).
FISH analysis reveals that a synthetic A tail of 48 adenosines is sufficient to relieve the nuclear accumulation of ribozyme-terminated RNA (Fig. 3A). An adenosine or homopolymer requirement is clearly visible, as nuclear accumulation persists in GFP 50 RZ, which has 50 nt of 3′ UTR sequence. A synthetic U75 tail results in an intermediate phenotype: Nuclear accumulation is still visible, but it is more diffuse and more cytoplasmic than with GFP RZ. No nuclear accumulation is observed for GFP A150 RZ, suggesting that a hyperadenylated tail does not cause RNA accumulation per se. This observation is also consistent with a report in which a defect in mRNA export could be uncoupled from concomitant hyperadenylation (Hector et al. 2002).
By primer extension analysis, steady-state RNA levels generally increase with longer synthetic A tail lengths (Fig. 3B). A similar increase is also observed for GFP protein production as assayed by α-GFP Western blotting (Fig. 3C). This result is likely due to a combination of factors, including RNA stability and translatability as well as export. Although the synthetic A tail length effects on RNA and protein levels did not correlate precisely with each other or with that for rescue of nuclear accumulation, we are not certain that these differences are meaningful. For example, the fact that protein levels peak earlier than RNA levels may be due to Western blotting nonlinearity (D. Belostotsky, pers. comm.). Nonetheless, a contribution of the polyA tail to mRNA stability and translation is clearly evident. Importantly, GFP 50 RZ yields GFP protein, in contrast to GFP RZ (Fig. 3C). This indicates that some fraction of unadenylated RNAs can access the cytoplasm, despite clear nuclear accumulation by FISH.
The export defect of ribozyme-terminated RNA is incomplete
We considered that our FISH results might fail to distinguish an export defect from cytoplasmic instability, as the latter might uncover underlying nuclear accumulation. Although the intensity of the GFP RZ nuclear signal made this possibility unlikely (see Fig. 2A), we tested it directly in two ways. We stabilized cytoplasmic RNAs in a strain deleted of the major cytoplasmic exoribonuclease XRN1 (Heyer et al. 1995; Johnson 1997), and we inserted a premature termination codon (PTC) to target RNAs to the nonsense-mediated decay (NMD) pathway and decrease cytoplasmic RNA levels.
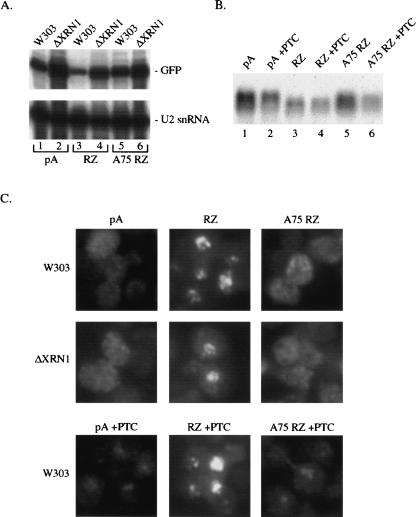
The levels of GFP pA, GFP RZ, and GFP A75 RZ substantially increase in the ΔXRN1 strain, consistent with a prominent role of Xrn1p in mRNA turnover (Fig. 4A). GFP RZ stabilization in particular indicates that a significant fraction of this RNA is exported to the cytoplasm. It is presumably difficult to detect by FISH, as it is relatively unstable and diffusely localized throughout the large cytoplasmic volume. However, nuclear accumulation of GFP RZ is still visible in the ΔXRN1 strain (Fig. 4C, middle panels). This is despite RNA levels that now approximate those of GFP pA and GFP A75 RZ in the wild-type strain (Fig. 4A, cf. lane 4 and lanes 1,5). We conclude that the FISH results are not simply due to differences in cytoplasmic stabilities of the different RNAs.
FIGURE 4.
FISH for RNAs stabilized by an XRN1 deletion or destabilized by NMD. (A) Primer extension analysis for GFP pA, GFP RZ, and GFP A75 RZ in the wild-type W303 and ΔXRN1 strains. Primer extension for U2 snRNA is also shown. (B) Northern blot analysis for GFP RNAs containing a premature termination codon (+PTC series). (C) FISH for GFP RNA in the W303 and ΔXRN1 strains and for the PTC-containing RNAs. The FISH analyses shown were performed at the same time. The panels for the +PTC series have been additionally software enhanced to better visualize the faint nuclear foci in GFP pA +PTC and GFP A75 RZ +PTC.
By Northern blot analysis, PTC introduction (+PTC) reduces the levels of GFP pA and GFP A75 RZ, consistent with NMD (Fig. 4B). The reduction is modest, however, indicating that these RNAs are not robust NMD substrates. Little to no effect on RNA level is seen with GFP RZ +PTC, likely due to inherent instability and/or a defect in translation of the GFP RZ RNA. PTC-introduction therefore generates more comparable levels of GFP pA, GFP RZ, and GFP A75 RNAs (Fig. 4B, cf. lanes 2, 4, and 6). By FISH, NMD-mediated destabilization of GFP pA and GFP A75 RZ reveals a nuclear signal, which is substantially less intense than that observed for GFP RZ +PTC (Fig. 4C, bottom panels). The modest reduction in RNA levels, together with the less punctate signature of the remaining cytoplasmic signal, allows for greater image manipulation that reveals this nuclear signal. The GFP RZ accumulation may therefore reflect an exaggeration of normal transcription site accumulation (see Discussion).
In a previous yeast study, it was reported that ribozyme-terminated RNA is efficiently exported in yeast (Duvel et al. 2002). This conclusion was largely on the basis of protein production and polysome profiles. We therefore carried out a similar analysis on GFP pA and GFP RZ. We also included GFP 50 RZ, which yields GFP protein (see Fig. 3C). Extracts were loaded onto 10%–50% sucrose gradients and fractions were analyzed by Northern blotting for GFP RNA (Fig. 5). The percentage of RNA in polysomes (fractions 7–12/fractions 1–12) is indicated to the right of each panel.
FIGURE 5.
Polysome association of ribozyme-terminated RNA. (Top) Representative profile of the absorbance at 254 nm (A254) across the 10%–50% sucrose gradient. Peaks for 40S small ribosomal subunits, 60S large ribosomal subunits, and 80S monosomes are indicated. Twelve fractions were collected. (Bottom) Northern blot analysis for GFP pA, GFP RZ, and GFP 50 RZ across sucrose gradients. The amount of each RNA in the polysome fractions (fractions 7–12) as a percentage across the entire gradient are given to the right of each panel.
GFP RZ is less polysome associated than GFP pA (36% vs. 80%, respectively) and this difference persists with GFP 50 RZ, although it is less pronounced (57%). In an independent experiment, the polysome-associated fractions of GFP pA, GFP RZ, and GFP 50 RZ were 71%, 32%, and 49%, respectively (data not shown). Taken together, the results indicate that a significant fraction of unadenylated ribozyme-terminated RNAs are cytoplasmic, and therefore exported. There is presumably no protein from GFP RZ (Fig. 3C) because the short distance between the stop codon and the RNA 3′ end interferes with productive translation.
Ribozyme-terminated RNA accumulation and synthetic A tail rescue after transcriptional induction
It was possible that our FISH results were influenced by the steady-state analysis, i.e., by the fact that the reporters are under the control of the constitutive TDH3 promoter. We therefore generated GFP constructs under the control of the galactose-inducible GAL1 promoter (GALGFP pA, GALGFP RZ, and GALGFP A75 RZ). These constructs are otherwise identical to those described above, except that they also contain the GAL1 5′ UTR and, in the case of GALGFP pA, 500 bp of GAL1 3′ region.
RNA analysis at time points after transcriptional induction is shown in Figure 6A. GALGFP RZ accumulates to a lower threshold than either GALGFP pA or GALGFP A75 RZ, similarly to the constitutive TDH3 promoter-driven constructs. The results of FISH analysis after transcriptional induction are shown in Figure 6B. Strong nuclear accumulation of GALGFP RZ is visible by 20 min after induction. In contrast, only faint nuclear foci are observed in some cells with GALGFP pA and GALGFP A75 RZ, which is clearly distinguishable from the accumulation of GALGFP RZ. The export defect of unadenylated RNA, as well as its rescue by a synthetic A tail, is therefore also detectable shortly after transcriptional induction.
FIGURE 6.
Transcriptional induction of ribozyme-terminated RNA and synthetic A tail rescue. (A) Primer extension analysis for GFP RNA in strains transformed with GAL1 promoter-driven GFP at time points after transcriptional induction. 3′ ends were directed either by a GAL1 polyadenylation signal (GALGFP pA), ribozyme (GALGFP RZ), or synthetic A tail (GALGFP A75 RZ). Constructs were introduced into yeast on 2μ high-copy plasmids. Primer extension for U2 snRNA is also shown. Time 0 represents uninduced cells. (B) FISH analysis for GFP RNA at time points after transcriptional induction.
A synthetic A tail-containing RNA escapes nuclear accumulation in 3′ end formation mutant strains
We next localized the galactose-inducible RNAs in temperature-sensitive strains for the 3′ end formation factors polyA polymerase (pap1-1) and Rna15p (rna15-2). Cells were grown without glucose at 25°C and temperature shifted to 37°C with superheated media containing galactose for 40 min prior to fixation and FISH analysis (Fig. 7A). Cells fixed prior to induction and containing the GALGFP pA construct are also shown. GALGFP pA becomes nuclear accumulated in both strains, as expected for RNA that is normally cleaved and polyadenylated, and the GALGFP RZ nuclear accumulation persists in these strains. However, little to no nuclear accumulation is observed for GALGFP A75 RZ in either strain, indicating that a synthetic A tail-containing RNA can bypass a temperature-sensitive block in 3′ end formation.
FIGURE 7.
Export of a synthetic A tail-containing RNA in 3′ end formation mutant strains. (A) FISH for GFP RNA in strains temperature sensitive for polyA polymerase (pap1-1) or a cleavage factor (rna15-2) transformed with the GAL1 promoter-driven constructs described in Figure 6. Cultures at 25°C were temperature shifted to 37°C with superheated media containing galactose for 40 min prior to fixation and FISH analysis. Uninduced cells (time 0) for GALGFP pA are also shown. (B) FISH analysis for GFP RNA in the wild-type W303 strain and a strain deleted for the nucleoporin Rip1p using the TDH3 promoter-driven constructs. Cultures at 30°C were temperature shifted to 42°C for 30 min prior to fixation and FISH analysis. The exposure time in B is one-eighth typical exposure times to accommodate the strong nuclear accumulation in the ΔRip1 strain.
To test whether this bypass capacity extends to a defect in a downstream mRNA export factor, RNAs were localized in a strain deleted for the nucleoporin RIP1, which has an mRNA export defect at 42°C (Saavedra et al. 1997; Stutz et al. 1997). The galactose-inducible reporters were not used in this experiment, as neither the export block in the ΔRIP1 strain nor induction is robust under galactose-inducing conditions at 42°C (data not shown). Instead, we used the constitutive TDH3 promoter-driven constructs described previously. Wild-type W303 and ΔRIP1 strains were grown at 30°C and shifted to 42°C with superheated media for 30 min prior to fixation and FISH analysis (Fig. 7B). GFP pA, GFP RZ, and GFP A75 RZ are all nuclear accumulated in the ΔRIP1 strain under these conditions. Therefore, a synthetic A tail cannot bypass a requirement for the late mRNA export factor Rip1p.
A synthetic A tail restores Pab1p association to ribozyme-terminated RNA
Given that the 3′ end formation bypass effects are mediated by a synthetic A tail, a logical prediction was that they result from restoration of Pab1p binding. We therefore tested whether GFP RZ was defective in Pab1p binding and whether any defect was rescued by a synthetic A tail. Reporter constructs were introduced into a strain in which the endogenous PAB1 gene had been C terminally tagged with a V5 epitope. We also used a strain similarly tagged for CBP20, the small subunit of the nuclear cap binding complex. Extracts from transformed strains were subjected to α-V5 immunoprecipitation, followed by RNA extraction and analysis by primer extension (Fig. 8). Primer extension for U2 snRNA was included as an internal control.
FIGURE 8.
Pab1p association is restored by a synthetic A tail. (A) Primer extension analysis for GFP RNA after α-V5 immunoprecipitation (IP) from extracts of transformed CBP20 or PAB1 V5 epitope-tagged strains. Primer extension for U2 snRNA is also shown. Input RNA from 1/100th of the extract used for IP is shown on the left. Only a typical input is shown as the levels of GFP pA, GFP RZ, and GFP A75 RZ were unaffected in these strains (data not shown). Primer extension after IP from the tagged extracts, and from untagged W303 extract from GFP pA transformed cells, is shown on the right. (B) Quantitation of the amounts of GFP pA, GFP RZ, and GFP A75 RZ coimmunoprecipitated with Cbp20V5p and Pab1V5p. The GFP/U2 ratios in the IPs were divided by the GFP/U2 ratios in the inputs, and the value for GFP pA was normalized to 1.0. Results from three independent experiments (expt) for CBP20V5 and two independent experiments for PAB1V5 are shown, as indicated.
Both the reporter RNAs and U2 snRNA are immunoprecipitated with Cbp20V5p and Pab1V5p, whereas neither RNA is appreciably immunoprecipitated from an untagged extract (Fig. 8A, W303 pA). U2 snRNA association with Cbp20p is not unexpected, given that this RNA has a mono-methylated intermediate. Association of U2 snRNA with Pab1p was less expected and may indicate a direct Pab1p association with U2 snRNP or an indirect association through pre-mRNA. Nonetheless, the U2 snRNA signal served as a normalization control for quantitating the amounts of the different GFP RNAs immunoprecipitated from the extracts (Fig. 8B). GFP RZ is disproportionately associated with Cbp20p, consistent with a larger fraction of this RNA being nuclear. In contrast, the percentage of GFP RZ that is Pab1p associated is reproducibly less than that of GFP pA. Moreover, a synthetic A tail restores Pab1p association to normal levels. Although reduced Pab1p association with GFP RZ may reflect the predominantly cytoplasmic localization of Pab1p and the more nuclear distribution of this RNA, we favor the explanation that deficient Pab1p association by immunoprecipitation reflects poor Pab1p recruitment within the nucleus.
DISCUSSION
We present evidence that replacing the normal 3′ end processing signals of a yeast RNA with a self-cleaving ribozyme yields RNA that is predominantly unadenylated and that accumulates near its site of synthesis. We further show that a stretch of 3′ DNA-encoded adenosine residues (a synthetic A tail) rescues this accumulation, both under steady-state conditions and early after transcriptional induction. Indeed, a synthetic A tail-containing RNA escaped nuclear accumulation when factors required for 3′ end processing were functionally inactivated, in contrast to a normally cleaved and polyadenylated RNA. These results demonstrate the contribution of a polyA tail to overcoming nuclear accumulation, and further indicate that this effect can occur independently of cleavage and polyadenylation.
A synthetic A tail of 48 adenosines was sufficient for rescue in our studies. This effect may be mediated by restoration of proper Pab1p binding. Consistent with this, unadenylated ribozyme-terminated RNA is deficient in Pab1p association, and this defect is rescued by a synthetic A tail. Interestingly, Pab1p has also been shown to bind non-polyA regions (Sachs et al. 1987; Burd et al. 1991; Kuhn and Pieler 1996), and moderate Pab1p affinity for homopolymeric uri-dines may account for the partial rescue we observe with a synthetic U tail. Binding to non-polyA RNA would also explain the partial Pab1p association with ribozyme-terminated RNA that is unadenylated. It is possible, however, that this association is due to a very short polyA tail on this RNA or to a subpopulation that is undetectably adenylated in our assays (Fig. 1). Indeed, more sensitive assays reveal some polyadenylation of ribozyme-terminated ends in yeast (Egli and Braus 1994; Duvel et al. 2003). The synthetic A tail rescue may not extend to higher eukaryotes, as 90 adenosines failed to rescue the distribution of ribozyme-terminated RNA in fractionation studies (Huang and Carmichael 1996). However, this stretch of adenosines is short by higher eukaryotic standards.
Pab1p is involved in cleavage and polyadenylation and is thought to coat the polyA tail during synthesis (Amrani et al. 1997a; Minvielle-Sebastia et al. 1997; Brown and Sachs 1998). There is additional genetic evidence that Pab1p functions within the nucleus to promote mRNP biogenesis and export, presumably in part during 3′ end formation (Chekanova et al. 2001; Chekanova and Belostotsky 2003). Nonetheless, it is noteworthy that a direct and prominent role for Pab1p in mRNA export has remained experimentally elusive, and our attempts to directly implicate Pab1p in rescuing unadenylated RNA accumulation by overexpression and tethering have also been unsuccessful (data not shown). Bypass suppressors of an inviable PAB1 deletion strain are in ribosome biogenesis and translation factors, indicating an essential cytoplasmic role (Sachs and Davis 1989). Furthermore, strains genetically depleted of Pab1p as well as bypass suppressed PAB1 deletion strains are not overtly export defective (Kadowaki et al. 1992; data not shown). Perhaps these results are due to a lack of competition, in which RNAs are still equally export competent in the absence of nuclear Pab1p. Alternatively, the export role of Pab1p is subtle, one that promotes but is not essential for proper mRNP formation and export.
Moreover, our results do not distinguish whether the synthetic A tail effects are mediated by Pab1p or by some other factor. Indeed, the mRNA export factor and poly(A)+ RNA binding protein Nab2p has been implicated in 3′ end formation, both in vivo and in vitro (Hector et al. 2002). Strains defective for Nab2p function accumulate hyperadenylated RNAs in the nucleus (Hector et al. 2002), like other strains mutated for general mRNA export factors (Jensen et al. 2001; Hilleren et al. 2001). Interestingly, Pab1p overexpression rescues the export defect of the nab2 strain, with no detectable shortening of polyA tails. This export defect may therefore be due to insufficient nuclear Pab1p under conditions where RNAs are globally hyperadenylated, which strengthens a role for Pab1p in promoting export and/or removing retention factors.
Despite clear nuclear accumulation by FISH, our results also indicate that a considerable fraction of unadenylated ribozyme-terminated RNA is cytoplasmic: (1) it can give rise to protein, (2) it is stabilized in the absence of Xrn1p, and (3) it cosediments with polysomes. Indeed, this polysome-associated fraction may in fact underestimate cytoplasmic RNA, as the absence of a polyA tail on this RNA is expected to reduce translational initiation. Our results are therefore consistent with another ribozyme-export study (Duvel et al. 2002), and they are also consistent with an earlier report that unadenylated RNAs are polysome associated in a pap1-1 mutant strain (Proweller and Butler 1994). As we directly assay RNA localization by FISH, however, we show that unadenylated RNA also has an export defect. Our results suggest that yeast nuclear RNA accumulation can occur with only a potentially modest effect on functional export.
Our preferred explanation is therefore that the ribozyme 3′ end causes a decreased rate of RNA movement away from its site of synthesis. In this view, aberrant mRNP formation increases the dwell time of RNA near the gene, which would offer the cellular machinery further opportunity to complete RNA processing and/or to degrade aberrant RNA. There may be little or even no steady-state effect on export once the RNA is clear of the transcription site. A more modest delay in transcription site escape may even characterize wild-type gene expression in yeast, as we observe nuclear accumulation of otherwise normal RNA if the cytoplasmic signal has been diminished. Similar observations have been made in higher eukaryotes, and mutation of either splicing or 3′ end formation signals cause these transcription site foci to disappear less rapidly after transcription inhibition (Custodio et al. 1999). As 3′ end formation is implicated in transcription termination (Birse et al. 1998), we cannot exclude that even the robust nuclear accumulation of GFP RZ reflects nascent RNA. We find this possibility unlikely, however, given the intensity of the foci and the fact that transcription termination difficulties should also apply to the synthetic A tail reporter, where nuclear accumulation is difficult to detect.
An emerging picture of mRNP formation is increasingly concerned with transcription site-associated events, where the removal of retention factors as well as the recruitment of export factors may be influenced by 3′ end formation during nascent RNA processing (Lei and Silver 2002; Gilbert and Guthrie 2004). We have previously suggested that the Rrp6p-containing nuclear exosome helps prevent unadenylated ribozyme-terminated RNA from freely diffusing away from the gene (Libri et al. 2002). However, more recent repeat experiments reinforce the view that this accumulation is substantially exosome independent (data not shown), indicating that the polyA tail functions more broadly than just to promote exosome escape. As Pab1p interacts directly with nucleoporins (Allen et al. 2002) and some active transcription sites even interact with nuclear pore components (Casolari et al. 2004), an unambiguous distinction between transcription site retention and RNA export may become increasingly difficult. Additional experimentation will hopefully clarify the mechanism(s) that retain aberrant RNA near its gene, as well as the possible roles of the polyA tail and Pab1p in this process.
MATERIALS AND METHODS
Plasmid construction
The parental plasmid, pRS304 2μ, was generated by cloning a PCR product of the Yep24 2μ region into the AatII site of pRS304. TDH3 promoter-driven constructs have 670 bp of TDH3 promoter and 5′ UTR sequence, followed by the GFP open reading frame (ORF) and either 500 bp TDH3 3′ region (GFP pA) or a self-cleaving hammerhead ribozyme element (GFP RZ; the ribozyme sequence is CCTGTCACCGGATGTGTTTTCCGGTCT GATGAGTCCGTGAGGACGAAACAGG). Synthetic tail constructs were generated by sequential PCR using primers containing the full synthetic tail sequence and partial ribozyme sequence, followed by PCR with a primer with the full ribozyme sequence. All constructs were sequenced for confirmation. GAL1 promoter-driven constructs are similar to those described above, except with 500 bp GAL1 promoter and 5′ UTR sequence replacing TDH3 sequence, and 500 bp GAL1 3′ region in GALGFP pA. TDH3 promoter-driven constructs with a premature termination codon (+PTC series) were generated by introducing a GAA → TAA mutation in codon six of GFP. For the colocalization studies (Fig. 2B), TDH3 promoter-driven GFP RZ was subcloned into pDB62, which contains 256 lacO sequences. pDB62 is a modified version of pAFS52 (Straight et al. 1996; Bressan et al. 2004).
Strains
Strains used in this study are described in Table 1. The ΔXRN1 strain was generated by replacing the XRN1 ORF with a kanamycin resistance cassette using standard methods (Longtine et al. 1998). PCR analysis was used to confirm the deletion. The CBP20V5 and PAB1V5 strains were generated by modifying methods used to introduce three hemagglutinin tags (3xHA) at the C terminus of endogenous genes. PCR products were generated by sequential PCR with a primer containing V5 epitope sequence (V5 sequence: GGTAAGCCTATCCCTAACCCTCTCCTCGGTCTCG ATTCTACG), which primed immediately downstream of the 3xHA tag in pFA6a-3HA-kanMX6 (Longtine et al. 1998), followed by PCR with primers that included either CBP20 or PAB1 sequences for targeted integration. Strains were confirmed by α-V5 Western blotting.
TABLE 1.
Yeast strains
| Strain | Genotype | Source |
| W303 | MAT a ade2-1 his3-11 leu2-3,112 trp1-1 ura3-1 can R1-100 | |
| ΔXRN1 | MAT a ΔXRN1::kanR, otherwise isogenic to W303 | This study |
| ΔRip1 | MAT a ΔRIP1::kanR, otherwise isogenic to W303 | Stutz et al. 1997 |
| pap1-1 | MAT a or αpap1-1 ade2-1 his2-11,15 leu2-3,112 trp1-1 ura3-1 or −52 | Patel and Butler 1992 |
| rna15-2 | MAT a rna15-2 ade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-1 | Amrani et al. 1997b |
| CBP20V5 | MAT a CBP20-(V5)-kanR, otherwise isogenic to W303 | This study |
| PAB1V5 | MAT a PAB1-(V5)-kanR, otherwise isogenic to W303 | This study |
| YDB073 | MAT a ade1 ade3::GAL-HO leu2 trp1:hisG ura3 HIS3::URA3pro::lacI-GFP-kanR | Bressan et al. 2004 |
Yeast cultures
Yeast cultures were grown overnight in the appropriate synthetic dropout medium, diluted the next day, and harvested at an OD600 of ~0.5–1.0 for analysis. For galactose inductions, cultures were grown overnight in media containing 2% lactate, 2% glycerol, and 0.025% glucose and diluted the next day into media containing 2% lactate and 2% glycerol. Mid-log cultures were induced by adding galactose to a final concentration of 2%. For temperature shift experiments, an equal volume of superheated media was added to rapidly shift cultures to the desired temperature. Where applicable, superheated media included 4% galactose.
RNA analysis
Total RNA was isolated from yeast by hot acid phenol extraction (Ausubel et al. 1987). Primer extension analysis was carried out using standard methods and primers specific to GFP (GGAACAG GTAGTTTTCCAGTAGTG) and U2 snRNA (GCCAAAAAATGT GTATTGTAA). Primer extension products were resolved by 5% denaturing gel electrophoresis. Binding to oligo dT was performed using Oligotex resin (Qiagen) and 250 μg total RNA according to the supplied protocol. 1/100th (a 2.5 μg equivalent) of the total and flowthrough samples and 1/10th (a 25 μg equivalent) of the bound fraction were analyzed by primer extension. Northern blot analysis for GFP 3′ end fragments (Fig. 1C) was carried out by 2.5% formaldehyde-agarose gel electrophoresis. Samples were treated with RNase H using an oligonucleotide to internally cleave GFP RNA (GAACGCTTCCATCTTCAATGTTGT) and, where applicable, oligo dT18. Membranes were probed with a radiolabeled oligonucleotide complementary to the GFP 3′ region (GCAG CCAGATCCTTTGTATAGTTCATCCATGCATG). Hybridization was performed using ExpressHyb (BD Biosciences), and membranes were washed with 2× SSC, 0.1% SDS twice for 15 min at 25°C and twice for 15 min at 42°C. Northern blot analysis for GFP RNA (Figs. 4B, 5) was carried out by 1% formaldehyde-agarose gel electrophoresis. Probe was generated by random prime labeling a PCR product of the entire GFP ORF (Stratage Prime-it II). Hybridization was performed using ExpressHyb, and membranes were washed twice with 2× SSC, 0.05% SDS for 20 min at 25°C and twice with 0.1× SSC, 0.1% SDS for 20 min at 50°C.
FISH and indirect immunofluorescence
FISH was carried out using four oligonucleotides spanning the GFP open reading frame (T* represents amino modifier dT; Glen Research): GT*GCCCAT TAACAT*CACCATCTAAT T*CAACA AGAAT*TG GGACAACT*CCAGT, GTACAT*AACCTTCGGGCAT*GGCACTCTT*GAAAAAGTCAT *GCCGTTTCAT*AT, GATTCCAT*TCTTTTGTT*TGTCTGCCAT*GATGTATACAT*T GTGTGAGTT*ATA, CCCAGCAGCT*GTTACAAACT*CAAGAAGGACCAT*GTGGTC T*CTCTTTTCGT*T,
coupled to Cy3 fluorochrome (Amersham). Sheroplasted cells were adhered to poly-L-lysine-treated 12-well slides (Cel-line/Erie Scientific Co.) for subsequent manipulation. A detailed protocol can be provided upon request, and is also available at http://www.bio.brandeis.edu/rosbashlab. Indirect immunofluorescence was performed subsequent to FISH manipulation and prior to adding mounting solution (1 mg/mL phenylenediamine, 90% glycerol, 100 ng/mL DAPI). Wells were washed once in 1× PBS, 0.1% Triton X-100 (wash buffer) for 5 min and incubated for 30 min in wash buffer containing 1% BSA. A 1:1000 dilution of α-GFP antibody (Clontech) in wash buffer was applied and slides were incubated for 1 h at 37°C. Wells were washed three times with wash buffer, 5 min each wash, and incubated with a 1:250 dilution of goat α-mouse FITC (Jackson Immuno Research Technologies) in wash buffer for 60 min at 37°C. Wells were washed as before and allowed to air dry prior to adding mounting solution. Cells were visualized using an Olympus IX70 inverted scope using Openlab software (Improvision). Image manipulations were performed using Adobe Photoshop.
Polysome profiles
Polysome profiles were carried out essentially as described (Arava et al. 2003). Gradients were prepared by underlaying 10%, 20%, 30%, 40%, and 50% sucrose solutions prepared in 20 mM Tris-HCl (pH 8.0), 140 mM KCl, 5 mM MgCl2, 0.5 mM DTT, 0.1 mg/mL cycloheximide, 0.5 mg/mL heparin. Gradients were made in SW40 Ti tubes and allowed to equilibrate overnight at 4°C. Extracts were prepared from 100-mL mid-log cultures. Cycloheximide was added to a final concentration of 0.1 mg/mL and cultures were chilled on ice for 5 min prior to cell pelleting and washing in lysis buffer (20 mM Tris-Cl at pH 8.0, 140 mM KCl, 1.5 mM DTT, 1% Triton X-100, 0.1 mg/mL cycloheximide, 1 mg/mL heparin). Cells were resuspended in 0.7 mL lysis buffer and broken by bead beating with four 20-sec pulses, keeping samples on ice in between. Extracts were prepared by centrifugation at 5 Krpm for 5 mins at 4°C and subsequent centrifugation of supernatants at 10 Krpm for 5 min at 4°C. Extracts were overlaid onto gradients and centrifuged at 35 Krpm for 160 min at 4°C. Fractions of 1 mL were collected using an ISCO Foxy Jr. fraction collector. Fractions were phenol/chloroform extracted and ethanol precipitated for Northern blot analysis using random primed GFP probe.
RNA immunoprecipitation
RNA immunoprecipitations were perfomed essentially as described (Vilardell et al. 2000). Cells from 100-mL mid-log cultures were pelleted and washed in lysis buffer (10 mM HEPES-KOH at pH 7.5, 1.5 mM MgCl2, 10 mM KCl, 1 mM DTT, 1 mM PMSF). Cells were resuspended in 500 μL lysis buffer and broken by bead beating with four 1-min pulses, keeping samples on ice in between. Extracts were prepared by centrifugation at 14 Krpm for 5 min at 4°C. One hundred fifty microliter extract and 850 μL IP buffer (100 mM NaCl, 50 mM Tris-HCl at pH 7.5, 2 mM MgCl2, 0.5 mM DTT, 0.05% Nonidet NP40) were added to prepared beads. α-V5 agarose congugate beads (SIGMA) were prepared by washing 20 μL bead slurry in 1 mL IP buffer supplemented with 100 μg/mL BSA and 100 μg/mL yeast tRNA, followed by 1 mL IP buffer containing 100 μg/μL yeast tRNA. Samples were incubated with beads at 4°C on a rotator for 1 h and washed four times for 10 min with 1 mL IP buffer. Beads were resuspended in 200 μL IP buffer supplemented with 10 μg yeast tRNA, and RNA was isolated by phenol/chloroform extraction and ethanol precipitation. For input samples, 1.5 μL undiluted extract was similarly extracted except that these were not supplemented with yeast tRNA. The entire yield from ethanol precipitation was analyzed by primer extension.
Acknowledgments
We are grateful to R. Singer and members of his laboratory for providing protocols and advice for fluorescent in situ hybridization. We also thank T.H. Jensen and D.A. Belostotsky for critical reading of the manuscript, R. Parker and D. Bressan for providing strains, F. LaRiviere for assistance with polysome profiles, and T. McCarthy for excellent technical assistance. This work was supported in part by NIH grant GM23549.
Article and publication are at http://www.rnajournal.org/cgi/doi/10.1261/rna.7166704.
REFERENCES
- Allen, N.P., Patel, S.S., Huang, L., Chalkley, R.J., Burlingame, A., Lutzmann, M., Hurt, E.C., and Rexach, M. 2002. Deciphering networks of protein interactions at the nuclear pore complex. Mol. Cell. Proteomics 1: 930–946. [DOI] [PubMed] [Google Scholar]
- Amrani, N., Minet, M., Le Gouar, M., Lacroute, F., and Wyers, F. 1997a. Yeast Pab1 interacts with Rna15 and participates in the control of the poly(A) tail length in vitro. Mol. Cell. Biol. 17: 3694–3701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amrani, N., Minet, M., Wyers, F., Dufour, M.E., Aggerbeck, L.P., and Lacroute, F. 1997b. PCF11 encodes a third protein component of yeast cleavage and polyadenylation factor I. Mol. Cell. Biol. 17: 1102–1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arava, Y., Wang, Y., Storey, J.D., Liu, C.L., Brown, P.O., and Herschlag, D. 2003. Genome-wide analysis of mRNA translation profiles in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. 100: 3889–3894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ausubel, F.M., Brent, R., Kingston, R., Moore, D.D., Seidman, J.G., Smith, J.A., and Struhl, K. (eds.). 1987. Current protocols in molecular biology, vol. 1. Wiley, New York.
- Baer, B.W. and Kornberg, R.D. 1983. The protein responsible for the repeating structure of cytoplasmic poly(A)-ribonucleoprotein. J. Cell Biol. 96: 717–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bear, D.G., Fomproix, N., Soop, T., Bjorkroth, B., Masich, S., and Daneholt, B. 2003. Nuclear poly(A)-binding protein PABPN1 is associated with RNA polymerase II during transcription and accompanies the released transcript to the nuclear pore. Exp. Cell Res. 286: 332–344. [DOI] [PubMed] [Google Scholar]
- Bienroth, S., Keller, W., and Wahle, E. 1993. Assembly of a processive messenger RNA polyadenylation complex. EMBO J. 12: 585–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birse, C.E., Minvielle-Sebastia, L., Lee, B.A., Keller, W., and Proudfoot, N.J. 1998. Coupling termination of transcription to messenger RNA maturation in yeast. Science 280: 298–301. [DOI] [PubMed] [Google Scholar]
- Bressan, D.A., Vazquez, J., and Haber, J.E. 2004. Mating type-dependent constraints on the mobility of the left arm of yeast chromosome III. J. Cell Biol. 164: 361–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodsky, A.S. and Silver, P.A. 2000. Pre-mRNA processing factors are required for nuclear export. RNA 6: 1737–1749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown, C.E. and Sachs, A.B. 1998. Poly(A) tail length control in Saccharomyces cerevisiae occurs by message-specific deadenylation. Mol. Cell. Biol. 18: 6548–6559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burd, C.G., Matunis, E.L., and Dreyfuss, G. 1991. The multiple RNA-binding domains of the mRNA poly(A)-binding protein have different RNA-binding activities. Mol. Cell. Biol. 11: 3419–3424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casolari, J.M., Brown, C.R., Komili, S., West, J., Hieronymus, H., and Silver, P.A. 2004. Genome-wide localization of the nuclear transport machinery couples transcriptional status and nuclear organization. Cell 117: 427–439. [DOI] [PubMed] [Google Scholar]
- Chekanova, J.A. and Belostotsky, D.A. 2003. Evidence that poly(A) binding protein has an evolutionarily conserved function in facilitating mRNA biogenesis and export. RNA 9: 1476–1490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chekanova, J.A., Shaw, R.J., and Belostotsky, D.A. 2001. Analysis of an essential requirement for the poly(A) binding protein function using cross-species complementation. Curr. Biol. 11: 1207–1214. [DOI] [PubMed] [Google Scholar]
- Custodio, N., Carmo-Fonseca, M., Geraghty, F., Pereira, H.S., Grosveld, F., and Antoniou, M. 1999. Inefficient processing impairs release of RNA from the site of transcription. EMBO J. 18: 2855–2866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daneholt, B. 1999. Pre-mRNP particles: From gene to nuclear pore. Curr. Biol. 9: R412–415. [DOI] [PubMed] [Google Scholar]
- Decker, C.J. and Parker, R. 1993. A turnover pathway for both stable and unstable mRNAs in yeast: Evidence for a requirement for deadenylation. Genes & Dev. 7: 1632–1643. [DOI] [PubMed] [Google Scholar]
- Deo, R.C., Bonanno, J.B., Sonenberg, N., and Burley, S.K. 1999. Recognition of polyadenylate RNA by the poly(A)-binding protein. Cell 98: 835–845. [DOI] [PubMed] [Google Scholar]
- Dower, K. and Rosbash, M. 2002. T7 RNA polymerase-directed transcripts are processed in yeast and link 3′ end formation to mRNA nuclear export. RNA 8: 686–697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duvel, K., Valerius, O., Mangus, D.A., Jacobson, A., and Braus, G.H. 2002. Replacement of the yeast TRP4 3′ untranslated region by a hammerhead ribozyme results in a stable and efficiently exported mRNA that lacks a poly(A) tail. RNA 8: 336–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duvel, K., Pries, R., and Braus, G.H. 2003. Polyadenylation of rRNA-and tRNA-based yeast transcripts cleaved by internal ribozyme activity. Curr. Genet. 43: 255–262. [DOI] [PubMed] [Google Scholar]
- Eckner, R., Ellmeier, W., and Birnstiel, M.L. 1991. Mature mRNA 3′ end formation stimulates RNA export from the nucleus. EMBO J. 10: 3513–3522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egli, C.M. and Braus, G.H. 1994. Uncoupling of mRNA 3′ cleavage and polyadenylation by expression of a hammerhead ribozyme in yeast. J. Biol. Chem. 269: 27378–27383. [PubMed] [Google Scholar]
- Gilbert, W. and Guthrie, C. 2004. The Glc7p nuclear phosphatase promotes mRNA export by facilitating association of Mex67p with mRNA. Mol. Cell 13: 201–212. [DOI] [PubMed] [Google Scholar]
- Hammell, C.M., Gross, S., Zenklusen, D., Heath, C.V., Stutz, F., Moore, C., and Cole, C.N. 2002. Coupling of termination, 3′ processing, and mRNA export. Mol. Cell. Biol. 22: 6441–6457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hector, R.E., Nykamp, K.R., Dheur, S., Anderson, J.T., Non, P.J., Urbinati, C.R., Wilson, S.M., Minvielle-Sebastia, L., and Swanson, M.S. 2002. Dual requirement for yeast hnRNP Nab2p in mRNA poly(A) tail length control and nuclear export. EMBO J. 21: 1800–1810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry, M.F., Mandel, D., Routson, V., and Henry, P.A. 2003. The yeast hnRNP-like protein Hrp1/Nab4 sccumulates in the cytoplasm after hyperosmotic stress: A novel Fps1-dependent response. Mol. Biol. Cell 14: 3929–3941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heyer, W.D., Johnson, A.W., Reinhart, U., and Kolodner, R.D. 1995. Regulation and intracellular localization of Saccharomyces cerevisiae strand exchange protein 1 (Sep1/Xrn1/Kem1), a multifunctional exonuclease. Mol. Cell. Biol. 15: 2728–2736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilleren, P., McCarthy, T., Rosbash, M., Parker, R., and Jensen, T.H. 2001. Quality control of mRNA 3′-end processing is linked to the nuclear exosome. Nature 413: 538–542. [DOI] [PubMed] [Google Scholar]
- Howe, K.J. 2002. RNA polymerase II conducts a symphony of pre-mRNA processing activities. Biochim. Biophys. Acta 1577: 308–324. [DOI] [PubMed] [Google Scholar]
- Huang, Y. and Carmichael, G.C. 1996. Role of polyadenylation in nucleocytoplasmic transport of mRNA. Mol. Cell. Biol. 16: 1534–1542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen, T.H., Patricio, K., McCarthy, T., and Rosbash, M. 2001. A block to mRNA nuclear export in S. cerevisiae leads to hyperadenylation of transcripts that accumulate at the site of transcription. Mol. Cell 7: 887–898. [DOI] [PubMed] [Google Scholar]
- Johnson, A.W. 1997. Rat1p and Xrn1p are functionally interchangeable exoribonucleases that are restricted to and required in the nucleus and cytoplasm, respectively. Mol. Cell. Biol. 17: 6122–6130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadowaki, T., Zhao, Y., and Tartakoff, A.M. 1992. A conditional yeast mutant deficient in mRNA transport from nucleus to cytoplasm. Proc. Natl. Acad. Sci. 89: 2312–2316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller, R.W., Kuhn, U., Aragon, M., Bornikova, L., Wahle, E., and Bear, D.G. 2000. The nuclear poly(A) binding protein, PABP2, forms an oligomeric particle covering the length of the poly(A) tail. J. Mol. Biol. 297: 569–583. [DOI] [PubMed] [Google Scholar]
- Kerwitz, Y., Kuhn, U., Lilie, H., Knoth, A., Scheuermann, T., Friedrich, H., Schwarz, E., and Wahle, E. 2003. Stimulation of poly(A) polymerase through a direct interaction with the nuclear poly(A) binding protein allosterically regulated by RNA. EMBO J. 22: 3705–3714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessler, S.H. and Sachs, A.B. 1998. RNA recognition motif 2 of yeast Pab1p is required for its functional interaction with eukaryotic translation initiation factor 4G. Mol. Cell. Biol. 18: 51–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krebber, H., Taura, T., Lee, M.S., and Silver, P.A. 1999. Uncoupling of the hnRNP Npl3p from mRNAs during the stress-induced block in mRNA export. Genes & Dev. 13: 1994–2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn, U. and Pieler, T. 1996. Xenopus poly(A) binding protein: Functional domains in RNA binding and protein–protein interaction. J. Mol. Biol. 256: 20–30. [DOI] [PubMed] [Google Scholar]
- Lei, E.P. and Silver, PA. 2002. Intron status and 3′-end formation control cotranscriptional export of mRNA. Genes & Dev. 16: 2761–2766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Libri, D., Dower, K., Boulay, J., Thomsen, R., Rosbash, M., and Jensen, T.H. 2002. Interactions between mRNA export commitment, 3′-end quality control, and nuclear degradation. Mol. Cell. Biol. 22: 8254–8266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine, M.S., McKenzie 3rd, A., Demarini, D.J, Shah, N.G., Wach, A., Brachat, A., Philippsen, P., and Pringle, J.R. 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14: 953–961. [DOI] [PubMed] [Google Scholar]
- Mangus, D.A., Evans, M.C., and Jacobson, A. 2003. Poly(A)-binding proteins: Multifunctional scaffolds for the post-transcriptional control of gene expression. Genome Biol. 4: 223.1–223.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minvielle-Sebastia, L., Preker, P.J., Wiederkehr, T., Strahm, Y., and Keller, W. 1997. The major yeast poly(A)-binding protein is associated with cleavage factor IA and functions in premessenger RNA 3′-end formation. Proc. Natl. Acad. Sci. 94: 7897–7902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muhlrad, D., Decker, C.J., and Parker, R. 1995. Turnover mechanisms of the stable yeast PGK1 mRNA. Mol. Cell. Biol. 15: 2145–2156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel, D. and Butler, J.S. 1992. Conditional defect in mRNA 3′ end processing caused by a mutation in the gene for poly(A) polymerase. Mol. Cell. Biol. 12: 3297–3304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Politz, J.C., Tuft, R.A., Pederson, T., and Singer, R.H. 1999. Movement of nuclear poly(A) RNA throughout the interchromatin space in living cells. Curr. Biol. 9: 285–291. [DOI] [PubMed] [Google Scholar]
- Proudfoot, N. and O’Sullivan, J. 2002. Polyadenylation: A tail of two complexes. Curr. Biol. 12: R855–857. [DOI] [PubMed] [Google Scholar]
- Proweller, A. and Butler, S. 1994. Efficient translation of poly(A)-deficient mRNAs in Saccharomyces cerevisiae. Genes & Dev. 8: 2629–2640. [DOI] [PubMed] [Google Scholar]
- Saavedra, C.A., Hammell, C.M., Heath, C.V., and Cole, C.N. 1997. Yeast heat shock mRNAs are exported through a distinct pathway defined by Rip1p. Genes & Dev. 11: 2845–2856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sachs, A.B. and Davis, R.W. 1989. The poly(A) binding protein is required for poly(A) shortening and 60S ribosomal subunit-dependent translation initiation. Cell 58: 857–867. [DOI] [PubMed] [Google Scholar]
- Sachs, A.B., Bond, M.W., and Kornberg, R.D. 1986. A single gene from yeast for both nuclear and cytoplasmic polyadenylate-binding proteins: Domain structure and expression. Cell 45: 827–835. [DOI] [PubMed] [Google Scholar]
- Sachs, A.B., Davis, R.W., and Kornberg, R.D. 1987. A single domain of yeast poly(A)-binding protein is necessary and sufficient for RNA binding and cell viability. Mol. Cell. Biol. 7: 3268–3276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samarsky, D.A., Ferbeyre, G., Bertrand, E., Singer, R.H., Cedergren, R., and Fournier, M.J. 1999. A small nucleolar RNA:ribozyme hybrid cleaves a nucleolar RNA target in vivo with near-perfect efficiency. Proc. Natl. Acad. Sci. 96: 6609–6614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shav-Tal, Y., Darzacq, X., Shenoy, S.M., Fusco, D., Janicki, S.M., Spector, D.L., and Singer, R.H. 2004. Dynamics of single mRNPs in nuclei of living cells. Science 304: 1797–1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straight, A.F., Belmont, A.S., Robinett, C.C., and Murray, A.W. 1996. GFP tagging of budding yeast chromosomes reveals that protein–protein interactions can mediate sister chromatid cohesion. Curr. Biol. 6: 1599–1608. [DOI] [PubMed] [Google Scholar]
- Stutz, F. and Rosbash, M. 1998. Nuclear RNA export. Genes & Dev. 12: 3303–3319. [DOI] [PubMed] [Google Scholar]
- Stutz, F., Kantor, J., Zhang, D, McCarthy, T., Neville, M., and Rosbash, M. 1997. The yeast nucleoporin rip1p contributes to multiple export pathways with no essential role for its FG-repeat region. Genes & Dev. 11: 2857–2868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takemura, R., Inoue, Y., and Izawa, S. 2004. Stress response in yeast mRNA export factor: reversible changes in Rat8p localization are caused by ethanol stress but not heat shock. J. Cell. Sci. 15: 4189–4197. [DOI] [PubMed] [Google Scholar]
- Tarun Jr., S.Z. and Sachs, A.B. 1996. Association of the yeast poly(A) tail binding protein with translation initiation factor eIF-4G. EMBO J. 15: 7168–7177. [PMC free article] [PubMed] [Google Scholar]
- Thomsen, R., Libri, D., Boulay, J., Rosbash, M., and Jensen, T.H. 2003. Localization of nuclear retained mRNAs in Saccharomyces cerevisiae. RNA 9: 1049–1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilardell, J., Chartrand, P., Singer, R.H., and Warner, J.R. 2000. The odyssey of a regulated transcript. RNA 6: 1773–1780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinciguerra, P. and Stutz, F. 2004. mRNA export: An assembly line from genes to nuclear pores. Curr. Opin. Cell Biol. 16: 285–292. [DOI] [PubMed] [Google Scholar]
- Wahle, E. 1991. A novel poly(A)-binding protein acts as a specificity factor in the second phase of messenger RNA polyadenylation. Cell 66: 759–768. [DOI] [PubMed] [Google Scholar]
- Wells, S.E., Hillner, P.E., Vale, R.D., and Sachs, A.B. 1998. Circularization of mRNA by eukaryotic translation initiation factors. Mol. Cell 2: 135–140. [DOI] [PubMed] [Google Scholar]
- Wilusz, C.J., Gao, M., Jones, C.L., Wilusz, J., and Peltz, S.W. 2001. Poly(A)-binding proteins regulate both mRNA deadenylation and decapping in yeast cytoplasmic extracts. RNA 7: 1416–1424. [PMC free article] [PubMed] [Google Scholar]
- Zhao, J., Hyman, L., and Moore, C. 1999. Formation of mRNA 3′ ends in eukaryotes: Mechanism, regulation, and interrelationships with other steps in mRNA synthesis. Microbiol. Mol. Biol. Rev. 63: 405–445. [DOI] [PMC free article] [PubMed] [Google Scholar]