Abstract
Tertiary stabilizing motifs (TSMs) between terminal loops or internal bulges facilitate folding of natural hammerhead ribozymes (hRz) under physiological conditions. However, both substrate and enzyme strands contribute nucleotides to the TSMs of trans-cleaving hRz, complicating the design of hRz that exploit TSMs to target specific mRNA. To overcome this limitation, we used SELEX to identify new, artificial TSMs that are less sensitive to sequence context. Nucleotides in loop II or in a bulge within the ribozyme strand of stem I were randomized, while the interaction partner was held constant. All nucleotides of the substrate pair with the ribozyme, minimizing their possible recruitment into the TSM, as such recruitment could constrain choice of candidate target sequences. Six cycles of selection identified cis-acting ribozymes that were active in 100 μM MgCl2. The selected motifs partially recapitulate TSMs found in natural hRz, suggesting that the natural motifs are close to optimal for their respective contexts. Ribozyme “RzB” showed enhanced thermal stability by retaining trans-cleavage activity at 80°C in 10 mM MgCl2 and at 70°C in 2 mM MgCl2. A variant of ribozyme “RzB” with a continuously paired stem 1 rapidly lost activity as temperature was increased. The selected motifs are modular, in that they permit trans-cleavage of several substrates in submillimolar MgCl2, including two substrates derived from the U5 genomic region of HIV-1. The new, artificial tertiary stabilized hRz are thus nearly independent of sequence context and enable for the first time the use of highly active hRz targeting almost any mRNA at physiologically relevant magnesium concentrations.
Keywords: hammerhead ribozymes, SELEX, thermal stability, magnesium ions, HIV-1 gene therapy, kinetic analysis
INTRODUCTION
Nucleic acids are versatile tools for selective gene knockdown. Ribozymes, antisense, and small interfering RNAs can disrupt RNA targets, while aptamers can antagonize protein and small molecule targets. Although siRNA is facile and effective against many cytoplasmic targets, it cannot yet target noncoding RNAs, introns, or other RNAs localized to the nucleus. In addition, several viruses have evolved mechanisms that disable RNA interference (Chen et al. 2004; Lakatos et al. 2004), thus making it difficult to develop effective antiviral drugs based on siRNA in those cases. Small nucleolytic ribozymes, such as the hammerhead ribozyme (hRz), are especially attractive in these contexts.
To fully exploit hRz inside cells, it is essential to overcome the limited availability of free Mg2+. Divalent metal ions such as Mg2+ are required for hammerhead catalysis at physiological ionic strength, serving both structural and catalytic roles. hRz that have been truncated to their minimal catalytic core structure typically require several millimolar to several tens of millimolar MgCl2 or equivalent divalent metal ion for optimal activity. Although the total Mg2+ concentration inside cells is in the millimolar range, the concentration of free Mg2+ ions appears to be substantially lower. For example, the kinetic behavior of a hairpin ribozyme in 2 mM MgCl2 more closely matched its intracellular kinetic behavior than when the in vitro reaction was carried out in 10 mM MgCl2 (Yadava et al. 2004). Most intracellular Mg2+ is chelated by ATP and other ligands, and the free Mg2+ concentration is estimated to be closer to 0.1–0.5 mM (Darnell et al. 1986).
Natural hRz form tertiary interactions that stabilize their catalytically active structures at submillimolar concentrations of MgCl2 (De la Peña et al 2003; Khvorova et al. 2003) and that greatly enhance intracellular cis-cleavage activity. The active fold of the SMα1 hammerhead ribozyme from Schistosoma mansoni (Ferbeyre et al. 1998; Vazquez-Tello et al. 2002) was recently demonstrated through fluorescence resonance energy transfer analysis to be stabilized by tertiary interactions between loop II and a symmetrical internal bulge in stem I. These interactions simplify divalent ion-dependent folding from a two-step process (with [Mg2+]50 values of ~0.1 and 2.0 mM) to a one-step process ([Mg2+]50 = 0.16 mM) (Penedo et al. 2004). In the hRz derived from the satellite RNA of the negative strand of tobacco ringspot virus (sTRSV) and from several related viroids, mutational analysis and computational modeling suggest that nucleotides at the ends of stems I and II form a parallel docking interaction held together by the hydrogen bonds of a trans Watson-Crick–Watson-Crick pair and a trans sugar–sugar pair (see Leontis and Westhof 2001 for interaction nomenclature; Khvorova et al. 2003).
It is attractive to exploit natural TSM to effect rapid trans-cleavage of intracellular RNA targets by expressed hRz. However, nucleotides corresponding to both the substrate and enzyme strands contribute to forming the TSMs of trans-cleaving hRz, complicating the design of hRz that exploit TSMs to target specific mRNA. We have used two separate strategies to solve this conundrum. In a separate work, we describe the dissection of the sTRSV hRz into substrate and ribozyme strands, such that the TSMs within the ribozyme strand remain intact (S.T. Greathouse and D.H. Burke, in prep.). The in vitro selection strategy used here yielded two structurally distinct hRz that are active in submillimolar MgCl2 (RzA and RzB families). The selected motifs allow stable cis-cleaving hRz to be converted into readily generalized, stable trans-cleaving hRz. Their ability to stabilize hRz structure is nearly independent of the sequence of the RNA targeted for cleavage. The stabilization is also evident under conditions of extreme temperature (T ≥70°C). These artificial tertiary motifs will greatly benefit the design of stabilized hRz for intracellular applications.
RESULTS
Selection of artificially stabilized hRz
Tertiary stabilized hRz were selected from partially randomized libraries designed to make substrate binding stable, reliable, and versatile. Stem I was extended beyond the tertiary interaction domain, dividing it into stem Ia (proximal to catalytic core) and stem Ib (distal to the core). In both Libraries A (N = 8; 48 = 65,536 species) and B (N = 6; 46 = 4096 species), one half of each pair of tertiary interaction modules was based on natural hammerhead sequences, with the other half being random sequence. For Library A (Fig. 1A), the SMα1 sequence was used for stem II, loop II, stem III, and most of stem Ia. Eight random nucleotides were included within the ribozyme strand of stem I at the same position where there is a four-base symmetric internal bulge in SMα1 (GUAC/UCCA in SMα1, NNNNNNNN/-in LibA; ribozyme strand in bold; Fig. 2; Ferbeyre et al. 1998; Vazquez-Tello et al. 2002). Library B (Fig. 1B) is based on the hRz from peach latent mosaic virus minus strand (PLMV) (Hernandez and Flores 1992). The six nucleotides of PLMV loop II were replaced with six random nucleotides. Because of the presence of stem Ib, the tri-nucleotide loop UAA that caps stem I in the PLMV hRz is repositioned as an internal bulge within the ribozyme strand of Library B.
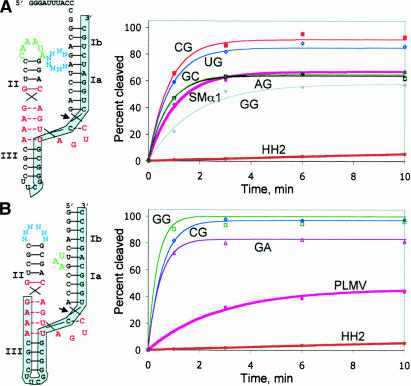
FIGURE 1.
Selection of stabilized cis-cleaving hRz. (A,B, left) Design of randomized hRz used in selection. (Red) catalytic core nucleotides; (green) constant TSM nucleotides; (cyan) randomized TSM nucleotides. Shaded segments are complementary to oligonucleotides used in reverse-transcription steps of selection to regenerate full-length transcription template. (A,B, right) Kinetic analyses of individual isolates selected from libraries A and B. Reactions were carried out at 37°C in 0.1 mM MgCl2. For library A, dinucleotide variants in the selected motif xxGCUACG are indicated by filled red squares (CG), filled gray diamonds (GG), open green triangles (GC), open black squares (AG), and open blue diamonds (UG). The GC and AG traces overlap almost exactly. For library B, dinucleotide variants in the selected motif UGxxAU are indicated by open green squares (GG), open blue diamonds (CG), and open plum triangles (GA). In both plots, control hRz are indicated with thick lines as filled brown circles (minimized ribozyme HH2) and filled pink circles (parental hRz SMα1 or PLMV, respectively).
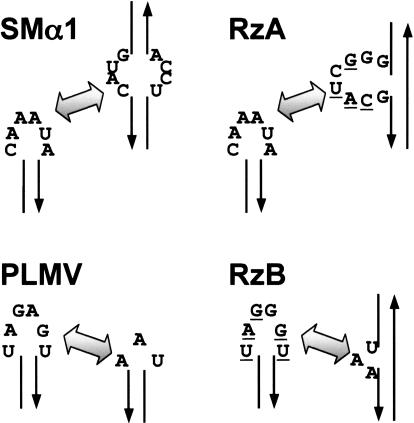
FIGURE 2.
Comparison of secondary structural contexts of tertiary interactions in natural ribozymes (left) vs. selected motifs (right). Underlined nucleotides in selected modules match sequence of the natural hRz on which selection was based.
After six cycles of selection, both libraries showed rapid self-cleavage in 0.1 mM MgCl2. Of the 33 sequences obtained from Library A, 12 contained the motif GGGCUACG as the eight-nucleotide selected element. The same motif was present in each of the remaining isolates with the following variations in the first two positions: CG(5), UG(4), AG(4), UU(2), GC(2), GU(1), GA(1), AA(1), and CA(1). Numbers in parentheses indicate the number of occurrences in the sequenced set. Representatives from each sequence group displayed cis-cleavage activity that was equivalent to that of the SMα1 hRz at 37°C in 0.1 mM MgCl2 (Fig. 1A). The observed nucleotide variations in these first two positions are, therefore, readily tolerated by the variants examined. Interestingly, the natural SMα1 bulge sequence (GUAC) was present in discontinuous form in the selected internal bulge motif in all isolates (xxGCUACG underlined, Fig. 2, top). Thus, the native tertiary interaction is at least partially recapitulated in a significantly altered structural context.
The 64 sequences obtained from Library B carried UGG GAU (40/64), UGCGAU (16/64), or UGGAAU (8/64) as the selected loop II element. These sequences are identical to the PLMV hexaloop consensus (UGARAU) in five of the six positions (underlined). Cis-cleavage by each construct was rapid (kobs >0.5 min−1) and converted nearly 100% of the substrate into product at 0.1 mM Mg2+, outperforming the parental PLMV ribozyme under these conditions (kobs ~0.1 min−1; 47% yield) (Fig. 1B). Thus, this population also recapitulates — and may somewhat improve upon — the natural tertiary interactions in PLMV, even though the secondary structural context of the UAA triloop was significantly altered in the selected hRz (Fig. 2, bottom). This may imply that the UAA element adopts similar configurations in both structural contexts (see Discussion).
Stabilization of trans-cleavage at submillimolar MgCl2
The dominant motifs from libraries A and B were engineered into trans-cleaving species by removing the loop at the end of stem III to generate ribozymes “RzA” and “RzB”. RzA cleaved its substrate at 1 mM Mg2+ with a rate of 1.71 min−1. This value is well above rates typically observed at this MgCl2 concentration for minimal hammerheads lacking TSM. However, both the rate and extent of cleavage diminished rapidly as MgCl2 was decreased to 0.5 mM or less (Table 1), and cleavage no longer followed first-order kinetic behavior. The parental SMα1 ribozyme on which Library A was based is very stable and has shown trans-cleavage of an mRNA target at 70°C both in vitro (10 mM MgCl2 at pH 8.0) and in the bacterium Thermus thermophilus (Vazquez-Tello et al. 2002). The sensitivity of RzA to submillimolar MgCl2 in our assays may reflect structural constraints imposed by the different sequence contexts of the tertiary interactions in SMα1 versus RzA (four unpaired nucleotides in each strand vs. eight single-stranded nucleotides in one strand), or it may indicate that the concentration of free Mg2+ in T. thermophilus is greater than 0.5 mM.
TABLE 1.
Kinetic parameters for cis- and trans-cleaving hRz
| Cis-reactionsb | ||||
| MgCl2, mM | Temp. °C | kobs min−1 | plateaua | |
| GG | 0.1 | 37 | 0.56 | 0.61 |
| CG | 0.1 | 37 | 1.2 | 0.91 |
| UG | 0.1 | 37 | 1.2 | 0.85 |
| AG | 0.1 | 37 | 1.3 | 0.64 |
| GC | 0.1 | 37 | 1.2 | 0.65 |
| HH2 | 0.1 | 37 | 0.041 | 0.16 |
| SMα1 | 0.1 | 37 | 0.93 | 0.67 |
| GG | 0.1 | 37 | 2.8 | 1.0 |
| CG | 0.1 | 37 | 1.8 | 0.97 |
| GA | 0.1 | 37 | 2.1 | 0.83 |
| HH2 | 0.1 | 37 | 0.042 | 0.16 |
| PLMV | 0.1 | 37 | 0.36 | 0.46 |
| Trans-reactions | ||||
| MgCl2, mM | Temp. °C | kobs min−1 | plateaua | |
| Observed initial cleavage rates (kobs) and fraction converted to product for constructs described in this study. Reported values are derived from averages of at least two measurements, which were usually within 5%–10% of each other. Uncertainties of fit to single-exponential kinetic models for calculated kobs were generally ±10% using the averaged data. Dashes indicate that no cleavage was observed under these conditions during 1 h reaction. | ||||
| aCalculated plateau value, f∞, as described in methods. | ||||
| bIndividual isolates are designated by the dinucleotide present at the variable positions within the selected motifs: xxGCUACG for LibA (top set) and UGxxAU for LibB (bottom set). | ||||
| cThese reaction conditions gave complex, multiphasic kinetics with rapid cleavage of 20%–70% of the input material, followed by nearly complete religation within 1–2 min. Reported rates are for initial cleavage only. | ||||
| RzA | 1.0 | 25 | 1.7 | 0.69 |
| 0.5 | 25 | 1.2 | 0.15 | |
| 0.1 | 25 | — | — | |
| RzA-4 | 10 | 37 | — | — |
| 20 | 25 | 1.3 | (c) | |
| 20 | 37 | 1.6 | (c) | |
| 50 | 25 | 3.5 | (c) | |
| 50 | 37 | 4.7 | (c) | |
| RzB | 0.5 | 25 | 2.0 | 0.56 |
| 0.1 | 25 | 1.4 | 0.47 | |
| 0.5 | 37 | 1.8 | 0.63 | |
| 0.1 | 37 | 0.04 | (c) | |
| RzB-1 | 0.5 | 25 | 2.2 | 0.39 |
| 0.1 | 25 | 0.28 | 0.28 | |
| 0.5 | 37 | 2.0 | 0.39 | |
| 0.1 | 37 | 0.20 | 0.29 | |
| RzB-2 | 0.5 | 25 | 1.3 | 0.38 |
| 0.1 | 25 | 1.3 | 0.09 | |
| 0.5 | 37 | 3.2 | 0.31 | |
| 0.1 | 37 | (c) | (c) | |
| RzB-3 | 0.5 | 25 | 1.9 | 0.46 |
| 0.1 | 25 | 1.2 | 0.06 | |
| 0.5 | 37 | 1.6 | 0.29 | |
| 0.1 | 37 | 1.3 | 0.05 | |
| RzB-4 | 0.5 | 25 | 1.9 | 0.56 |
| 0.1 | 25 | 1.2 | 0.45 | |
| 0.5 | 37 | 1.5 | 0.51 | |
| 0.1 | 37 | 0.59 | 0.55 | |
| RzB-5 | 0.5 | 25 | — | — |
| 0.5 | 37 | — | — | |
| 10 | 25 | 1.1 | 0.53 | |
| 10 | 37 | ≈1.2 | ≈0.6 | |
RzB was much more active than RzA under all conditions tested, converting 75% of substrate to product within 15 sec of adding MgCl2 to a final concentration of 1 mM (data not shown). Cleavage activity remained vigorous even at 0.5 and 0.1 mM MgCl2 (kobs = 2.0 and 1.4 min−1), covering the full range of estimated free intracellular Mg2+ concentration. Cleavage at 37°C was nearly threefold faster than at 25°C, when the reaction was carried out at 0.5 mM MgCl2; however, at 0.1 mM MgCl2, only the 25°C reaction showed efficient cleavage (Table 1). Deletion of the TSM from stem I to generate a continuous helix (ribozyme “RzB-0”) obliterated activity at submillimolar Mg2+, confirming that these nucleotides are responsible for stabilizing the ribozyme to very low MgCl2. The exact reason(s) for why the trans-cleaving RzB is superior to that of RzA is not yet clear and needs detailed investigation. However, it is possible that the size of the internal bulge within the substrate-binding arm (8 nt in RzA vs. 3 nt in RzB) may influence the overall kinetics of cleavage. Because of the superior performance of RzB at submillimolar Mg2+, this ribozyme was chosen for further analysis.
Thermal stabilization by selected TSM
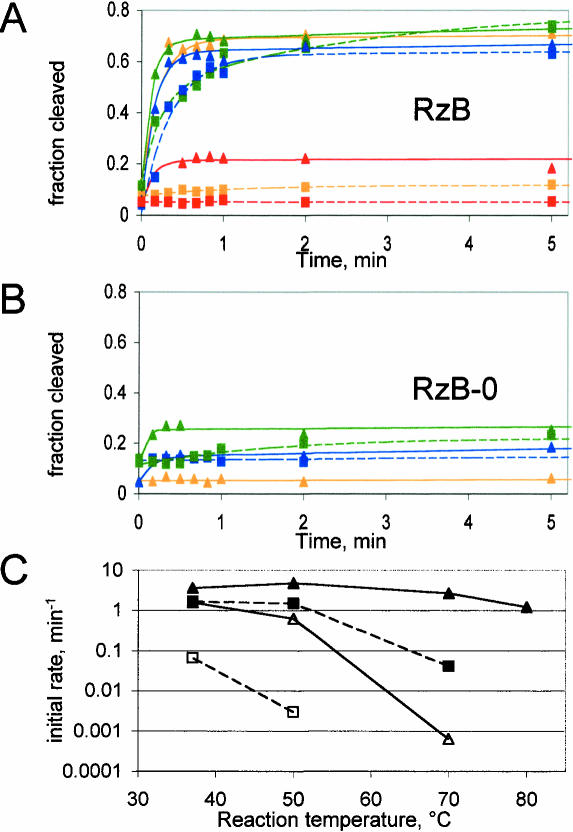
The data above demonstrate that the artificial TSM carried within the RzB family stabilizes the hRz in such a way as to retain trans-cleavage activity at low concentrations of MgCl2 where the ribozyme would otherwise become inactive. We reasoned that the selected TSM might similarly stabilize against thermal denaturation. To test this hypothesis, the trans-cleavage activities of ribozymes RzB and RzB-0 were studied as a function of temperature and magnesium concentration. In 10 mM MgCl2, RzB cleaved its substrate to similar extents and at similar rates at 37, 50, and 70°C. At 80°C, the extent of cleavage drops three- to fourfold, possibly due to melting of the tertiary interaction or melting of the ribozyme–substrate pairing (Fig. 3A). Either way, retention of substantial hammerhead activity at 80°C is remarkable and reinforces the profound impact of TSM’s on hammerhead integrity and enzymology. In 2 mM MgCl2, RzB was consistently slower than in 10 mM MgCl2. Activity at this MgCl2 concentration is equivalent at 37 and 50°C, with barely any cleavage seen at 70°C, and none at all at 80°C (Fig. 3A). In contrast to RzB, ribozyme RzB-0 rapidly lost activity as temperatures were raised above 37°C, becoming undetectable at 70°C even in 10 mM MgCl2 (Fig. 3B). The ratio of the rate for RzB to that of RzB-0 increases dramatically at higher temperatures, with more than 1000-fold difference between the two at 70°C (Fig. 3C). The TSM is therefore essential for the observed thermal stabilization.
FIGURE 3.
Thermal stabilization by selected tertiary motifs. (A) Kinetic profiles of ribozyme RzB in 10 mM (triangles, solid lines) or 2 mM (squares, dashed lines) MgCl2 at four temperatures as follows: (blue) 37°C; (green) 50°C; (orange) 70°C; (red) 80°C. (B) Kinetic profiles of ribozyme RzB-0, following same symbols as in A. (C) Temperature dependence of initial rate, calculated as described in Materials and Methods. (Closed symbols) RzB; (open symbols) RzB-0; (triangles with solid lines) 10 mM MgCl2; (squares with dashed lines) 2 mM MgCl2. Reactions that gave no measurable rate are not plotted (e.g., RzB at 80°C in 2 mM MgCl2).
Substrate independence of the selected motifs
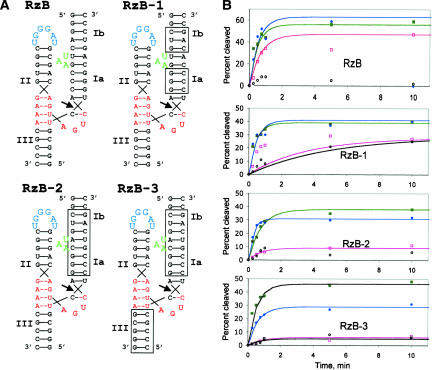
For the nucleotide modules selected here to be broadly useful in stabilizing hRz for intracellular applications, it is essential that they not be highly sensitive to the substrate sequence context in which they were selected. Three variations of RzB were constructed that progressively reversed each base pair in substrate-binding stems I and III. RzB-1 retains the base pairs that immediately flank the tertiary interaction element, while reversing most of the rest of stem I. RzB-2 reverses all of stem I, while RzB-3 reverses the pairing in both stems I and III (Fig. 4A). Each of these constructs is kinetically well-behaved at 25°C and cleaves its respective substrate rapidly and efficiently at 0.5 mM MgCl2 (kobs = 1.3–2.2 min−1) (Fig. 4B; Table 1). Similar rates and extents of cleavage are observed at 37°C. Even at 0.1 mM MgCl2, RzB-1 retained vigorous cleavage activity, although RzB-2 and RzB-3 were inactive under these conditions (Fig. 4B). Thus, while ribozymes based on RzB show little sequence context dependence at moderately low MgCl2 concentrations representative of physiological conditions (0.5 mM), there is some sequence context dependence under the most stringent conditions (0.1 mM MgCl2).
FIGURE 4.
Substrate independence of stabilized ribozymes. (A) Predicted secondary structures of ribozymes RzB, RzB-1, RzB-2, and RzB-3. Nucleotides that differ from those in RzB are boxed. (B) Plots of cleavage kinetics at 25°C, 0.5 mM MgCl2 (filled green squares); 25°C, 0.1 mM MgCl2 (open pink squares); 37°C, 0.5 mM MgCl2 (filled blue circles); and 37°C, 0.1 mM MgCl2 (open black circles). For RzB and RzB-2, the low reactivity at 37° in 0.1 mM MgCl2 did not fit to a simple kinetic scheme, so these traces are not shown. Rates and maximal fraction cleaved (k∞) reported in Table 1 for these two data sets were calculated from the initial points. All reactions were performed at pH 7.5.
Cleavage of an HIV-1 genome-derived RNA target
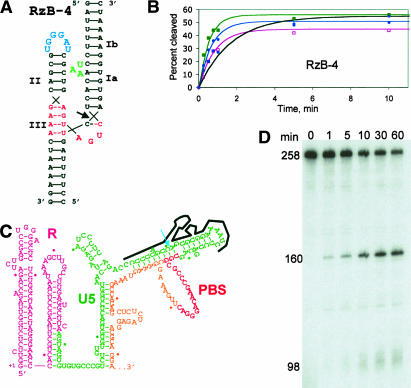
Ribozyme RzB-4 was designed to target a site within the U5 genomic region of HIV-1 that is 20 nt 5′ of the binding site for the tRNA3Lys primer (Fig. 5A). A synthetic 23-nt RNA substrate corresponding to this site was converted to cleaved product by RzB-4 at both moderate and stringent MgCl2 concentrations, and at both 25 and 37°C (Fig. 5B). Even under the most stringent conditions tested, 0.1 mM Mg2+ and 37°C, the initial cleavage rate was 0.6 min−1, with more than half of the input substrate converted into product within 5 min (Table 1). An analogous hRz based on RzA, designated RzA-4, displayed no cleavage at low MgCl2 and complex, multiphasic kinetics at high MgCl2 (Table 1). The expected hRz secondary structure is predicted by mfold to be the fourth most stable conformation, suggesting that the inactivity of this sequence may reflect the formation of alternative secondary structures rather than an intrinsic deficiency of the TSM. Removing the UAA tertiary module from stem I of RzB-4 to form ribozyme RzB-5 abolished activity at submillimolar MgCl2 concentrations (Table 1). The target site for RzB-4 lies within a region of HIV-1 RNA that is believed to have significant secondary structure (Fig. 5C). A 258-nt RNA corresponding to the 5′ end of HIV-1 genomic RNA was generated by transcribing a template encoding the R, U5, PBS, and an artificial 3′ flanking regions. When RzB-4 was annealed to this transcript in the presence of an oligonucleotide to mimic binding of the natural tRNA3Lys primer for HIV-1 replication, 60%–80% of the input 258-nt substrate was cleaved into 160- and 98-nt products in 0.5 mM MgCl2 at 37°C (Fig. 5D). This hRz and others based on RzB could thus be further studied as a potential therapeutic tool for blocking HIV replication or for other intracellular applications.
FIGURE 5.
Cleavage of HIV-1 RNA at submillimolar MgCl2. (A) Predicted structure of ribozyme RzB-4 annealed to a 23-nt substrate derived from the genomic RNA of HIV-1 strain HXB2. (B) Plots of substrate cleavage by RzB-4 under same conditions as in Figure 4. (C) Predicted secondary structure the HIV-1 genomic 5′ leader, based on comparison between SIV and HIV sequences (Rivzi and Panganiban 1993). Note that other secondary structures have also been proposed for this region. The first 242 nt of the 258-nt transcript are shown. Annealed RzB-4 is shown schematically, with cleavage site indicated by the arrow. (D) Cleavage reaction of internally radiolabeled, 258-nt synthetic genomic transcript in 0.5 mM MgCl2 at 37°C (pH 7.5), in the presence of a DNA oligonucleotide to mimic binding of the natural tRNA3Lys replication primer. Reaction times are given in minutes above the lanes. Cleavage products at 160 and 98 nt are indicated.
DISCUSSION
Developing ribozymes for intracellular applications requires that they function at maximum capacity under the ionic conditions found within cells, while simultaneously retaining versatility of design. While the realization last year that natural ribozymes are stabilized through tertiary interactions resolved the question of how to achieve cis-cleavage activity in low concentrations of magnesium, natural TSMs are not fully versatile because nucleotides from both strands of helix I contribute to tertiary stabilization. For example, when the SMα1 ribozyme from schistosomes is divided into two strands to effect trans-cleavage of one strand by the other, the substrate strand carries an essential unpaired UCCA that lies across helix I from the tertiary interacting nucleotides 6 nt downstream of the cleavage site. The cleavage specificity of hRz is normally described as NUH∧ (H = A,C,U; ∧ = cleavage site), such that a random sequence is densely packed with potential recognition sites. Even the preferred site, GUC∧, is found on average every 64 nt in a random sequence. Under low magnesium or high temperature conditions, including the TSM in the cleavage site, definition expands the SMα1 recognition specificity to GUC∧N6UCCA. Such a site is found within the human synaptobrevin gene (GenBank accession no. U30291). When Vazquez-Tello et al. (2002) studied cleavage of a fragment of this gene both in vitro (10 mM MgCl2 at pH 8.0) or expressed within the bacterium T. thermophilus, the SMα1 derivative showed efficient trans-cleavage in both contexts. However, the extra found nucleotides in the expanded sequence definition of the cleavage site decrease the frequency with which appropriate sites will be found by a factor of 44 = 256-fold, potentially limiting the direct applicability of SMα1 to a small subset of desirable target sites.
The present work demonstrates that artificial tertiary interactions for trans-cleaving ribozymes can be selected from diverse libraries (RzA and RzB). The stabilization afforded by RzB shows little sensitivity to sequence context under moderately stringent conditions (0.5 mM MgCl2, 25°C) and is active in at least three sequence contexts (RzB, RzB-1, and RzB-4) under highly stringent conditions (0.1 mM MgCl2, 37°C). Because the effectiveness of the TSM in RzB is relatively unconstrained by substrate sequence, RzB may not be subject to the context limitations noted above for SMα1. RzB is thus a logical starting point for the construction of new ribozymes for use in gene knockdown experiments for therapeutic applications or for studies of biological mechanisms. It may also be possible to achieve highly efficient, regulated cleavage by combining these tertiary stabilization motifs into an RNA that also carries modules for allosteric regulation by oligonucleotides (e.g., the TRAP design), by small molecules (e.g., cAMP or theophylline), or by proteins (Koizumi et al. 1999; Koizumi and Breaker 2000; Sekella et al. 2000; Burke et al. 2002; Wang and Sen 2002; Saksmerprome and Burke 2003, 2004).
Isolates from both selections are remarkably similar to the natural tertiary interacting modules on which they were based (Fig. 2). The natural sequences may thus be close to optimal within the contexts of their respective interaction partners. Furthermore, the structural interactions formed by, and the functional consequences of, the selected TSMs may be similar to those of the natural TSMs. The structural framework into which a given TSM is embedded are expected to constrain the positions of the interacting nucleotides both in terms of distance from the catalytic core and rotation about the helical axis. The similarity between the selected and the natural TSM sequences therefore further suggests that nucleotides presented within an asymmetric bulge and those in a loop at the end of a helix can assume overall similar conformations.
It is not yet clear how tertiary stabilization by natural or artificial sequence elements diminishes the divalent magnesium requirement for hRz, although we speculate that one or more of the structural roles for Mg2+ may be taken over by the TSM. Minimal hRz exhibit proficient magnesium-independent cleavage at high concentrations of monovalent ions (Murray et al. 1998; Curtis and Bartel 2001), although even in 2 M Li+, the reaction is stimulated by Mg2+ ions that appear to bind with low affinity (Inoue et al. 2004). Discrete Mg2+ binding sites have been suggested in analyses by crystallography, thiophilic metal ion rescue, EPR, and NMR (Scott et al. 1996; Peracchi et al. 1997; Hansen et al. 1999; Hunsicker and DeRose 2000; Hoogstraten et al. 2002; Wang et al. 2004). In contrast, recent analysis based on single-molecule FRET analysis (Rueda et al. 2003) and isothermal titration calorimetry (Mikulecky et al. 2004) suggest that Mg2+ wields its effects through diffuse electrostatic binding rather than through interactions at specific sites.
There are at least three mechanisms by which TSMs could exert their influence over hRz function: through their effects on static hRz structure, through their effects on global hRz conformational dynamics, or through their effects on local conformation within the catalytic core. First, whether binding of Mg2+ is through interactions at specific sites or through diffuse binding, the addition of low concentrations of Mg2+ at low ionic strength induces helices I and II to move closer together than their orientation in the absence of Mg2+, as evidenced by FRET transfer efficiency using tethered fluorophores (Bassi et al. 1999; Rueda et al. 2003), by hydroxyl radical footprinting (Hampel and Burke 2003), and by transient electric birefringence (Amiri and Hagerman 1996). The TSM could similarly “staple” helices I and II together in a global conformation that is similar to that of the catalytically competent ground state. Second, time-resolved FRET studies suggest that conformational mobility of helices I and II strongly affect the kinetic behavior of minimal hRz. Mutations that increase mobility of these helices with respect to each other correlate with faster kinetics at low mM Mg2+ and saturation at lower MgCl2 concentrations than for more rigid hRz (Rueda et al. 2003). Frequency of access to the catalytically competent conformation is proposed to be rate limiting in that context. Rather than using increased mobility to get into a catalytically competent helical orientation, a TSM may decrease the conformational mobility of a tertiary stabilized hRz to avoid depopulating an already-competent orientation. Noninvasive dynamic studies are needed to test this hypothesis. Third, Dunham et al. (2003) observed near-in-line orientation in the hRz active site between the nucleophilic 2′ oxygen and the scissile phosphate when a single nucleotide extending from stem I crosses over to pair with its complement within stem II, leading those investigators to propose that TSMs may influence orientation and reactivity at the active site by constraining the helical rotation of stem I, in addition to their effects on the angle between stems I and II. Finally, as shown in Figure 3, the TSM in RzB allows this hRz to retain activity at temperatures as high as 80°C. The fact that this activity is seen only in 10 mM MgCl2 and not in 2 mM MgCl2 demonstrates that the stabilizing contributions of TSMs and divalent magnesium are additive. Rather than considering the TSM as replacing divalent magnesium, these results indicate that the two factors can contribute simultaneously and synergistically to stabilization of hammerhead ribozyme structure.
MATERIALS AND METHODS
Ribozymes, RNA substrate, and oligonucleotides
RNA and DNA oligonucleotides were synthesized by Dharmacon and by Integrated DNA Technologies, respectively. Ribozymes were synthesized using a modified method of ACE protection chemistry (Dharmacon), or they were transcribed in vitro from synthetic templates, radiolabeled, and purified as described (Burke et al. 2002). The integrity of synthetic ribozymes was confirmed by mass spectrometry and purified by denaturing gel electrophoresis. The DNA template for transcription of the human immunodeficiency virus type 1 (HIV-1) substrate was prepared from synthetic oligonucleotides based on the sequence of strain HXB2 (GenBank entry KO3455; courtesy of Jay Kissel, Indiana University).
Selection in vitro
For selection “A”, 2 nmol of synthetic single-stranded DNA template A (5′-gggatttacccGgcagNNNNNNNNatccagCTGATGAgtcc CAAATAggacGAAacgccTTCGggcgtC∧ctggatctgc) was made double stranded by PCR. Regions of alternating upper and lower case indicate nucleotides encoding secondary structural elements in the transcribed product, and cleavage site is indicated by “∧”. PCR primers were “Forward T7-A” (5′-taatacgactcactatagggatttaccc-GGCAG-3′) and “Reverse RTA” (5′-GCAGATCCAG GACGC-CCG-3′). T7 RNA polymerase promoter is singly underlined, and nucleotides from the 3′ cleavage fragment that are restored during reverse transcription (see below) are doubly underlined. For selection “B”, 2 nmol of DNA template B (5′-gggactTAAgcccactgat gagtcgcnnnnnngcgacGAAacgccTTCGggcgtc∧tgggcagtccc-3′) using primers T7-B (5′-taatacgactcactatagggacTtaagcccactg-3′) and RT-B (5′-GGGACTGCCCAGACGCCCGAAGGCGTTTC-3′). Double-stranded DNA was purified by Qiagen column.
In vitro transcription was carried out at 37°C for 1 h in the presence of 400 μM antisense oligos in 200-μL reaction volumes containing 0.2 nmol of DNA template. Addition of antisense oligo was crucial for inhibiting ribozyme self cleavage during transcription. For libraries A and B, antisense oligo sequences were 5′-GTCCTATTTGGGACTCATCAGCTGGAT-3′and 5′-GCGACTC ATCAGTGGGCTTAAGTCCC-3′, respectively. Self-cleavage reactions were carried out in 100 μM MgCl2, 50 mM Tris (pH 7.0) at 37°C, gradually decreasing the time of reaction from 5 min initially to 30 sec in the sixth and seventh selection cycles. The 5′ cleavage fragments were separated from uncleaved RNA by denaturing PAGE. The 5′ cleavage fragments (containing the random sequence region flanked by constant sequence) were recovered from excised gel slices and annealed to primer “RT-A” or “RT-B” noted above. Upon reverse transcription, these primers restore the nucleotides corresponding to the 3′ cleavage fragment (see Fig. 1). PCR amplification with primers that restored the T7 promoter then yielded a transcription template with which to generate RNA for the next selection cycle. PCR products after seven cycles of selection were cloned for sequencing. Because the extent of RNA cleavage exceeded 80%, further selection under these conditions would not improve significantly the catalytic properties of ribozymes, and the libraries were cloned into plasmids for sequencing.
Determination of apparent rate constants kobs
A total of 10 pmol of end-labeled substrate RNA was mixed with 100 pmol ribozyme in Tris-HCl (pH 7.5) (final 50 mM in reaction), heated to 90°C for 1 min, then allowed to cool slowly to the reaction temperature. After equilibrating at either 25 or 37°C for ~5 min, one-tenth of the sample was removed as a zero time point and quenched in an equal volume of stop buffer (95% formamide, 20 mM EDTA, 0.5% bromophenol blue, 0.5% xylene cyanol). Reactions were initiated by addition of MgCl2 to the desired concentration. Aliquots were removed at indicated time intervals and quenched in stop buffer. Samples were analyzed by denaturing (7 M urea) 12% PAGE. Cleaved and uncleaved substrate bands in the gels were quantified using the ImageQuant software from Molecular Dynamics. Most kobs values were determined as a nonlinear best fit of the data to the single-exponential equation ft = f0 + (f∞− f0) (1−exp−kt), where ft = fraction cleaved at a given time (product/(product + substrate)), f0 = zero point correction (essentially zero), f∞ = estimated plateau value at infinite time, and kobs = first-order rate constant (kobs = k2 + k−2). Those datasets that could not be adequately modeled as single-exponential processes were fit to a double-exponential process using the equation ft = f0 + (f∞−f0)[(1−a · exp−kat −(1−a) · exp−kbt)], where ka and kb are the kobs values for the two exponentially decaying processes, and “a” is the fraction of the active ribozyme that partitions into the faster of the two processes. For kinetic analysis of cis-cleavage by individual isolates from the selected libraries, PCR products corresponding to individual isolates were purified and transcribed as above, resuspended in 2 μM EDTA, diluted into water, and refolded in pH buffer, whereupon the reactions were initiated by addition of MgCl2. For cleavage of 258 nt HIV-1 genomic RNA analog, 0.2 μM substrate was folded in the presence of 6 μM DNA oligo 5′-TCCCTGTTC GGGCGCCACTGCTAGA-3′ to simulate annealing and initial extension of the 3′ end of the tRNA3Lys primer used by the virus in replication. A total of 2 μM RzB-4 was also included during the annealing step. Components were equilibrated at 37°C, and reactions were initiated by addition of MgCl2.
Activity profile at elevated temperatures
For kinetic analysis of trans-cleaving ribozymes RzB and RzB-0 at elevated temperatures, 2 μM of the ribozyme strand was unfolded with ~10–20 nM of 5′-radiolabeled substrate strand in 50 mM Tris•Cl (pH 7.5), and 10 μM EDTA at 90°C for 2 min. Reannealing was done at 70°C for 2 min, followed by a slow ramp down to 30°C at −3°C/min. After 10 min at 30°C, the samples were heated to the final temperature of the kinetic assay for 10 min and reactions were initiated by adding MgCl2 to the concentrations indicated. Initial cleavage rates reported in Figure 3 were calculated by taking the first derivative of the double exponential equation above and solving for t = 0 to yield: rate = f∞•· a · ka + f∞•· (1−a) · kb.
Acknowledgments
We thank Steven Rhee for insightful comments on the manuscript, Jay Kissel for preparing transcription template for the HIV-1 genomic substrate, and Sugata Roy Chowdhury for technical assistance. This work was supported by NIH Grant AI45344 and a Packard Interdisciplinary Science Award to D.H.B., and by a graduate research fellowship from the Royal Thai Government to V.S. NSF grant DBI-0244815 supported purchase of the Molecular Dynamics Typhoon PhosphorImager.
Abbreviations
TSM, tertiary stabilizing motif
hRz, hammerhead ribozyme
HIV-1, human immunodeficiency virus type 1
Article and publication are at http://www.rnajournal.org/cgi/doi/10.1261/rna.7159504.
REFERENCES
- Amiri, K. and Hagerman, P. 1996. The global conformation of an active hammerhead RNA during the process of self-cleavage. J. Mol. Biol. 261: 125–134. [DOI] [PubMed] [Google Scholar]
- Bassi, G., Mollegaard, N., Murchie, A., and Lilley, D. 1999. RNA folding and misfolding of the hammerhead ribozyme. Biochemistry 38: 3345–3354. [DOI] [PubMed] [Google Scholar]
- Burke, D.H., Ozerova, N.D.S., and Nilsen-Hamilton, M. 2002. Allosteric hammerhead TRAP ribozymes. Biochemistry 41: 6588–6594. [DOI] [PubMed] [Google Scholar]
- Chen, J., Li, W.X., Xie, D., Peng, J.R., and Ding, S.W. 2004. Viral virulence protein suppresses RNA silencing-mediated defense but upregulates the role of microRNA in host gene expression. Plant Cell 16: 1302–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis, E. and Bartel, D. 2001. The hammerhead cleavage reaction in monovalent cations. RNA 7: 547–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darnell, J., Lodish, H., and Baltimore, D. 1986. Molecular cell biology. W.H. Freeman, New York.
- De la Peña, M., Gago, S., and Flores, R. 2003. Peripheral regions of natural hammerhead ribozymes greatly increase their self-cleavage activity. EMBO J. 22: 5561–5570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunham, C., Murray, J., and Scott, W. 2003. A helical twist-induced conformational switch activates cleavage in the hammerhead ribozyme. J. Mol. Biol. 332: 327–336. [DOI] [PubMed] [Google Scholar]
- Ferbeyre, G., Smith, J., and Cedergren, R. 1998. Schistosome satellite DNA encodes active hammerhead ribozymes. Mol. Cell. Biol. 18: 3880–3888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hampel, K. and Burke, J. 2003. Solvent protection of the hammerhead ribozyme in the ground state: Evidence for a cation-assisted conformational change leading to catalysis. Biochemistry 42: 4421–4429. [DOI] [PubMed] [Google Scholar]
- Hansen, M., Simorre, J., Hanson, P., Mokler, V., Bellon, L., Beigelman, L., and Pardi, A. 1999. Identification and characterization of a novel high affinity metal-binding site in the hammerhead ribozyme. RNA 5: 1099–1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez, C. and Flores, R. 1992. Plus and minus RNAs of peach latent mosaic viroid self-cleave in vitro via hammerhead structures. Proc. Natl. Acad. Sci. 89: 3711–3715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoogstraten, C., Grant, C., Horton, T., DeRose, V., and Britt, R. 2002. Structural analysis of metal ion ligation to nucleotides and nucleic acids using pulsed EPR spectroscopy. J. Am. Chem. Soc. 124: 834–842. [DOI] [PubMed] [Google Scholar]
- Hunsicker, L. and DeRose, V. 2000. Activities and relative affinities of divalent metals in unmodified and phosphorothioate-substituted hammerhead ribozymes. J. Inorg. Biochem. 80: 271–281. [DOI] [PubMed] [Google Scholar]
- Inoue, A., Takagi, Y., and Taira, K. 2004. Importance in catalysis of a magnesium ion with very low affinity for a hammerhead ribozyme. Nucleic Acids Res. 32: 4217–4223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khvorova, A., Lescoute, A., Westhof, E., and Jayasena, S.D. 2003. Sequence elements outside the hammerhead ribozyme catalytic core enable intracellular activity. Nat. Struct. Biol. 10: 708–712. [DOI] [PubMed] [Google Scholar]
- Koizumi, M. and Breaker, R. 2000. Molecular recognition of cAMP by an RNA aptamer. Biochemistry 39: 8983–8992. [DOI] [PubMed] [Google Scholar]
- Koizumi, M., Soukup, G.A., Kerr, J.N., and Breaker, R.R. 1999. Allosteric selection of ribozymes that respond to the second messengers cGMP and cAMP. Nat. Struct. Biol. 6: 1061–1071. [DOI] [PubMed] [Google Scholar]
- Lakatos, L., Szittya, G., Silhavy, D., and Burgyan, J. 2004. Molecular mechanism of RNA silencing suppression mediated by p19 protein of tombusviruses. EMBO J. 23: 876–884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leontis, N.B. and Westhof, E. 2001. Geometric nomenclature and classification of RNA base pairs. RNA 7: 499–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikulecky, P., Takach, J., and Feig, A. 2004. Entropy-driven folding of an RNA helical junction: An isothermal titration calorimetric analysis of the hammerhead ribozyme. Biochemistry 43: 5870–5881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray, J.B., Seyhan, A.A., Walter, N.G., Burke, J.M., and Scott, W.G. 1998. The hammerhead, hairpin and VS ribozymes are catalytically proficient in monovalent cations alone. Chem. Biol. 5: 587–595. [DOI] [PubMed] [Google Scholar]
- Penedo, J., Wilson, T., Jayasena, S., Khvorova, A., and Lilley, D. 2004. Folding of the natural hammerhead ribozyme is enhanced by interaction of auxiliary elements. RNA 10: 880–888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peracchi, A., Beigelman, L., Scott, E., Uhlenbeck, O., and Herschlag, D. 1997. Involvement of a specific metal ion in the transition of the hammerhead ribozyme to its catalytic conformation. J. Biol. Chem. 272: 26822–26826. [DOI] [PubMed] [Google Scholar]
- Rizvi, T. and Panganiban, A. 1993. Simian immunodeficiency virus RNA is efficiently encapsidated by human immunodeficiency virus type 1 particles. J. Virol. 67: 2681–2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rueda, D., Wick, K., McDowell, S., and Walter, N. 2003. Diffusely bound Mg2+ ions slightly reorient stems I and II of the hammerhead ribozyme to increase the probability of formation of the catalytic core. Biochemistry 42: 9924–9936. [DOI] [PubMed] [Google Scholar]
- Saksmerprome, V. and Burke, D.H. 2003. Structural flexibility and the thermodynamics of helix exchange constrain attenuation and allosteric activation of hammerhead ribozyme TRAPs. Biochemistry 42: 13879–13886 [DOI] [PubMed] [Google Scholar]
- ———. 2004. Deprotonation favors productive folding in allosteric TRAP hammerhead ribozymes. J. Mol. Biol. 341: 685–694. [DOI] [PubMed] [Google Scholar]
- Scott, W., Murray, J., Arnold, J., Stoddard, B., and Klug, A. 1996. Capturing the structure of a catalytic RNA intermediate: The hammerhead ribozyme. Science 274: 2065–2069. [DOI] [PubMed] [Google Scholar]
- Sekella, P., Rueda, D., and Walter, N. 2002. A biosensor for theophylline based on fluorescence detection of ligand-induced hammerhead ribozyme cleavage. RNA 8: 1242–1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vazquez-Tello, A., Castán, P., Moreno, R., Smith, J.M., Berenguer, J., and Cedergren, R. 2002. Efficient trans-cleavage by the Schistosoma mansoni SMalpha1 hammerhead ribozyme in the extreme thermophile Thermus thermophilus. Nucleic Acids Res. 30: 1606–1612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, D. and Sen, D. 2002. Rationally designed allosteric variants of hammerhead ribozymes responsive to the HIV-1 Tat protein. Comb. Chem. High Throughput Screen 5: 301–312. [DOI] [PubMed] [Google Scholar]
- Wang, G, Gaffney, B.L., and Jones, R.A. 2004. Differential binding of Mg2+, Zn2+ and Cd2+ at two sites in a hammerhead ribozyme motif, determined by 15N NMR. J. Am. Chem. Soc. 126: 8908–8909. [DOI] [PubMed] [Google Scholar]
- Yadava, R.S, Mahen, E.M., and Fedor, M.J. 2004. Kinetic analysis of ribozyme-substrate complex formation in yeast. RNA 10: 863–879. [DOI] [PMC free article] [PubMed] [Google Scholar]