Abstract
We present an efficient method of introducing fluorophore labels at selected locations in a large RNA. The method is based on specific and highly efficient hybridization between a fluorophore-containing DNA oligonucleotide and a modular hairpin loop replacing a functionally unimportant hairpin loop in the RNA. We demonstrate its feasibility using a 255-nucleotide RNA derived from the catalytic domain of RNase P from Bacillus subtilis. Hybridization of the DNA oligonucleotide to the modular hairpin loop minimally perturbs the structure and function of this RNA. This labeling scheme should be applicable in studies of RNA conformational dynamics by ensemble and single molecule fluorescence methods.
Keywords: labeling, hybridization, FRET, single molecule
INTRODUCTION
Site-specific fluorophore labeling of RNA is extremely useful in studies of its structure, function, and dynamics. Fluorescence resonance energy transfer (FRET) (Weiss 2000) can be used to measure structural parameters—distances between two points (Stryer and Haugland 1967; Yang and Soll 1974) and fluctuations—allowing one to establish folding rates, pathways, and free energy landscapes (Ha et al. 1999; Xie 2001; McKinney et al. 2003; Zhuang and Rief 2003; Xie et al. 2004). Fluorescence emission from one or several fluorophores can be used to establish the absolute position of the single molecules with application for DNA (or RNA) sequence mapping (Gordon et al. 2004; Qu et al. 2004). Fluorescence correlation spectroscopy (FCS) has been employed to establish the diffusion constant and hydrodynamic radius to establish the (folded) size of RNA (Bonnet et al. 1998; Pljevaljcic et al. 2004). In all cases, site-specific attachment of fluorophores with specific fluorescence or environment-sensitive properties is a requirement.
Fluorescent labels can be introduced site-specifically by using fluorescent nucleotide analogs through chemical synthesis (Stryer and Haugland 1967; Haughland 2002), covalent attachment of reactive fluorophores to a specific modified base (Yang and Soll 1974), or to a thiophosphate group in the RNA backbone (Ozaki and McLaughlin 1992). Although quite useful for small RNAs (fewer than 100 residues), each of these methods has drawbacks for large RNAs. Fluorescent analog tags can be incorporated into short synthetic sequences, but generally cannot be incorporated by RNA polymerase during transcription. Posttranscriptional modifications are quite useful for systems in which an appropriate labeling site is fortuitously modified in vivo, but cannot be generalized to in vitro transcripts. The covalent coupling reactions are often inefficient, so that substantial effort is required to remove unlabeled RNA.
Several other methods take advantage of the unique reactivity of the RNA termini. The 5′ terminus of a transcript can be selectively labeled during transcription by adding fluorophore-labeled nucleotide analogs into the transcription mixture (Fang et al. 1999). These analogs cannot, however, be incorporated into the middle of a transcript because labeling is specific to the 5′ end. Another approach involves enzymatically functionalizing the termini of a transcript either by reacting the 5′ hydroxyl to a thiophosphate for subsequent coupling to reactive fluorophores or by introducing a labeled nucleotide to the 3′ end (kits are available through Vector Laboratories, Burlingame, CA). Although useful, these approaches can only incorporate fluorophores at the termini of the molecule, which often are not the most desirable labeling sites for the intended study.
A popular way of introducing site-specific RNA modifications or labels is the “Moore-Sharp” method using a DNA oligonucleotide splint and T4 DNA ligase (Moore and Sharp 1992). In principle, this method allows specific fluorophore-labeling at any site in a large RNA. The RNA structure, however, often competes with the hybridization of the DNA oligonucleotide splint so that the labeling efficiency at different sites can be highly variable. For this reason, this method has not been used extensively for biophysical studies where the amount of sample needed is significantly greater than functional studies that require only radioactively labeled RNA.
This work presents a method that efficiently incorporates one or more fluorophores in a large RNA for biophysical studies. The catalytic domain (C-domain) of the Bacillus subtilis RNase P, an independently folding ribozyme consisting of 255 nt (Fang et al. 1999, 2002; Xie et al. 2004) is chosen as a model system. As shown in Figure 1A, the approach involves enlarging a native hairpin loop by inserting 10 nt to allow specific hybridization of a DNA oligo-nucleotide of appropriate sequence. A particular loop sequence results in specific and very efficient hybridization of its complementary DNA oligonucleotide. Proper folding of the ribozyme is demonstrated through footprinting, catalytic activity, and FRET signals.
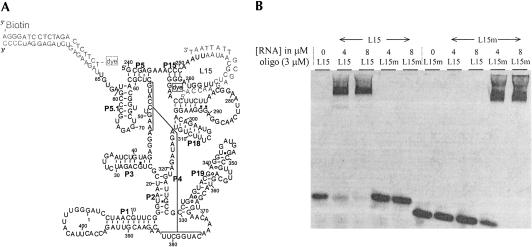
FIGURE 1.
(A) Secondary structure representation of the C-domain with the enlarged L15 loop (L15-RNA). The complete loop sequence is 5′-AAAUUAAUAACGCUCUUGGUA, and the sequence of the complementary DNA oligonucleotide is 5′-Alexa488-ACCAAGAGCGTTAT TAAT. A second fluorophore can be attached through hybridization of a 21-mer DNA oligonucleotide containing Cy3 and biotin to the 3′ extension. The enlarged L15 loop (residues 247–264), which includes the three CG base pairs, is termed the “L15-module” in this work. (B) Hybridization specificity demonstrated by nondenaturing gel electrophoresis. The sequence of the L15m loop is 5′-AAAUAUAUUUCGGACUACGUA (differences between L15 and L15m sequences are underlined), and the complementary DNA oligonucleotide is 5′ Alexa488-ACGTAGTCCGAAATATAT. The oligonucleotide concentration was fixed at 3 μM and the RNA concentrations varied between 4 and 8 μM. The gel was imaged by fluorescence detection of the oligonucleotides. The RNA–DNA hybrid is shifted relative to free oligonucleotide.
RESULTS AND DISCUSSION
DNA oligonucleotide hybridization to RNA is an efficient means of attaching a fluorophore to a specific location in an RNA (Zhuang et al. 2000; Liphardt et al. 2001; Xie et al. 2004). All applications, however, have relied on oligo-nucleotide hybridization to extensions to the 5′ or 3′ termini of the RNA, or cotranscriptional incorporation of a modified nucleotide at the 5′ end. Incorporating a fluorophore at an internal position using this strategy may be achieved by hybridizing the 5′ or 3′ fluorophore-containing DNA oligonucleotide to the loop of an RNA hairpin that is not involved in the formation of the tertiary structure.
Extensive studies on the tertiary folding properties of the C-domain of RNase P from B. Subtilis conclude that five hairpin loops, L15, L18, L19, L1, and L3 (Fig. 1) are not directly involved in the formation of the tertiary structure (Haas et al. 1994; Massire et al. 1998; Fang et al. 2001). With respect to length, only L1 among these five loops is large enough to complement a DNA oligonucleotide with sufficient stability for effective labeling, i.e., the DNA–RNA helix should be retained under all ionic conditions used in folding studies. When an 18-mer DNA oligonucleotide complementary to residues 396–409 + 1–4 (L1) is used, however, <5% of this DNA oligonucleotide hybridizes to the RNA as assayed by nondenaturing gel electrophoresis (data not shown). Previous chemical modification studies have shown that the L1 loop alone is somewhat structured (Odell et al. 1998), which offers an explanation for inefficient hybridization.
To test an alternative loop sequence for efficient hybridization, 10 additional nucleotides are inserted into the loop L15 (termed L15-RNA, Fig. 1A). This insertion sequence is chosen to minimize overlap and complementarity with the rest of the C-domain sequence. For sequence specificity the hybrid duplex should have 12 or more base pairs. Our 18-mer overshoots this lower limit to enhance specificity and stability (Kd for this 18 base pair duplex is calculated to be ~10−13 M in 1 M NaCl at 25°C) (Sugimoto et al. 1995). Hybridization efficiency is tested by mixing L15-RNA and a fluorescently labeled DNA oligonucleotide complementary to 18 of the 21 nt in the loop at 50 mM NaCl, which is the weakest ionic condition used in the present study. Hybridization products, detected by fluorescence, are shifted relative to the free oligonucleotides on nondenaturing gels. Hybridization proceeds nearly to completion at similar molar ratio of RNA to DNA oligonucleotide (Fig. 1B).
To test hybridization specificity, L15-RNA is mutated by eight sequence changes in the enlarged L15 loop and the mutant RNA (L15m-RNA) hybridized with DNA oligonucleotides complementary to the original or to the mutated L15 loop sequence (Fig. 1B). These eight substitutions are chosen to conserve the GC-content in the hybrid region and are the only sequence differences between L15-RNA and L15m-RNA. In this case, no crosshybridization is detected and hybridization between two RNAs, and their respective complementary oligonucleotides is preserved. The L15 oligonucleotide is shifted only in the presence of the L15-RNA, so is the L15m oligonucleotide and the L15m-RNA. The lack of crosshybridization also suggests that the DNA oligonucleotide does not bind elsewhere in the C-domain.
Mg2+ and Pb2+ cleavage studies are performed to further demonstrate localization of the DNA oligonucleotide in the L15-RNA (Fig. 2A). A significant fraction of the enlarged L15 loop is cleaved by Mg2+ (lane 1) and by Pb2+ (lane 3) in the absence of DNA oligonucleotide suggesting that this loop by itself does not form significant structures. Cleavage by Mg2+ and Pb2+ is considerably diminished in the presence of the DNA oligonucleotide (lanes 2,4) indicating that the DNA oligonucleotide binds to this loop region. Cleavage patterns show that most, if not all 18 nt are base-paired in the loop and the P15 stem remains intact. This result suggests that no significant strain is introduced into the stem of the hairpin by the RNA–DNA duplex in the loop.
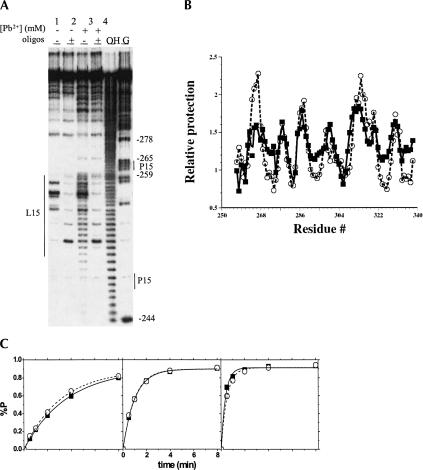
FIGURE 2.
(A) Detection of DNA oligonucleotide hybridization to the enlarged L15 region in L15-RNA by Mg2+ (left two lanes) and Pb2+ cleavage (middle two lanes). Reactions were carried out at 37°C, 25 mM MOPS, pH 7.0, 10 mM Mg2+, 4 μM RNA, 16 μM each of two DNA oligonucleotides complementary to L15 and the 3′ extension, and 0 (-) or 0.15 mM Pb(OAc)2 (+). Lanes marked OH— and G represent L15–RNA subjected to base hydrolysis and nuclease T1 digest, respectively. The 18-nucleotide region complementary to the DNA oligo is indicated by a thin line on the left. The P15 stem is indicated by two thin lines on the right. (B) Comparable tertiary folding of the L15–RNA in the presence (squares and solid line) and absence (circles and dashed line) of the L15 DNA oligonucleotide measured by hydroxyl radical protection. (C) Catalytic activity using an in vitro selected substrate under single turnover conditions in the absence (filled squares) and presence (open circles) of the L15 DNA oligo-nucleotide. The ribozyme concentration was 0.08 μM, 0.4 μM, and 2 μM from left to right.
Tertiary folding of the L15-RNA hybridized with DNA oligonucleotide is compared to the wild-type C-domain by hydroxyl radical protection (Fig. 2B) and catalytic activity using an in vitro selected substrate (Fig. 2C; Pan and Jakacka 1996). Hydroxyl radical footprinting determines the location of nucleotides and the fraction of the RNA protected against solvent exposure, and is an indicator of tertiary contacts present in the RNA structure (Celander and Cech 1991; Cate et al. 1996). Both L15 RNA and the wild-type C-domain have the same protection pattern, suggesting that they have very similar, if not identical tertiary structures. Quantitatively, the extent of protection is somewhat different for each RNA; such differences have also been observed when comparing the protection between two homologous, naturally occurring C-domains (Fang et al. 2001). Hybridization of the 18-mer DNA oligonucleotide to the L15 loop does not affect catalytic efficiency (Fig. 2C). These results support the conclusion that hybridization of the DNA oligonucleotide to this extended loop, termed “L15 module,” does not interfere with tertiary folding in a significant manner.
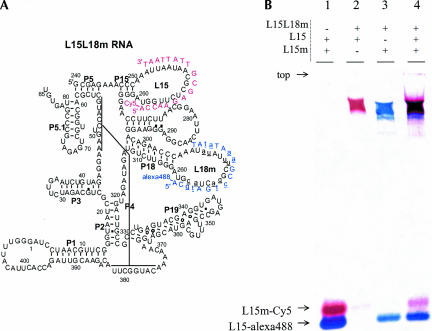
The utility of this 27-nt hairpin loop as a module that allows efficient fluorophore labeling to other loop regions in the C-domain is explored (Fig. 3). Besides L15, four other hairpin loops in this ribozyme, L18, L19, L1, and L3, are not involved in tertiary folding. These loops are therefore candidates for replacement with the “L15 module.” Two such 27-nt modules are available, the original “L15-module” (cccAAAUUAAUAACGCUCUUGGUAggg), and its octal mutant “L15m-module” (cccAAAUAUAUUUCGGACUA CGUAggg). The L15 and L15m modules may be inserted simultaneously into the same RNA. Figure 3A shows a C-domain construct that includes the L15 labeling loop, and in which L18 is replaced by the L15m labeling loop (L18m). Figure 3B shows that this construct hybridizes to both L15 and L15m complementary DNA oligonucleotides. Because of the previously demonstrated specificity from constructs containing a singly modified L15 loop (Fig. 1B), hybridization to this doubly modified C-domain is implicitly specific. Hybridization of L15L18m RNA to both oligonucleotides (lane 4) proceeds with the same efficiency as the individual hybridization events (lanes 2 and 3), indicating that simultaneous hybridization reactions do not interfere with one another.
FIGURE 3.
(A) Secondary structure representation of a C-domain construct containing both L15 and L15m modules in the L15 and L18 loops. (B) Hybridization of two fluorescently labeled oligonucleotides to this C-domain with two modified loops. The RNA concentration is 10 μM and the DNA oligonucleotide concentrations are 5 μM each. Hybridization of both oligo-nucleotides to the RNA proceeds with the same efficiency as each separate hybridization reaction.
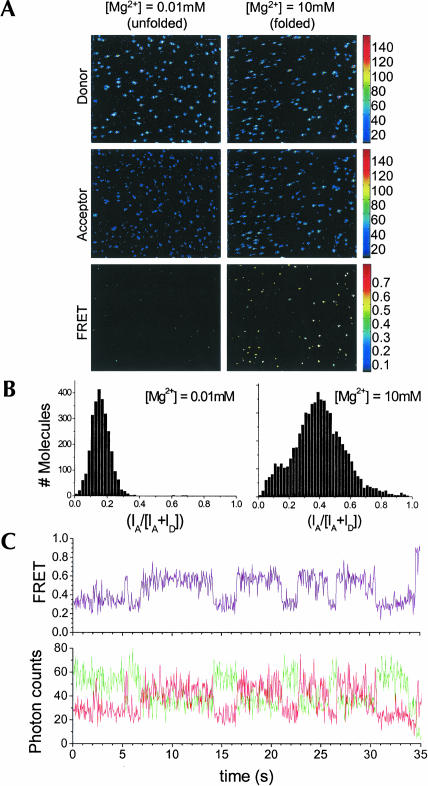
This labeling method allows convenient positioning of fluorophores for single molecule FRET studies of conformational changes and fluctuations of the catalytic domain. Here, L15-RNA is labeled through hybridization to two DNA oligonucleotides: an Alexa 488-labeled oligonucleo-tide to the L15 module, and a biotinylated and Cy3-labeled oligonucleotide to the 21-mer 3′ extension (Fig. 1A). This molecule is immobilized on a BSA-streptavidin functionalized surface (Ha et al. 1999) and examined under unfolded (0.01 mM Mg2+) and folded (10 mM Mg2+) conditions (Fig. 4A). Fluorescence intensity is spectrally separated into donor and acceptor channels in order to calculate the FRET efficiency for each individual molecule. The efficiency of energy transfer, EFRET, is defined as the fraction of total fluorescence due to the acceptor fluorophore: EFRET = IA/(ID + IA), where IA is the intensity of the acceptor fluorescence. In the unfolded form at low [Mg2+], the two dyes are largely separated in this RNA; the distance-dependent FRET signal is at a minimum and fluorescence is detected predominantly from the donor. In the folded form at high [Mg2+], the two dyes are closer, allowing more efficient energy transfer from donor to acceptor by means of Förster dipole–dipole coupling (Weiss 2000). Fluorescence emission is detected from both donor and acceptor fluorophores. Consistent with these images, histograms of FRET efficiencies at low and high [Mg2+] show that the mean FRET values increase from 0.17 to 0.40 (Fig. 4B). A sample equilibrium time trajectory of the donor and acceptor fluorescence at an intermediate Mg2+ concentration shows distinctly anticorrelated signal changes in the two channels, a definitive signature of FRET (Fig. 4C). This result indicates that individual molecules labeled by means of the L15 module are readily applicable to study conformational fluctuations over time.
FIGURE 4.
Single molecule measurements of the L15–RNA. The RNA is hybridized to the 18-mer L15 DNA oligonucleotide containing FRET donor and the 21-mer DNA oligonucleotide containing FRET acceptor and biotin complementary to the extension at the 3′ end. The RNA is immobilized on a coverslip. Both images (A) and histograms (B) under unfolded (0.01 mM Mg2+) and folded (10 mM Mg2+) conditions are shown. (C) Time trajectory for an individual molecule at an intermediate Mg2+ concentration showing anticorrelation of the donor and acceptor fluorescence.
In conclusion, this work demonstrates specific and highly efficient hybridization of labeled DNA oligonucleotides to insertion sequences within a large RNA molecule. For the C-domain, these loop extensions and hybridized DNA segments minimally perturb its folded structure. It is shown that two modular hairpin loops can be used to replace other loops in a wild-type sequence. This “plug-and-play” method of fluorescent labeling should allow easy search through possible labeling positions for structural, functional, and dynamic studies of large RNAs that contain structurally or functionally unimportant hairpin loops.
MATERIALS AND METHODS
RNA constructs
The catalytic domain of the B. Subtilis RNase P RNA was modified by inserting 10 nt into the L15 loop between residues 255 and 256 (these insertion nucleotides are numbered 255a–j) to produce the L15-RNA. For FRET studies, the 3′ end of the C-domain was extended by 21 nt. RNA transcripts were obtained by using standard in vitro transcription protocols with T7 RNA polymerase (Milligan and Uhlenbeck 1989) using Sma I cut plasmid DNA as the transcription template. RNA transcripts were purified on denaturing polyacrylamide gels containing 7 M urea, extracted from the gel by crush-and-soak in 50 mM KOAc/200 mM KCl, pH 7, precipitated in ethanol and stored in water at −20° C.
Hybridization of DNA oligonucleotides
RNA was renatured by heating at 85°C to 90°C for 2 min in Tris-HCl (pH 8.1) buffer alone, followed by equilibration for 3 min at room temperature. NaCl was added and the sample heated at 37°C for 5 min. DNA oligonucleotides were added and the solution incubated at 37°C for 30 more minutes to complete hybridization. The hybridization conditions were 20 mM Tris-HCl, pH 8.1, 50 mM NaCl, 2–16 μM RNA and DNA oligonucleotides. Hybridization products for Figures 1B and 3B were imaged by fluorescence detection on a Storm Phosphorimager (Molecular Dynamics).
Pb2+ Cleavage and hydroxyl radical protection
5′ 32P-labeled RNA was renatured by heating at 85°C to 90°C for 2 min in MOPS buffer followed by incubation at room temperature for 3 min. MgCl2 was added to a final concentration of 10 mM and the mixture was incubated for 10 min at 37°C. To this prefolded RNA at 4 μM, complementary DNA oligonucleotide was added to 16 μM and the hybridization reaction allowed to proceed for 30 min at 37°C. Pb2+ cleavage was performed at 37°C in 25 mM MOPS, pH 7.0, 10 mM MgCl2, 0.15 mM Pb(OAc)2 for 60 min. The reaction was stopped by the addition of an equal volume of 10 M Urea and 100 mM EDTA, separated on denaturing polyacrylamide gels and imaged with a Fuji phosphorimager.
RNA prepared as described above (4 μM RNA and 16 μM oligonucleotides) were subjected to Fe(II)-EDTA footprinting as described previously (Pan 1995) in 20 mM trisHCl, pH 7.5, 10 mM MgCl2, 1 mM Fe(NH4)2(SO4)2 and 1.2 mM EDTA, at 37°C for 30 min. The reaction mixture was separated on denaturing polyacrylamide gel containing 7 M urea and the fraction products quantified with a Fuji phosphorimager.
Catalytic activity
L15 RNA hybridized to the 3′ extension oligonucleotide in the absence or presence of L15 oligonucleotide was prepared as described above in 50 mM trisHCl, pH 8, and 25 mM MgCl2. Low amount (<5 nM) of the 5′ 32P-labeled substrate was renatured as described (Pan and Jakacka 1996). The cleavage reaction was initiated by mixing an equal volume of renatured ribozyme and substrate at a final ribozyme concentration of 0.08, 0.4, and 2 μM. The final reaction condition was 50 mM Tris HCl, pH 8, 25 mM MgCl2 and 1 mM spermine. The reaction was incubated at 37°C and aliquots at varying time points were mixed with an equal volume of 9 M urea and 100 mM EDTA to stop the cleavage reaction. The mixture was separated on denaturing polyacrylamide gels and the percent product quantified by phosphorimaging.
Single molecule imaging
L15 RNA hybridized to the L15 and 3′ oligonucleotides was purified on 12% nondenaturing gels, extracted by the crush and soak method, and immobilized on a coverslip prepared with biotinylated BSA and streptavidin surface (Ha et al. 1999). Individual molecules were visualized with an epifluorescence inverted microscope (Xie et al. 2004). The donor and acceptor emission signals were separated by a set of dichroic mirrors and band pass filters. To enhance the lifetime of the fluorophores, the imaging buffer (20 mM Tris, 50 mM NaCl, 0.01 mM Mg2+) also contained 5%β-mercaptoethanol.
Acknowledgments
This work was supported by a grant from the NIH (GM067961). We thank Dr. B. Golden for cloning the L15-RNA plasmid.
Abbreviations
C-domain, the catalytic domain of the B. subtilis RNase P RNA consisting of residues 240–409 + 1–85
FRET, fluorescence resonance energy transfer
L15-RNA, C-domain containing 10 nucleotides insertion in the L15 loop and a 21-nt extension 3′ to residue #85
L15m-RNA, L15-RNA with eight nucleotides changes in the enlarged L15 loop
L15-module, cccAAAUUAAUAACGCUCUUGGUAggg
L15m-module, cccAAAUAUAUUUCGGACUACGUAggg
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.7180305.
REFERENCES
- Bonnet, G., Krichevsky, O., and Libchaber, A. 1998. Kinetics of conformational fluctuations in DNA hairpin-loops. Proc. Natl. Acad. Sci. 95: 602–8606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cate, J.H., Gooding, A.R., Podell, E., Zhou, K., Golden, B.L., Kundrot, C.E., Cech, T.R., and Doudna, J.A. 1996. Crystal structure of a group I ribozyme domain: Principles of RNA packing. Science 273: 1678–1685. [DOI] [PubMed] [Google Scholar]
- Celander, D.W. and Cech, T.R. 1991. Visualizing the higher order folding of a catalytic RNA molecule. Science 251: 401–407. [DOI] [PubMed] [Google Scholar]
- Fang, X., Pan, T., and Sosnick, T.R. 1999. A thermodynamic framework and cooperativity in the tertiary folding of a Mg2+-dependent ribozyme. Biochemistry 38: 16840–16846. [DOI] [PubMed] [Google Scholar]
- Fang, X.W., Golden, B.L., Littrell, K., Shelton, V., Thiyagarajan, P., Pan, T., and Sosnick, T.R. 2001. The thermodynamic origin of the stability of a thermophilic ribozyme. Proc. Natl. Acad. Sci. 98: 4355–4360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang, X.W., Thiyagarajan, P., Sosnick, T.R., and Pan, T. 2002. The rate-limiting step in the folding of a large ribozyme without kinetic traps. Proc. Natl. Acad. Sci. 99: 8518–8523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon, M.P., Ha, T., and Selvin, P.R. 2004. Single-molecule high-resolution imaging with photobleaching. Proc. Natl. Acad. Sci. 101: 6462–6465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha, T., Zhuang, X., Kim, H.D., Orr, J.W., Williamson, J.R., and Chu, S. 1999. Ligand-induced conformational changes observed in single RNA molecules. Proc. Natl. Acad. Sci. 96: 9077–9082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas, E.S., Brown, J.W., Pitulle, C., and Pace, N.R. 1994. Further perspective on the catalytic core and secondary structure of ribonuclease P RNA. Proc. Natl. Acad. Sci. 91: 2527–2531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haughland, R.P. 2002. Handbook of fluorescent probes and research products. Molecular Probes, Eugene, OR.
- Liphardt, J., Onoa, B., Smith, S.B., Tinoco, I.J., and Bustamante, C. 2001. Reversible unfolding of single RNA molecules by mechanical force. Science 292: 733–737. [DOI] [PubMed] [Google Scholar]
- Massire, C., Jaeger, L., and Westhof, E. 1998. Derivation of the three-dimensional architecture of bacterial ribonuclease P RNAs from comparative sequence analysis. J. Mol. Biol. 279: 773–793. [DOI] [PubMed] [Google Scholar]
- McKinney, S.A., Declais, A.C., Lilley, D.M., and Ha, T. 2003. Structural dynamics of individual Holliday junctions. Nat. Struct. Biol. 10: 93–97. [DOI] [PubMed] [Google Scholar]
- Milligan, J.F. and Uhlenbeck, O.C. 1989. Synthesis of small RNAs using T7 RNA polymerase. Methods Enzymol. 180: 51–62. [DOI] [PubMed] [Google Scholar]
- Moore, M.J. and Sharp, P.A. 1992. Site-specific modification of pre-mRNA: The 2′-hydroxyl groups at the splice sites. Science 256: 992–997. [DOI] [PubMed] [Google Scholar]
- Odell, L., Huang, V., Jakacka, M., and Pan, T. 1998. Interaction of structural modules in substrate binding by the ribozyme from Bacillus subtilis RNase P. Nucleic Acids Res. 26: 3717–3723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozaki, H. and McLaughlin, L.W. 1992. The estimation of distances between specific backbone-labeled sites in DNA using fluorescence resonance energy transfer. Nucleic Acids Res. 20: 5205–5214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan, T. 1995. Higher order folding and domain analysis of the ribozyme from Bacillus subtilis ribonuclease P. Biochemistry 34: 902–909. [DOI] [PubMed] [Google Scholar]
- Pan, T. and Jakacka, M. 1996. Multiple substrate binding sites in the ribozyme from Bacillus subtilis RNase P. EMBO J. 15: 2249–2255. [PMC free article] [PubMed] [Google Scholar]
- Pljevaljcic, G., Millar, D.P., and Deniz, A.A. 2004. Freely diffusing single hairpin ribozymes provide insights into the role of secondary structure and partially folded states in RNA folding. Biophys. J. 87: 457–467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu, X., Wu, D., Mets, L., and Scherer, N.F. 2004. Nanometer-localized multiple single-molecule fluorescence microscopy. Proc. Natl. Acad. Sci. 101: 11298–11303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stryer, L. and Haugland, R.P. 1967. Energy transfer: A spectroscopic ruler. Proc. Natl. Acad. Sci. 58: 719–726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugimoto, N., Nakano, S., Katoh, M., Matsumura, A., Nakamuta, H., Ohmichi, T., Yoneyama, M., and Sasaki, M. 1995. Thermodynamic parameters to predict stability of RNA/DNA hybrid duplexes. Biochemistry 34: 11211–11216. [DOI] [PubMed] [Google Scholar]
- Weiss, S. 2000. Measuring conformational dynamics of biomolecules by single molecule fluorescence spectroscopy. Nat. Struct. Biol. 7: 724–729. [DOI] [PubMed] [Google Scholar]
- Xie, X.S. 2001. Single molecule approach to enzymology. Single Mol. 2: 229–236. [Google Scholar]
- Xie, Z., Srividya, N., Sosnick, M.T.R., Pan, T., and Scherer, N.F. 2004. Single-molecule studies highlight conformational heterogeneity in the early folding steps of a large ribozyme. Proc. Natl. Acad. Sci. 101: 534–539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, C.H. and Soll, D. 1974. Studies of transfer RNA tertiary structure of singlet-singlet energy transfer. Proc. Natl. Acad. Sci. 71: 2838–2842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuang, X. and Rief, M. 2003. Single-molecule folding. Curr. Opin. Struct. Biol. 13: 88–97. [DOI] [PubMed] [Google Scholar]
- Zhuang, X., Bartley, L.E., Babcock, H.P., Russell, R., Ha, T., Herschlag, D., and Chu, S. 2000. A single-molecule study of RNA catalysis and folding. Science 288: 2048–2051. [DOI] [PubMed] [Google Scholar]