Abstract
Formation of the P3-P7 pseudoknot structure, the core of group I ribozymes, requires long-range base pairing. Study of the Tetrahymena ribozyme appreciates the hierarchical folding of the large, multidomain RNA, in which the P3-P7 core folds significantly slower than do the other domains. Here we explored the formation of the P3-P7 pseudoknot of the Candida ribozyme that has been reported to concertedly fold to the catalytically active structure with a rate constant of 2 min−1. We demonstrate that pseudoknot formation occurs during the rapid ribozyme compaction, coincident with formation of many tertiary interactions of the ribozyme. A low physiological concentration of magnesium (1.5 mM) is sufficient to fully support the pseudoknot formation. The presence of nonnative intermediates containing an unfolded P3-P7 region is evident. However, catalysis-based analysis shows these nonnative intermediates are stable and fail to convert to the catalytically active structure, suggesting that rapid pseudoknot formation is essential for folding of the active ribozyme. Interestingly, RNAstructure predicts no stable Alt P3 structure for the Candida ribozyme, but two stable Alt P3s for the Tetrahymena ribozyme, explaining the dramatic difference in folding of the P3-P7 core of these two ribozymes. We propose that rapid formation of the P3-P7 pseudoknot represents a folding strategy ensuring efficient production of the catalytically active structure of group I ribozymes, which sheds new light on the mechanism of effective ribozyme folding in vivo.
Keywords: RNA folding, group I ribozyme, P3-P7 pseudoknot, fast folding pathway, antisense oligonucleotide hybridization
INTRODUCTION
Rapid compaction of a large multidomain RNA results in distinct intermediates containing native or nonnative interactions; these intermediates follow either the trap-free or kinetically trapped pathway to reach the catalytically active structure (Russell et al. 2002a; Woodson 2002; Sosnick and Pan 2003). The tendency of an RNA to fold into native or nonnative intermediates at the early folding stage is proposed to be affected by the length and the primary sequence of a RNA molecule, as well as the ability of the readily formed local secondary structures in promoting the subsequent tertiary interactions (Go 1984; Brion and Westhof 1997; Thirumalai et al. 2001; Woodson 2002).
Although it is much more challenging for RNA molecules to form long-range interactions than to form local base pairing, long-range interactions are widely used in nature to organize the three-dimensional structure of large multidomain RNAs (Michel and Ferat 1995; Brion and Westhof 1997; Massire et al. 1997). Group I ribozymes represent a large family of complex RNA molecules containing multiple domains, in which folding of the P3-P7 catalytic core involves formation of a pseudoknot structure consisting of long-range base pairing. The P3-P7 pseudoknot structure is essential in substrate binding, ribozyme catalysis (Cech 1990), and assembling the different domains of the ribozyme (Michel and Westhof 1990; Golden et al. 1998). For example, formation of the P7 helix brings the P4-P6 and P3-P7 domains into close proximity, forming the catalytic core structure. Formation of the P3 helix allows a close arrangement of the P1-P2 domain consisting of the 5′ splice site to the core structure. Studies with the Tetrahymena ribozyme reveal that one strand of the P3 helix readily mis-folds to form a stable local secondary structure Alt P3, making the formation of the native P3-P7 pseudoknot energetically unfavorable (Zarrinkar and Williamson 1994; Pan and Woodson 1998; Sclavi et al. 1998; Treiber et al. 1998; Pan et al. 2000). The misfolding problem also exists in folding of the full-length P RNA of the ribonucleoprotein RNase P, another well-studied large multidomain RNA (Pan et al. 1999; Sosnick and Pan 2003).
Almost all group I ribozymes possess nine conserved base-paired helices. These ribozymes catalyze the same type of transesterification reactions (Cech 1990), but substantial differences have been revealed in folding of different group I introns (Buchmueller et al. 2000; Zhuang et al. 2000; Treiber and Williamson 2001a; Rangan et al. 2003; Xiao et al. 2003). As demonstrated by the single-molecule study, a small fraction of the Tetrahymena ribozyme folds to the active structure directly with the rate constants of 1 sec−1 and 1 min−1, while the majority of the ribozyme is trapped in the metastable and stable intermediates in which folding of the P3-P7 pseudoknot requires several minutes (Zhuang et al. 2000). On the other hand, although no kinetic traps exist in folding of the bI5 intron, the native intermediates are inefficiently converted to the active ribozyme under physiological concentrations of magnesium; this problem is naturally resolved by the action of an RNA binding protein called CBP2 (Buchmueller et al. 2000). Recent work on the Azoarcus ribozyme demonstrates that nearly half of the ribozyme molecules reach the stable tertiary structure in which the P3-P7 pseudoknot folds in less than 100 msec (Rangan et al. 2003). Our study of a Candida ribozyme has shown that the majority of the ribozyme molecules fold to the catalytically active structure without being trapped in the energetically disadvantaged conformations (Xiao et al. 2003). The presence of the fast folding pathway in forming the catalytically active structure of group I ribozymes suggests that folding of the large multidomain RNAs could be cooperative and efficient.
Candida ribozyme is an excellent model in characterizing the ensemble RNA folding through the trap-free pathway. In this work, we studied the formation of the P3-P7 pseudoknot structure in order to elucidate the mechanisms of this folding pathway. Our study with both Ca.L-11 ribozyme, a trans-acting ribozyme slightly shorter than the wild-type self-splicing group I intron from the 26S rRNA gene of the Candida albicans, and the intron precursor reveals that rapid formation of the native P3-P7 pseudoknot in the presence of 1.5 mM or higher concentrations of magnesium commits the folding intermediates to the catalytically active structure through the trap-free pathway.
RESULTS
The P3-P7 pseudoknot forms during rapid ribozyme compaction
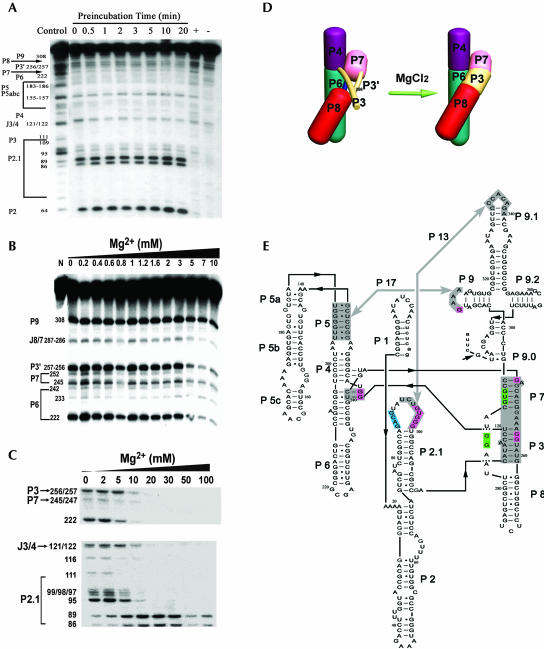
T1 ribonuclease is highly effective in cleaving the accessible unpaired guanosine residues over a broad range of ionic strengths and temperatures (data not shown), and therefore has been proven to be a valuable tool in monitoring the formation of RNA secondary structure. Because the tertiary structure of large complex RNA, such as group I ribozyme, involves extensive long-range base pairing, this base pairing is subjected to the detection by T1 ribonuclease as well (Zhang and Leibowitz 2001; Rangan et al. 2003; Xiao et al. 2003). As shown in Figure 1A, the Candida ribozyme was folded in the presence of 10 mM Mg2+ for 0–20 min at 37°C; T1 ribonuclease was then added to conduct 1-min cleavage of the ribozyme. The sample in the control lane was folded and cleaved in the absence of magnesium. As we previously reported (Xiao et al. 2003), magnesium-mediated compaction of the ribozyme occurred rapidly, as demonstrated by the same strong protection of G residues from T1 cleavage for the sample receiving no pre-incubation at 37°C as for that receiving a 20-min preincubation.
FIGURE 1.
Analysis of the rapid formation of the P3-P7 pseudoknot by T1 ribonuclease footprinting. (A) Candida ribozyme folding at the indicated periods of time. The 5′-end labeled Ca.L-11 ribozyme was folded at 10 mM of Mg2+, then cleaved by T1 ribonuclease (0.1 unit/μL) for 1 min and analyzed as previously described (Xiao et al. 2003). (Control lane) The sample was folded and cleaved in Tris-HCl (pH 7.5) without magnesium. (Lane +) The sample was cleaved by T1 in the presence of urea. (Lane −) The sample received no T1 ribonuclease. (B) Folding of the wild-type Candida ribozyme at magnesium concentrations ranging from 0 to 10 mM. (C) Folding of the Candida ribozyme containing 116 A → G mutation in the presence of 0–20 mM of MgCl2. (D) Illustration of the magnesium-dependent folding of the P3-P7 pseudoknot. (E) The illustrated secondary structure of the Candida ribozyme Ca.LSU, including a short exon sequence that forms P1 with the IGS. The major tertiary interaction sites described in the text and the P3-P7 pseudoknot are shaded in gray. Many G residues in these regions are strongly protected by magnesium, shaded in pink. Some magnesium-induced T1 cleavage sites are shaded in blue. The antisense oligo-induced cleavage sites are shaded in green.
Magnesium stabilizes base pairs in the local helical structure and the tertiary structure (Zhang and Leibowitz 2001). We examined whether the P3-P7 pseudoknot of the Candida ribozyme forms during magnesium-mediated ribozyme compaction by T1 footprinting. Strikingly, G256 and G257 in the P3 helix and G245 in the P7 helix of the Candida ribozyme became protected from T1 cleavage during the rapid ribozyme compaction. Meanwhile, many G residues involved in tertiary interactions including long-range base pairing and base triples became protected (Fig. 1A). In the case of G121 and G122 forming base triples with P6, G308 interacting with P5 by a tetraloop-receptor interaction and G95, G97–99 in P2.1 pairing with L9.1 to form the P13 helix (Lehnert et al. 1996), they were all strongly cleaved by T1 in the absence of magnesium, while strongly protected with magnesium present. Therefore, we propose that the P3-P7 pseudoknot and many tertiary interactions are rapidly formed during the magnesium-induced ribozyme compaction. This suggestion is consistent with the recent report that RNA tertiary structures in the collapsed and native states of the bI5 ribozyme core are indistinguishable, although only the latter is catalytically active (Buchmueller and Weeks 2003).
Detailed examination of the P3-P7 pseudoknot formation showed that base pairing of most of the P7 stem occurred in the absence of magnesium. However, the formation of the first base pair of the P7 stem, G245-C295, requires the presence of magnesium (Fig. 1B), probably because this orphan pair is separated from the other continuous pairs by the bulged A and thus is less stable. Strikingly, P3 regions were largely unpaired in the absence of magnesium. G256 and G257 were strongly cleaved by T1, suggesting that at least a half of the P3 stem is not formed (Fig. 1B,E). Mutation of A116 in the P3 stem to G did not affect the splicing activity of the Candida ribozyme (data not shown); T1 analysis revealed that G116 was moderately sensitive to T1 cleavage in the absence of magnesium, suggesting that the other half of the P3 stem is not stably paired either (Fig. 1C). Magnesium at low concentrations strongly protected mutant G116 from T1 cleavage, while higher concentrations dramatically protected G256 and G257 (Fig. 1B,C), proving that formation of the P3 stem requires magnesium (Fig. 1D).
Using an antisense oligonucleotide to study the formation of the functional P3-P7 pseudoknot
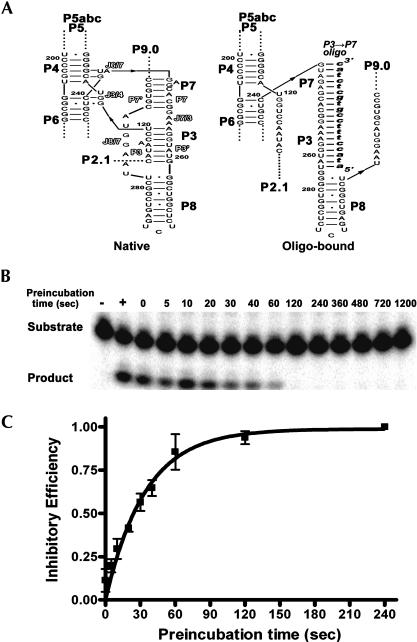
We previously reported that magnesium concentration required to fold a stable ribozyme structure resistant to T1 cleavage varies according to the amount of T1 ribonuclease used in a cleavage reaction (Xiao et al. 2003); thus the magnesium requirement for formation of the stable P3-P7 pseudoknot resistant to T1 cleavage (Fig. 1B,C) does not necessarily correlate with that for formation of the functional pseudoknot structure. Antisense oligonucleotide hybridization (Zarrinkar and Williamson 1994; Treiber and Williamson 2001b) was used in this study to assess the magnesium concentrations supporting the functional P3-P7 pseudoknot structure. The P3 → P7 oligo, an antisense oligonucleotide pairing with the P3′-J7/3-P7-J6/7 region of the Candida ribozyme, was used to target the unformed P3-P7 structure, while the ribozyme activity was used as readout for the formation of the functional P3-P7 pseudo-knot. Pairing of the P3 → P7 oligo with the unformed P3-P7 region of the ribozyme results in a DNA–RNA duplex consisting of 19 base pairs, which is expected to be more stable than the native P3-P7 pseudoknot structure. However, formation of either the P3-P7 pseudoknot or the mis-folded DNA–RNA duplex should exclude the formation of the other (Fig. 2A).
FIGURE 2.
Analysis of P3-P7 pseudoknot formation by the P3 → P7 oligo. (A) The secondary structure of the native P3-P7 pseudoknot and the misfolded DNA–RNA duplex proposed to form upon oligonucleotide binding (italicized lowercase). In addition to the double-stranded helices, the junctions between helices and each of the two single-stranded segments forming P3 and P7 helices are indicated (outlined fonts). (B) The Candida ribozyme Ca.L-11 was preincubated with the P3 → P7 oligo and 10 mM Tris-HCl (pH 7.5) for the indicated times, then the cleavage reaction was performed for 1 min in 10 μL reactions containing 15 nM of Ca.L-11 ribozyme, 0.1 mM GTP, 5 mM MgCl2, 50 mM Tris-HCl (pH 7.5), 15 nM of 5′-end radiolabeled substrate, and 5 μM P3 → P7 oligo. (Lane +) Complete reaction with H2O added instead of the P3 → P7 oligo. (Lane −) Control reaction containing no ribozyme. (C) The inhibitory efficiency was plotted against the pre-incubation time, fitting to the equation Y = Ymax[1 − e −k(t−t0)]. The inhibitory efficiency (Y) indicates the ability of antisense oligo to bind the ribozyme and to inhibit the ribozyme activity at each time point, which was calculated from the equation [Frac(t) − Frac(+)]/Frac(+). Frac(t) indicates the fraction of the cleaved substrate (product/input substrate) at each time point, and Frac(+) indicates the cleaved fraction in the positive control sample receiving no antisense oligo. Three sets of data were plotted for each point.
To test whether the P3 → P7 oligo binds to the unformed P3-P7 structure to form the stable DNA–RNA duplex, the oligo was preincubated with the unfolded trans-acting ribozyme in the absence of magnesium at 37°C; reactions were subjected to 1-min substrate cleavage after different times of preincubation. The formation of misfolded duplex is expected to retain the ribozyme in the inactive conformation even if Mg2+ and GTP are subsequently added during the 1-min substrate cleavage. Hence, the time-dependent reduction of the ribozyme activity reflects the kinetics of stable binding of the antisense oligo to the unfolded ribozyme (Fig. 2B,C). In the presence of 5 μM P3 → P7 oligo, formation of the stable DNA–RNA duplex occurred at a rate constant of 0.03 sec−1. Nonetheless, oligonucleotide binding is yet much slower than Mg2+-mediated ribozyme compaction.
We then asked if the antisense oligo binds to and unfolds the native ribozyme containing the folded P3-P7 pseudo-knot structure. The trans-acting ribozyme and precursor RNA were prefolded in 5 mM or 2 mM Mg2+, respectively, to reach the native conformations, then a 20-min preincubation with 2–5 μM of the antisense oligo was conducted followed by the activity analysis. Note that the optimal Mg2+ concentration for self-splicing of the Candida ribozyme under the assay condition described is 1.5–2.5 mM (data not shown). The 20-min preincubation was run to allow the binding to reach equilibrium. The ribozymes were fully active, as was the control sample receiving no antisense oligo (lanes in II of Fig. 3A, lanes in III of Fig. 3B), strongly supporting our hypothesis that the native P3-P7 pseudo-knot is not accessible to the antisense oligo hybridization. Magnesium-mediated rapid compaction of the group I ribozyme leads to the native and nonnative intermediates (Thirumalai et al. 2001). If nonnative intermediates containing unfolded P3-P7 structure result from compaction, the antisense oligonucleotide is expected to bind to these intermediates. If these nonnative intermediates are capable of slowly folding to the catalytically active structure, the ribozyme activity is expected to decrease in the presence of the P3 → P7 oligo. If the nonnative intermediates are not present or are incapable of folding to the catalytically active structure, the ribozyme activity would not be altered by the presence of the P3 → P7 oligo.
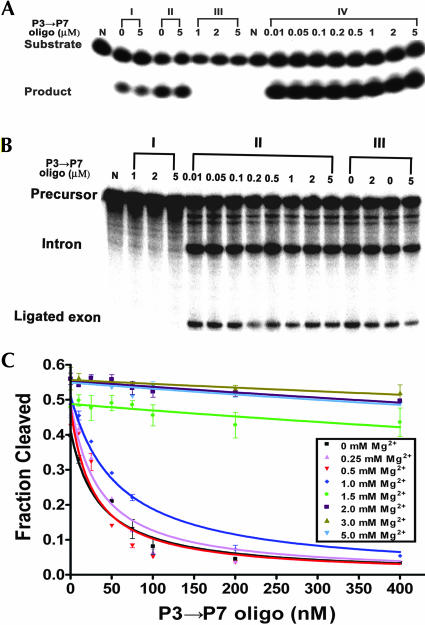
FIGURE 3.
Magnesium-dependent formation of the functional P3-P7 pseudoknot. (A) The Ca.L-11 ribozyme samples in I were preincubated with 2 mM Mg2+ for 10 min, then incubated in the presence or absence of 5 μM P3 → P7 oligo for 20 min. A 1-min substrate cleavage was followed in a 10 μL reaction containing 25 nM of Ca.L-11 ribozyme, 0.1 mM GTP, 2 mM MgCl2, 50 mM Tris-HCl (pH 7.5), 15 nM of 5′-end radiolabeled substrate, and the indicated concentrations of antisense oligo. Samples in II were treated the same as in I except that 5 mM Mg2+ were used. The ribozyme samples in III were preincu-bated with 5 μM P3 → P7 oligo and 10 mM Tris-HCl (pH 7.5) for 10 min in the absence of magnesium. The 1-min trans-cleavage reaction was performed as in II. The ribozyme samples in IV were directly subjected to a 1-min cleavage assay as in II without any pretreatment. (Lanes N) The negative control containing no ribozyme. (B) The precursor RNA in I was preincubated with the indicated concentrations of the P3 → P7 oligo and 10 mM Tris-HCl (pH 7.5) for 10 min, then the standard self-splicing reactions including 2 mM Mg2+ were performed for 40 min. (II) The standard self-splicing reactions as in I were performed for the precursor RNA receiving no pretreatment. (III) The precursor RNA was preincubated with 2 mM Mg2+ for 10 min, then incubated with 0, 2, or 5 μM P3 → P7 oligo for 20 min, followed by a standard self-splicing reaction. (Lane N) The negative control receiving no GTP. (C) The P3 → P7 oligo and the pure Ca.L-11 ribozyme RNA (5 nM) were preincubated for 20 min at 37°C in the presence of varying concentrations of magnesium. Then, a mixture containing substrate RNA, GTP, and the required amount of MgCl2 was added to initiate a 1-min cleavage assay under the standard reaction condition. The cleaved fraction (product/input substrate) was plotted against the oligonucleotide concentration, fitting to a one-site binding equation, Y = Ymax[1 − X/(KI + X)]. In this equation, Y is the cleaved fraction that reflects the amount of the “native” ribozyme free from oligonucleotide binding; Ymax is the maximal cleaved fraction at each magnesium concentration.
As shown in Figure 3A,B, the ribozyme activity was not affected by the presence of up to 5 μM oligo in the direct self-splicing and trans-cleavage reactions in which Mg2+, GTP, and antisense oligo were added simultaneously. The control samples receiving a 20-min preincubation with 1–5 μM P3 → P7 oligo completely inhibited the ribozyme activity. These results confirmed that formation of the functional P3-P7 pseudoknot is rapid, faster than the antisense oligonulceotide hybridization (Fig. 3A,B). Meanwhile, these results disprove the presence of folding intermediates that lack the readily formed P3-P7 pseudoknot and are capable of slowly folding to the catalytically active structure.
We noted that the presence of 2 mM Mg2+ in the direct self-splicing and trans-cleavage reactions completely protected the ribozyme from being inhibited by the antisense oligo (Fig. 3A,B), suggesting that formation of the functional P3-P7 pseudoknot requires lower concentrations of magnesium than formation of the structurally stable one resistant to T1 cleavage (Fig. 1B,C).
Formation of the functional P3-P7 pseudoknot requires low physiological concentrations of magnesium
To detail the magnesium requirement to fully support the functional pseudoknot formation, the trans-acting ribozyme was incubated with varying concentrations of anti-sense oligonucleotide and magnesium prior to the 1-min cleavage reaction. Mg2+ at 5 mM was present in each sample during the 1-min substrate cleavage to keep the same reaction condition for each sample. When the ribozyme RNA was folded at low concentrations of magnesium, the P3 → P7 oligo effectively inhibited the substrate cleavage reaction in a dose-dependent manner, consistent with the presence of “loose” ribozyme conformations permitting efficient binding of the antisense oligonucleotide (Fig. 3C).
The presence of Mg2+ ≥ 1.5 mM essentially eliminated the inhibitory effect, consistent with a rapid formation of the native P3-P7 pseudoknot at 1.5 mM or higher concentrations of magnesium, precluding the ribozyme from being bound by the oligonucleotide. Because a folding time of 20 min was run to allow the oligonucleotide binding and ribozyme folding to reach equilibrium prior to catalysis, the lack of the inhibitory effect further supports the absence of folding intermediates with an unformed P3-P7 structure capable of slowly folding to the catalytically active structure.
The magnesium requirement for the pseudoknot formation is significantly lower than that required for maximal activity of the Ca.L-11 ribozyme (6 mM), suggesting that further magnesium binding is involved in promoting tertiary interactions and/or structural rearrangements that are essential for forming the fully active ribozyme.
Rapid ribozyme compaction results in stable nonnative intermediates
Our results show that, unlike the ribozyme of the Tetrahymena ribozyme, the Candida ribozyme tends to not fold to the catalytically active structure through slow conversion of the metastable nonnative intermediates. However, we did not yet prove whether any nonnative intermediate(s) lacking the P3-P7 pseudoknot structure result from the rapid ribozyme compaction. During the T1 footprinting analysis, we noticed that substantial T1 cleavage remained at G245, G256, and G257 in the P3-P7 region in the presence of 10 mM Mg2+, suggesting that a fraction of ribozyme molecules might be rapidly converted to intermediates containing the stable unpaired P3-P7 structure (Fig. 1A,B).
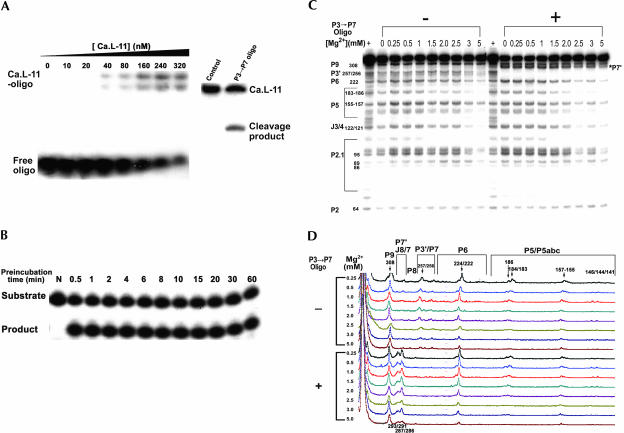
We then addressed this question by gel-shift analysis and RNase H experiment in the presence of the P3 → P7 oligo. Gel-shift experiments showed that a significant amount of radiolabeled antisense oligo was bound by the ribozyme in the presence of 3 mM Mg2+. RNase H experiments showed that a substantial fraction of ribozyme was accessible to the antisense oligo (Fig. 4A). To further confirm that the non-native intermediates containing unfolded P3-P7 structure do not slowly fold to the native structure, ribozyme folding at 5 mM Mg2+ was conducted up to 60 min, followed by a 1-min substrate cleavage assay. As shown in Figure 4B, no activity difference was observed after prolonged folding of the ribozyme, consistent with the nonnative intermediates being stable and failing to convert to the catalytically active structure.
FIGURE 4.
Detection of the nonnative folding intermediates containing the unfolded P3-P7 structure. (A) Gel-shift analysis of the binding of P3 → P7 oligo to the ribozyme (left). Varying concentrations of the pure Ca.L-11 ribozyme RNA and 50 nM of the 5′-end labeled oligonucleotide were preincubated in 50 mM Tris-HCl (pH 7.5) and 3 mM Mg2+ for 20 min, and were then analyzed by native gel electrophoresis. Detection of the oligo binding by RNase H cleavage (right). Magnesium, P3 → P7 oligo, and RNase H were simultaneously added to the reaction containing the 5′-end labeled Ca.L-11 ribozyme to initiate the folding, oligo binding, and cleavage at the same time. The control sample received no antisense oligo. A 5% PAGE-8 M urea gel was run to separate the products. (B) Ca.L-11 ribozyme was preincubated with 5 mM Mg2+ and 10 mM Tris-HCl (pH 7.5) for the indicated periods of time, then the 1-min cleavage assay was performed in a 10 μL reaction containing 25 nM ribozyme, 0.1 mM GTP, 5 mM MgCl2, 50 mM Tris-HCl (pH 7.5), 15 nM of 5′-end radiolabeled substrate, and 5 μM P3 → P7 oligo. (C) T1 ribonuclease footprinting analysis of the antisense oligo binding. The 5′-end labeled Ca.L-11 RNA (~2 ng) was preincubated in the presence or absence of antisense oligonucleotide and different concentrations of Mg2+ for 20 min. All samples were then cleaved by T1 ribonuclease (0.05 U/μL) for 1 min. The G ladder was made by cleaving the denatured Ca.L-11 RNA (Zhang and Leibowitz 2001). One representative gel run for 1.5 h is shown here, with the selected G residues being numbered and the corresponding helices of the ribozyme being indicated. (D) Analysis of the T1 sensitivity on the 10-h gel (Xiao et al. 2003).
T1 footprinting showed that the oligonucleotide precisely bound to the target site on the Candida ribozyme, completely protecting all of the accessible G residues in the P3-P7 region from T1 cleavage regardless of the magnesium concentrations (Fig. 4C,D). The binding at low concentrations of magnesium is readily explained by the “loose” ribozyme conformations that are accessible to antisense oligo-nucleotide. However, the binding at magnesium concentrations sufficient for pseudoknot formation is consistent with the presence of stable nonnative intermediates containing unfolded P3-P7 structure. Because all the T1 accessible G sites in the pseudoknot region are completely susceptible to the antisense oligonucleotide hybridization, we suggest that these two methods detect the same nonnative intermediates.
Simultaneously with the protection of G residues in the oligonucleotide binding target, the originally protected G291 and G293 complementary to the oligonucleotide target sequence in P7, as well as the G286 and G287 in J8/7, became accessible to T1 cleavage upon oligonucleotide binding. Note that T1 cleavage of the G sites whose sensitivity was induced by oligonucleotide binding decreased with increasing magnesium concentrations, correlating well with the magnesium-dependent folding of the P3-P7 pseudoknot that protects the P3-P7 region from T1 cleavage (Figs. 1E, 4C).
Interestingly, except for the G residues in the P3-P7 region and J8/7, T1 cleavage in the presence of antisense oligonucleotide at all the other G residues was the same as in its absence (Fig. 4C), suggesting that binding of antisense oligonucleotide to the unfolded P3-P7 region does not alter the entire ribozyme structure. Since in the absence or the presence of low concentrations of magnesium the RNA is expected to be in relatively loose conformations, it is conceivable that binding of antisense oligonucleotide in one region does not affect the structure in another region. In the presence of high concentrations of magnesium, the lack of structural alteration in the regions outside of P3-P7 of the nonnative intermediates upon oligonucleotide binding provides further evidence that the nonnative intermediates are stable collapsed states. Catalysis-based analysis showed that these stable nonnative intermediates of the Candida ribozyme barely fold to the catalytically active structure, which significantly differs from the meta-stable intermediates of the Tetrahymena ribozyme.
Minimal mispairing of the Candida ribozyme
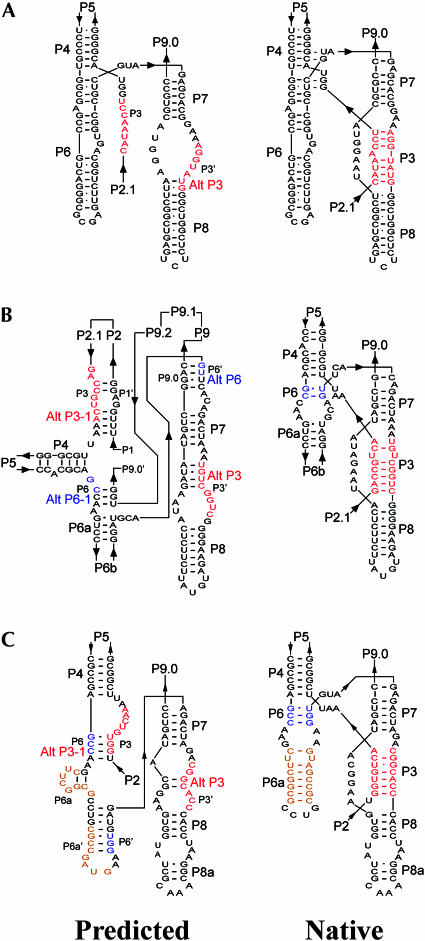
Trapping most of the Tetrahymena ribozyme molecules in misfolded intermediates has been attributed to the much slower folding of the P3-P7 pseudoknot than formation of the loop–loop interactions that tether the overall structure of the ribozyme (Treiber et al. 1998; Pan et al. 2000; Treiber and Williamson 2001b). The presence of the Alt P3 structure serves as a major barrier to forming the P3-P7 pseudoknot of that ribozyme (Pan and Woodson 1998; Pan et al. 2000; Woodson 2002). As a theoretical approach to reveal the structural details accounting for the higher tendency of the Candida ribozyme to fold rapidly into intermediates containing the native P3-P7 pseudoknot, the secondary structure with minimal free energy was predicted using the computer program RNAstructure 3.71 derived from the Mfold program (Zuker 2003). Although the predicted secondary structure does not necessarily represent the native secondary structure of the ribozyme, mispairing in this minimal free energy structure may suggest the tendency of a given large complex RNA to fold into nonnative intermediates.
In the predicted structure of the Candida ribozyme, all the core helices except for P3 are precisely formed (Fig. 5A). No stable Alt P3 structure was predicted. Instead, most of the bases involved in pairing of the native P3 were unpaired, and thus free to readily form correct tertiary interactions upon folding. Only a minimal Alt P3 including two base pairs adjacent to P8 was predicted. Note that the predicted base pairing in P9, P5abc, and part of P2.1 better correlated with our structural data using T1 ribonuclease footprinting; the secondary structure of the Candida ribozyme shown in Figure 1 reflects this modified structure. The minimal mis-pairing of the important helical structures of the Candida ribozyme in the predicted structure explains the smooth transition in rapid folding of most ribozyme molecules to a structure containing native P3-P7 pseudoknot, which is then concertedly converted to the catalytically active structure (Xiao et al. 2003).
FIGURE 5.
Mispairing of some core helices of the Candida ribozyme (A), Tetrahymena ribozyme (B), and Azoarcus ribozyme (C). The core helices in the predicted secondary structures with minimal free energy are shown on the left, and the native secondary structure of the core helices are shown on the right. The regions with difference between these two structures are colored, with each color indicating the same sequence element in the two structures.
In contrast, RNAstructure predicts that the Tetrahymena ribozyme tends to misfold at many regions of the core elements. In addition to Alt P3, the possible Alt P6, Alt P6-1 and Alt P3-1 were also identified (Fig. 5B). The tendency toward severe mispairing of P3 and P6 structural elements correlates with the extremely slow folding of the P3-P7 pseudoknot and the presence of multiple metastable and stable misfolded intermediates responsible for the kinetic traps that hinder correct folding of the ribozyme (Thirumalai et al. 2001; Treiber and Williamson 2001a; Woodson 2002; Onoa et al. 2003). The predicted secondary structure of Azoarcus ribozyme contains the completely misfolded P6 and two alternative P3s that are less stable than those of the Tetrahymena ribozyme (Fig. 5C), explaining the fast folding of about a half of the ribozyme molecules to the native structure (Rangan et al. 2003).
Hence, consistency of base pairings between local secondary structures and tertiary interactions correlates well with the tendency of the large RNA molecules to fold through a trap-free pathway, i.e., cooperative folding of the whole molecule to the active structure. Because of the importance of the P3-P7 pseudoknot in the folding of the catalytically active ribozyme, the tendency of forming mis-paired structure in this region may serve as a criterion in predicting the folding pathway of a particular group I ribozyme in vitro.
DISCUSSION
Extensive study of the Tetrahymena ribozyme has established the hierarchical scheme of the folding of the large multidomain RNA. Ensemble folding experiments demonstrate that P5abc folds first, acting as a nucleation site for rapid folding of the P4-P6 domain. Folding of the P3-P7 core of the ribozyme requires minutes, functioning as a rate-limiting step (Zarrinkar and Williamson 1994; Pan and Woodson 1998; Sclavi et al. 1998; Treiber et al. 1998; Pan et al. 2000; Woodson 2002). Single molecule study and ensemble folding experiments show that the major population of the Tetrahymena ribozyme folds via the kinetically trapped pathway (Rook et al. 1999; Russell and Herschlag 1999; Zhuang et al. 2000; Thirumalai et al. 2001). Hence, the hierarchical folding scheme describes the events in the kinetically trapped pathway. Although kinetic traps and other rate-limiting steps are encountered when this large RNA folds in vitro (Treiber and Williamson 2001a; Sosnick and Pan 2003), folding of the large RNA with multiple domains is presumably cooperative and efficient in vivo with the help of protein factors (Brion and Westhof 1997; Schroeder et al. 2002), ensuring the required functionality of the RNA in living cells. A model RNA that folds ideally in vitro would be essential for elucidating the cooperative folding mechanism of the large multidomain RNA without the complication of the need for protein factors.
Our previous work has demonstrated that most of the Candida ribozyme molecules uniformly fold to the catalytically active structure though a single folding pathway with a rate constant of ~2 min−1 (Xiao et al. 2003), making this an ideal system for studying the mechanism of the rapid folding of a large multidomain RNA.
Cooperative folding of the P3-P7 pseudoknot and the whole ribozyme
We used T1 cleavage analysis and oligonucleotide hybridization experiments to demonstrate that the folding of the P3-P7 pseudoknot of the Candida ribozyme occurs during rapid ribozyme compaction in the presence of an adequate amount of magnesium, coincident with the formation of many other tertiary interactions of the ribozyme. Similar to the prediction of RNAstructure 3.71, the P7 stem readily forms (except for the first G-C pair) in the absence of magnesium, while formation of the P3 stem is not energetically favorable and requires magnesium. We propose that the folding of the P3-P7 pseudoknot and other tertiary structures of the ribozyme are cooperative in order to commit the ribozyme to the trap-free pathway, as opposed to the hierarchical folding scheme used in the folding of the group I ribozyme through the kinetically trapped pathway.
As little as 1.5 mM magnesium fully supports folding of the P3-P7 pseudoknot of the Candida ribozyme. This magnesium requirement is similar to that required for the optimal self-splicing activity of the ribozyme (Miletti and Leibowitz 2000; Zhang and Leibowitz 2001), suggesting that the folding of the native P3-P7 correlates with the natural catalytic activity of the ribozyme. However, this magnesium requirement is much lower than that required for the maximal activity of the first transesterification reaction mimicked by the Ca.L-11 catalyzed substrate cleavage (Xiao et al. 2003), indicating that the tertiary interactions stabilized by high magnesium concentrations accelerate the first transesterification reaction, but hinder the self-splicing reaction, which involves two reactions at a single guanosine binding site (Cech 1990).
Rapid formation of the P3-P7 pseudoknot ensures an effective folding of the catalytically active structure
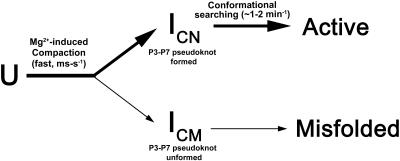
As revealed by the combination of analyses of catalytic activity, gel shift, RNase H, and ribonuclease T1 footprinting with an antisense oligonucleotide, a small fraction of the Candida ribozyme rapidly assumes a collapsed nonnative state containing unfolded P3-P7 structure, which is readily captured by the antisense oligonucleotide and T1 ribonuclease. The major RNA fraction rapidly assumes the collapsed nativelike conformation containing the correct P3-P7 pseudoknot. The native intermediates are uniformly converted to the catalytically active structure with a rate constant of ~2 min−1, while the nonnative intermediates show little tendency to convert to the catalytically active structure (Fig. 6).
FIGURE 6.
Scheme for the folding of the Candida ribozyme.
The P3-P7 pseudoknot constitutes the catalytic center of group I ribozymes. Formation of the pseudoknot involves the assembly of different domains of the group I ribozyme into a well-organized structure capable of catalyzing two consecutive transesterification reactions. Apparently, the P3-P7 pseudoknot functions as a key structure in regulating the folding pathways of the group I ribozymes. In the case of the Tetrahymena ribozyme, some RNA segments involved in the formation of the P3-P7 pseudoknot have a strong tendency to form a local base-paired structure, such as Alt P3 and Alt P3-1, which prevents the formation the P3-P7 pseudoknot. Consequently, the majority of Tetrahymena ribozyme adopts the strategy of hierarchical folding of different domains, in which folding of the P3-P7 core occurs last. This folding scheme is very inefficient in production of the catalytically active ribozyme. For example, about 10%–20% of the Tetrahymena ribozyme folds to the catalytically active structure with rate constants of 1 sec−1 and 1 min−1, while the major population takes up to hours to reach the catalytically active conformation (Russell and Herschlag 1999; Zhuang et al. 2000). In contrast, all the RNA segments involved in formation of the P3-P7 pseudo-knot of the Candida ribozyme are either correctly base paired (P7 strands) or remain unpaired (P3 strands), allowing the rapid formation of the P3-P7 pseudoknot structure during the ribozyme compaction. As a result, the major population of the Candida ribozyme is committed into the native intermediates, converting to the catalytically active structure with a rate constant of 2 min−1 (Xiao et al. 2003; this study). Clearly, rapid formation of the P3-P7 pseudo-knot structure is a productive way of folding the catalytically active structure of group I ribozymes.
These findings also suggest that the tendency for forming the correct P3-P7 pseudoknot is a critical criterion in predicting the folding pathways of different group I ribozymes. Moreover, the tendency for mispairing of some other important local secondary structures, such as the P6 helix, suggests additional energy barriers in folding of the native structures of the active ribozyme.
Because of the difficulty in conversion of the metastable or stable nonnative RNA intermediates to the active structure, preventing the unfolded RNA from entering the kinetically trapped pathway may be the most effective way to guarantee high catalytic activity of large multidomain RNA in vitro and in living cells. For example, Russell et al. (2002b) showed that the Tetrahymena ribozyme prefolded at high concentrations of Na+ greatly enhances catalytic activity. RNA chaperones may also facilitate large RNA folding by blocking entry to the kinetically trapped pathway (Schroeder et al. 2002). Other mechanisms for the effective folding of multidomain RNA to the catalytically active structure are subject to further elucidation.
MATERIALS AND METHODS
RNA preparation and nucleic acid labeling
Ca.L-11 ribozyme RNA and Ca.LSU precursor RNA were synthesized by in vitro transcription and purified as described (Zhang and Leibowitz 2001; Xiao et al. 2003). The precursor RNA preparation was slightly modified. The T7 promoter was placed at the 5′ end of the sense strand, and the resulting precursor RNA is 608 nt containing 129 nt of 5′ exon, 379 nt of intron, and 100 nt of 3′ exon. The T7 polymerase used in this study was from Promega, rNTPs was from Roche and [α-32P] GTP was from PerkinElmer-NEN. The Ca.L-11 RNA, antisense oligonucleotide, and substrate RNA Ca/sub-11 (GCUCUA5) (Dharmacon) were labeled at the 5′ end with [γ-32P] ATP (PerkinElmer-NEN) as described (Xiao et al. 2003). The Ca.LSU precursor RNA was uniformly labeled with [α-32P] GTP (Zhang and Leibowitz 2001).
RNase T1 footprinting
Ca.L-11 ribozyme (~2 ng) labeled at the 5′ end with [γ-32P] ATP was used for T1 analysis as previously described (Xiao et al. 2003). To study the binding sites of the antisense oligonucleotide on the ribozyme, the labeled ribozyme was preincubated with the concentrations of magnesium 10% higher than that indicated and 444 nM of the antisense oligonucleotide in 27 μL of 55.6 mM Tris-HCl (pH 7.5) at 37°C for 20 min, then samples were chilled on ice and ribonuclease T1 (3 μL of 0.5 unit/μL) was added. Hence, final concentrations of magnesium were as indicated, and those of the antisense oligonucleotide and Tris-HCl were 400 nM and 50 mM, respectively. T1 cleavage reactions were run for 1 min at 37°C, and stopped by adding 20 μL phenol-chloroform (1:1) immediately followed by vigorous vortexing and centrifugation sequentially. Supernatant was removed and analyzed as described previously (Xiao et al. 2003).
Trans-cleavage assay
The standard trans-cleavage reaction was performed in 10 μL reactions containing 5 nM of Ca.L-11 ribozyme, 0.1 mM GTP, 5 mM MgCl2, 50 mM Tris-HCl (pH 7.5), and 10 nM of 5′-end radiolabeled substrate, in the presence of varying oligonucleotide concentrations. Cleavage products were analyzed on 15% poly-acrylamide-8 M urea gels that were exposed both to X-ray film and a phosphor screen for visualization and analysis using the variable scanner Typhoon 9200 (Amersham-Pharmacia Biotech), respectively (Xiao et al. 2003). Data were plotted using GraphPad Prism program.
Self-splicing assay
The standard self-splicing reaction was performed in 10 μL reactions and analyzed as described previously except that RNasin was omitted (Zhang and Leibowitz 2001). Signals on X-ray films were analyzed by a Kodak 290 digital camera and 1D 3.5 image analysis software.
Gel-shift analysis
Varying concentrations of the purified Ca.L-11 ribozyme were incubated with 50 nM of radiolabeled oligonucleotide in 10 μL of 50 mM Tris-HCl (pH 7.5) containing the indicated concentrations of magnesium for 20 min at 37°C. Note that Mg2+ and oligo were added together to a RNA solution in Tris-HCl (pH 7.5). All samples were chilled on ice and an equal volume of the loading dye containing 50% glycerol was added, followed by electrophoresis on 5% native polyacrylamide gels in 1× TBE buffer (0.09 M Tris-borate at pH 7.5 and 2 mM EDTA) at ~20 V/cm for about half an hour at room temperature. Gels were then exposed to phosphor screens and analyzed by a Typhoon 9200 variable scanner.
RNase H assay
The 5′-end labeled Ca.L-11 ribozyme was heated to 95°C for 1 min in 10 mM Tris-HCl (pH 7.5), followed by an equilibration for 3 min at 37°C to allow the formation of the secondary structure. Magnesium-induced tertiary folding was started simultaneously with the oligo hybridization by adding folding buffer containing magnesium, P3 → P7 oligo, and RNase H (Zarrinkar and Williamson 1994). The reaction concentration of each component is 5 μM oligo, 50 mM Tris-HCl (pH 7.5), 10 mM MgCl2, 10 mM NaCl, 0.1 mM EDTA, 1 mM dithiothreitol, and 0.4 U/μL of RNase H (Takara, Dalian).
Prediction of the secondary structures of the group I ribozymes
RNAstructure (version 3.71) was used to predict the secondary structures of the Tetrahymena, Candida, and Azoarcus ribozymes containing the full-length intron sequence and a short 5′ exon sequence that forms P1.
Acknowledgments
We thank Michael J. Leibowitz (University of Medicine and Dentistry of New Jersey) and Paul deMena for their critical reading of the manuscript. This work is supported by National Nature Sciences Foundation Committee of China through grants 30040033 and 30170213 and by Wuhan University through grant 0000028 (Y.Z.).
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.7145105.
REFERENCES
- Brion, P. and Westhof, E. 1997. Hierarchy and dynamics of RNA folding. Annu. Rev. Biophys. Biomol. Struct. 26: 113–137. [DOI] [PubMed] [Google Scholar]
- Buchmueller, K.L and Weeks, K.M. 2003. Near native structure in an RNA collapsed state. Biochemistry 42: 13869–13878. [DOI] [PubMed] [Google Scholar]
- Buchmueller, K.L., Webb, A.E., Richardson, D.A., and Weeks, K.M. 2000. A collapsed non-native RNA folding state. Nat. Struct. Biol. 7: 362–366. [DOI] [PubMed] [Google Scholar]
- Cech, T.R. 1990. Self-splicing of group I introns. Annu. Rev. Biochem. 59: 543–568. [DOI] [PubMed] [Google Scholar]
- Go, N. 1984. The consistency principle in protein structure and pathways of folding. Adv. Biophys. 18: 149–164. [DOI] [PubMed] [Google Scholar]
- Golden, B.L., Gooding, A.R., Podell, E.R., and Cech, T.R. 1998. A preorganized active site in the crystal structure of the Tetrahymena ribozyme. Science 282: 259–264. [DOI] [PubMed] [Google Scholar]
- Lehnert, V., Jaeger, L., Michel, F., and Westhof, E. 1996. New loop–loop tertiary interactions in self-splicing introns of subgroup IC and ID: A complete 3D model of the Tetrahymena thermophila ribozyme. Chem. Biol. 3: 993–1009. [DOI] [PubMed] [Google Scholar]
- Massire, C., Jaeger, L., and Westhof, E. 1997. Phylogenetic evidence for a new tertiary interaction in bacterial RNase P RNAs. RNA 3: 553–556. [PMC free article] [PubMed] [Google Scholar]
- Michel, F. and Ferat, J.L. 1995. Structure and activities of group II introns. Annu. Rev. Biochem. 64: 435–461. [DOI] [PubMed] [Google Scholar]
- Michel, F. and Westhof, E. 1990. Modeling of the three-dimensional architecture of group I catalytic introns based on comparative sequence analysis. J. Mol. Biol. 216: 585–610. [DOI] [PubMed] [Google Scholar]
- Miletti, K.E. and Leibowitz, M.J. 2000. Pentamidine inhibition of group I intron splicing in Candida albicans correlates with growth inhibition. Antimicrob. Agents Chemother. 44: 958–966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onoa, B., Dumont, S., Liphardt, J., Smith, S.B., Tinoco Jr., I., and Bustamante, C. 2003. Identifying kinetic barriers to mechanical unfolding of the T. thermophila ribozyme. Science 299: 1892–1895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan, J. and Woodson, S.A. 1998. Folding intermediates of a self-splicing RNA: Mispairing of the catalytic core. J. Mol. Biol. 280: 597–609. [DOI] [PubMed] [Google Scholar]
- Pan, T., Fang, X., and Sosnick, T. 1999. Pathway modulation, circular permutation and rapid RNA folding under kinetic control. J. Mol. Biol. 286: 721–731. [DOI] [PubMed] [Google Scholar]
- Pan, J., Deras, M.L., and Woodson, S.A. 2000. Fast folding of a ribozyme by stabilizing core interactions: Evidence for multiple folding pathways in RNA. J. Mol. Biol. 296: 133–144. [DOI] [PubMed] [Google Scholar]
- Rangan, P., Masquida, B., Westhof, E., and Woodson, S.A. 2003. Assembly of core helices and rapid tertiary folding of a small bacterial group I ribozyme. Proc. Natl. Acad. Sci. 100: 1574–1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rook, S.M., Treiber, D.K., and Williamson, J.R. 1999. An optimal Mg2+ concentration for kinetic folding of the Tetrahymena ribozyme. Proc. Natl. Acad. Sci. 96: 12471–12476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell, R. and Herschlag, D. 1999. New pathways in folding of the Tetrahymena group I RNA enzyme. J. Mol. Biol. 291: 1155–1167. [DOI] [PubMed] [Google Scholar]
- Russell, R., Millett, L.S., Tate, M.W., Kwok, L.W., Nakatani, B., Gruner, S.M., Mochrie, S.G., Pande, V., Dniach, S., Herschlag, D., et al. 2002a. Rapid compaction during RNA folding. Proc. Natl. Acad. Sci. 99: 4266–4271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell, R., Zhuang, X., Babcock, H.P., Millett, I.S., Doniach, S., Chu, S., and Herschlag, D. 2002b. Exploring the folding landscape of a structured RNA. Proc. Natl. Acad. Sci. 99: 155–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder, R., Grossberger, R., Pichler, A., and Waldsich, C. 2002. RNA folding in vivo. Curr. Opin. Struct. Biol. 12: 296–300. [DOI] [PubMed] [Google Scholar]
- Sclavi, B., Sullivan, M., Chance, M.R., Brenowitz, M., and Woodson, S.A. 1998. RNA folding at millisecond intervals by synchrotron hydroxyl radical footprinting. Science 279: 1940–1943. [DOI] [PubMed] [Google Scholar]
- Sosnick, T.R. and Pan, T. 2003. RNA folding: Models and perspectives. Curr. Opin. Struct. Biol. 13: 309–316. [DOI] [PubMed] [Google Scholar]
- Thirumalai, D., Lee, N., Woodson, S.A., and Klimov, D.K. 2001. Early events in RNA folding. Annu. Rev. Phys. Chem. 52: 751–762. [DOI] [PubMed] [Google Scholar]
- Treiber, D.K. and Williamson, J.R. 2001a. Beyond kinetic traps in RNA folding. Curr. Opin. Struct. Biol. 11: 309–314. [DOI] [PubMed] [Google Scholar]
- ———. 2001b. Concerted kinetic folding of a multidomain ribozyme with a disrupted loop-receptor interaction. J. Mol. Biol. 305: 11–21. [DOI] [PubMed] [Google Scholar]
- Treiber, D.K., Rook, M.S., Zarrinkar, P.P., and Williamson, J.R. 1998. Kinetic intermediates trapped by native interactions in RNA folding. Science 279: 1943–1946. [DOI] [PubMed] [Google Scholar]
- Woodson, S.A. 2002. Folding mechanisms of group I ribozymes: Role of stability and contact order. Biochem. Soc. Trans. 30: 1166–1169. [DOI] [PubMed] [Google Scholar]
- Xiao, M., Leibowitz, M.J., and Zhang, Y. 2003. Concerted folding of a Candida ribozyme into the catalytically active structure posterior to a rapid RNA compaction. Nucleic Acids Res. 31: 3901–3908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarrinkar, P.P. and Williamson, J.R. 1994. Kinetic intermediates in RNA folding. Science 265: 918–924. [DOI] [PubMed] [Google Scholar]
- Zhang, Y. and Leibowitz, M.J. 2001. Folding of the group I intron ribozyme from the 26S rRNA gene of Candida albicans. Nucleic Acids Res. 29: 2644–2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuang, X.W., Bartley, L.E., Babcock, H.P., Russell, R., Ha, T., Herschlag, D., and Chu, S. 2000. A single-molecule study of RNA catalysis and folding. Science 288: 2048–2051. [DOI] [PubMed] [Google Scholar]
- Zuker, M. 2003. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 31: 3406–3415. [DOI] [PMC free article] [PubMed] [Google Scholar]