Abstract
The A3243G mutation within the human mitochondrial (hs mt) tRNALeu(UUR) gene is associated with maternally inherited deafness and diabetes (MIDD) and other mitochondrial encephalopathies. One of the most pronounced structural effects of this mutation is the disruption of the native structure through stabilization of a high-affinity dimeric complex. We conducted a series of studies that address the structural properties of this tRNA dimer, and we assessed its formation under physiological conditions. Enzymatic probing was used to directly define the dimeric interface for the complex, and a discrete region of the D-stem and loop of hs mt tRNALeu(UUR) was identified. The dependence of dimerization on magnesium ions and temperature was also tested. The formation of the tRNA dimer is influenced by temperature, with dimerization becoming more efficient at physiological temperature. Complexation of the mutant tRNA is also affected by the amount of magnesium present, and occurs at concentrations present intracellularly. Terbium probing experiments revealed a specific metal ion-binding site localized at the site of the A3243G mutation that is unique to the dimer structure. This metal ion-binding site presents a striking parallel to dimeric complexes of viral RNAs, which use the same hexanucleotide sequence for complexation and feature a similarly positioned metal ion-binding site within the dimeric structure. Taken together, these results indicate that the unique dimeric complex formed by the hs mt tRNALeu(UUR) A3243G mutant exhibits interesting similarities to biological RNA dimers, and may play a role in the loss of function caused by this mutation in vivo.
Keywords: tRNA structure, tRNA dimer, enzymatic probing, metal ion binding
INTRODUCTION
Over the past 15 years, clinical studies have linked a variety of degenerative diseases to mutations within the human mitochondrial (hs mt) genome (Schon et al. 1997). Surprisingly, the pathogenic mutations that lead to these diseases are predominantly located in genes that encode transfer RNAs (tRNAs). Considering that these genes represent a small fraction of the hs mt genome, this is a significant finding, indicating that hs mt tRNA function is highly sensitive to point mutations. Pathogenic mutations have been discovered in nearly all hs mt tRNAs (the sole exception being hs mt tRNAArg) and are dispersed throughout all domains of the secondary structure (Fig. 1A ▶; http://www.mitomap.org). Hs mt tRNAs may be particularly susceptible to deactivating mutations because their structures exhibit low thermodynamic stability (Helm et al. 2000).
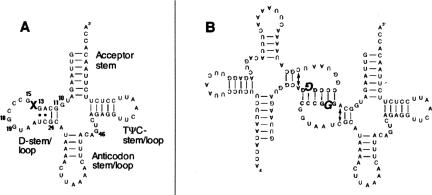
FIGURE 1.
Secondary structure of hs mt tRNALeu(UUR). (A) Cloverleaf structure of hs mt tRNALeu(UUR). The “X” designates the location of the A3243G (A14G) mutation. (B) Schematic representation of the dimeric complex formed by the pathogenic A3243G hs mt tRNALeu(UUR) mutant. The mutated base is shown in bold italics.
The hs mt tRNALeu(UUR) gene contains 20 of the 92 documented pathogenic tRNA mutations and therefore represents a hotspot where many disease-related base substitutions occur. The A3243G mutation within hs mt tRNALeu(UUR) is one of the most prevalent pathogenic mutations and is associated with maternally inherited diabetes and deafness (MIDD) and mitochondrial myopathy, encephalopathy, lactic acidosis and stroke-like episodes (MELAS) (Goto et al. 1990; van den Ouweland et al. 1992; Kadowaki et al. 1994; Maechler and Wollheim 2001; Maassen 2002). The A3243G mutation disrupts the conserved tertiary interaction between A14 and U8, an important contact that serves to stabilize the L-shaped tertiary fold. Position 14 is highly conserved (> 90%) as an adenine in tRNAs (Sprinzl et al. 1998), which underscores the structural importance of this nucleotide. Thus, the A14G mutant may not function correctly with tRNA processing and maturation enzymes, and accordingly, defects have been observed in a variety of biochemical pathways (Jacobs and Holt 2000; Florentz 2002; Wittenhagen et al. 2003). Cellular studies investigating the A3243G mutant have revealed deficiencies in RNA processing (Masucci and Schon 1996; Rossmanith and Karwan 1998; Koga et al. 2003), aminoacylation (El Meziane et al. 1998; Janssen et al. 1999; Borner et al. 2000; Chomyn et al. 2000; Wittenhagen and Kelley 2002; Park et al. 2003), post-transcriptional tRNA modification (Helm et al. 1999; Yasukawa et al. 2000), and translation (Chomyn et al. 1992; King et al. 1992; Schon et al. 1992; Flierl et al. 1997).
The A3243G mutation also induces a profound change in tRNA quaternary structure (Wittenhagen and Kelley 2002). The mutation promotes the formation of a dimeric complex through the introduction of a palindromic hexanucleotide sequence of 5′-GGGCCC-3′ within the D stem/loop. This single base change facilitates dimer formation (Fig. 1B ▶), whereas the wild-type (WT) sequence (5′-GAGCCC-3′) does not possess the self-complementary hexamer required for complexation. The formation of the dimer and the disruption of U8:A14 base pair both contribute to the significantly attenuated aminoacylation rates observed for the A3243G mutant.
Dimerization or aggregation of naturally occurring tRNAs in vitro is a known phenomenon, but typically occurs under nonphysiological conditions (Loehr and Keller 1968; Yang et al. 1972; Kholod 1999; Madore et al. 1999). Unmodified tRNA transcripts, as well as mature tRNAs, can undergo complexation at low temperatures or with high salt concentrations (Kholod 1999). The aminoacylation of tRNA dimers has been investigated and shown to be inefficient or completely eliminated due to the loss of synthetase identity elements upon aggregation (Schleich and Goldstein 1964; Zachau 1968; Hampel et al. 1971).
Dimerization of RNA also occurs in vivo and is an essential step in viral replication (Panganiban and Fiore 1988). The genomic RNA of retroviruses is packaged as a dimeric complex held together by short self-complementary sequences, referred to as dimerization initiation sites (DISs) (Paillart et al. 1994; Skripkin et al. 1994; Laughrea and Jette 1996). Structural studies of the DISs of various retroviral RNAs have shown that stable dimers are formed by base-pairing interactions occurring between the DIS nucleotides displayed at the ends of helical segments (Fosse et al. 1996; Muriaux et al. 1996; Ennifar et al. 2001). Extensive local or global reorganization of the sequence does not occur in these complexes; instead, a simple hybridization of a short stretch of looped nucleotides in each molecule stabilizes the genomic dimer.
Here, we describe structural analyses of a dimeric complex of the A3243G mutant of hs mt tRNALeu(UUR). Dimerization is dependent on both temperature and magnesium concentration, but is an efficient process under physiological conditions. Enzymatic probing studies confirm that the D stem and loop represent the dimeric interface, and that a global structural rearrangement is not involved in complexation. Using terbium cleavage, the presence of a metal-binding site at the mutated position within the dimeric complex was detected that is not present in the native structure. The results described suggest interesting similarities between the structure of this hs mt tRNALeu(UUR) dimer and complexes formed by viral RNAs.
RESULTS AND DISCUSSION
Previous studies investigating the A3243G hs mt tRNALeu(UUR) dimer revealed that the complex is strictly dependent on the D stem/loop nucleotides 13–17a (Fig. 1A ▶; Wittenhagen and Kelley 2002). These nucleotides were identified using an oligonucleotide interference assay and through mutational studies where the dimerization interface within the tRNA mutant was systematically modified. Given that the complexation of A3243G hs mt tRNALeu(UUR) may relate to the pathogenic effects of this mutation, more direct information concerning the properties of the aberrant structure was desired. A series of experiments were therefore conducted to investigate the extent of structural rearrangement within the dimer, and to investigate the role of metal ions and temperature in facilitating complexation.
Enzymatic probing
To obtain direct information about the structure of the A3243G dimer, a series of enzymatic probing experiments were undertaken. For these studies, enzymes that cleave unpaired nucleotides in the tRNA structure were used in order to identify the region(s) of the hs mt tRNALeu(UUR) A3243G mutant involved in dimerization and to assess the overall integrity of the structure. Previous enzymatic probing studies of the wild-type hs mt tRNALeu(UUR) structure revealed several unstructured regions including the anticodon and D stem, but supported the overall predicted secondary structure and the formation of a folded tertiary structure (Sohm et al. 2003; Roy et al. 2004). We therefore conducted enzyme probing experiments with the goal of identifying differences in the structure of the A3243G mutant. The two enzymes used were nuclease S1 and RNase T2, which are both specific for single-stranded RNA. The selection of the enzymes was based on the goal of identifying regions of the A3243G mutant tRNA that exhibited a different type of structure than that of the wild-type sequence.
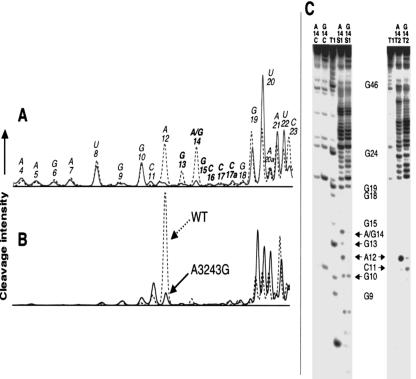
Cleavage of the A3243G tRNALeu(UUR) dimer using nuclease S1 (Fig. 2A ▶) and RNase T2 (Fig. 2B ▶) revealed a loss of enzyme accessibility within the predicted dimer interface. This loss of accessibility indicates that the nucleotides in this region are incorporated into double-stranded structures. Results obtained from polyacrylamide gel electrophoresis (PAGE) analysis of the cleavage reactions (Fig. 2C ▶) were used to generate normalized histograms (Fig. 2A,B ▶) representing cleavage intensity as a function of nucleotide position in each tRNA sample. Relative to the wild-type structure, significant losses of cleavage for the mutant are observed at positions 12–14 with nuclease S1 and position 12 with RNase T2 (a very pronounced cleavage site in the wild-type tRNA). Nucleotides 11 and 19–22, which flank the dimeric interface of the mutant, show an increase in susceptibility toward the probes for the A3243G tRNA, likely because of the loss of the conserved U8:A14 tertiary interaction. These findings indicate that the dimeric complex involves the hybridization of a self-complementary hexamer, which is favored in the presence of pathogenic A3243G mutation. This hybridization event does not appear to have an effect on the global structure of the tRNA itself, as the cleavage patterns obtained from the probing data are comparable in areas of wild-type and mutant A3243G hs mt tRNALeu(UUR) outside of the proposed dimeric interface. There are small changes in the levels of cleavage in other regions of the tRNA structure (e.g., the anticodon stem), but the variation is much less pronounced than that observed for the nucleotides discussed above, and the general pattern of cleavage is similar for the wild-type and mutated tRNA. It is noteworthy that there is a low, but reproducible, level of spontaneous cleavage at certain sites (e.g., U20) in the mutant tRNA that is not observed with the wild-type tRNA (Fig. 2C ▶).
FIGURE 2.
Enzymatic probing of the wild-type (WT) and mutant A3243G hs mt tRNALeu(UUR). (A) Normalized histograms generated from PAGE analysis of enzymatic cleavage of wild-type (dashed) and mutant A3243G (solid) hs mt tRNALeu(UUR) using nuclease S1 under nondenaturing conditions. Positions are numbered according to the consensus tRNA numbering system, and bold nucleotides are located within the proposed dimer interface. (B) Histograms generated from PAGE analysis of enzymatic cleavage of wild-type (dashed) and mutant A3243G (solid) hs mt tRNALeu(UUR) using RNase T2. (C) 20% denaturing PAGE of nuclease S1 and RNase T2 structural probing experiments. Sample lanes marked A14 represent probing of the wild-type hs mt tRNALeu(UUR), while those labeled G14 represent probing results for the mutant A3243G (A14G) hs mt tRNALeu(UUR). Lanes marked T1 represent the guanine ladder performed on wild-type hs mt tRNALeu(UUR). Lanes marked C are control samples that were not incubated with nucleases. Positions where losses in cleavage efficiency are observed due to dimerization in the mutant structure are identified with arrows. Multiple trials (> 3) of all probing experiments were performed to ensure the reproducibility of the trends shown.
Dimerization efficiency modulated by temperature and Mg2+
Magnesium ions play a critical role in stabilizing RNA tertiary structure (Laing et al. 1994). Metal cations can be involved in direct interactions with phosphates, ribose hydroxyls, and base functionalities, and provide stability to the tertiary fold. Mg2+ ions are known to be necessary for dimerization of viral RNAs in vitro, and structures of these complexes elucidated using X-ray crystallography have revealed that specific metal ion binding sites are present within the DIS (Marquet et al. 1991; Ennifar et al. 2001). Initial studies of the hs mt A3243G tRNALeu(UUR) dimer indicated that Mg2+ was necessary for complexation (Wittenhagen and Kelley 2002). To determine whether the complex bound Mg2+ at physiologically relevant concentrations of the cation, we performed a systematic analysis of the Mg2+ dependence.
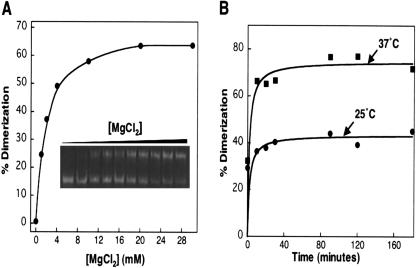
As shown in Figure 3A ▶, millimolar concentrations of Mg2+ promoted dimerization of the A3243G tRNALeu(UUR) mutant. Native gel electrophoresis analysis of samples of the tRNA mutant incubated with different concentrations of MgCl2 showed a pronounced dependence of the amount of dimer formed on the salt concentration. Appreciable dimerization was observed at magnesium concentrations as low as 2 mM, with 50% dimerization observed at 4 mM Mg2+, which is noteworthy considering that physiological magnesium levels are estimated to be ~5 mM (Fosse et al. 1996).
FIGURE 3.
Native gel analysis of the temperature and magnesium dependence on the formation of the A3243G tRNA dimer. (A) Mg2+ dependence of tRNA dimer formation. (Inset) 12% PAGE gel run under native conditions with varying [Mg2+] added to samples during annealing ([Mg2+] = 0, 1, 2, 4, 6, 8, 10, 20, 30, and 40 mM). (B) Time course of dimer formation at 25°C and at 37°C. Multiple trials (> 3) were performed to ensure the reproducibility of the trends shown.
Monovalent cations do not induce the dimerization of A3243G tRNALeu(UUR) efficiently. Concentrations of Li+, Na+, and K+ of 10 mM did not produce dimerization, and concentrations up to 0.5 M of these cations only yielded small amounts of dimer (10%–20%) (data not shown). These results are in contrast to the highly cation-sensitive dimerization of HIV-1 RNA that is suggested to involve quartets of adenine(s) and guanine(s) residues (Marquet et al. 1991). Since the A3243G dimer appears to be stabilized by Watson-Crick interactions, this may explain the lack of cation dependence observed.
The impact of temperature on the formation of the A3243G tRNALeu(UUR) mutant was also investigated. Native gel analysis revealed that the complex is more efficiently produced at 37°C versus 25°C (Fig. 3B ▶). Indeed, the observation of this complex at 37°C may indicate that it is sufficiently stable to exist in vivo. Increased dimerization at 37°C may result from an increased collision frequency, or may reflect a need for partial denaturation of the D stem for dimerization. Given that the D-stem would be predicted to exhibit low thermodynamic stability, raising the temperature from 25°C to 37°C could impact its structure.
Lanthanide probing of metal-binding sites within the dimeric interface
Lanthanide metals are useful probes of metal binding centers in RNA (Hargittai and Musier-Forsyth 2000). The affinity of lanthanide metals for metal binding sites in RNA is 600 to 10,000 higher than magnesium. Once bound, lanthanide metals facilitate phosphodiester backbone scission, allowing for the identification of magnesium binding sites (Matsumura and Komiyama 1997; Hargittai and Musier-Forsyth 2000). We have employed this probing technique to determine whether unique metal-binding sites within the dimeric interface of the mutant A3243G hs mt tRNALeu(UUR) exist.
Both the wild-type and mutant A3243G hs mt tRNALeu(UUR) transcripts were probed with terbium under nondenaturing conditions at 25°C over a tRNA concentration gradient (75–600 nM) spanning the Kd of the dimer (150 nM) (Wittenhagen and Kelley 2002) to identify metal-binding sites (Fig. 4 ▶). Samples were analyzed using PAGE as shown in Figure 4A ▶ and the cleavage results are shown graphically in Figure 4B ▶. The results for the A3243G hs mt tRNALeu(UUR) revealed an increase in terbium cleavage at the site of the A to G mutation that was most pronounced when tRNA concentrations greater than the Kd of the complex were present, while significant cleavage within the dimeric interface was not observed for wild-type hs mt tRNALeu(UUR) at low or high tRNA concentrations. The increase of terbium cleavage at position 14 for the A3243G hs mt tRNALeu(UUR) strongly suggests the formation of an additional metal-binding site at the site of the mutation that may stabilize the dimeric complex. Similar results were obtained using lead (Behlen et al. 1990) to probe the metal-binding sites (data not shown), providing additional evidence for the effects described.
FIGURE 4.
Terbium cleavage results for wild-type and mutant A3243G hs mt tRNALeu(UUR). Reactions were performed on both tRNA transcripts under nondenaturing conditions. (A) Gel analysis of terbium probing of wild-type and A3243G hs mt tRNALeu(UUR). Probing was performed over a tRNA concentration gradient of 75, 150, 300, and 600 nM tRNA, and samples were analyzed using 20% denaturing PAGE. Lanes representing the control samples for both the wild-type and mutant A3243G tRNAs are labeled A and G, respectively. Lane T1 is a guanine ladder prepared with wild-type tRNALeu(UUR), while Alk represents an alkaline ladder. Lanes representing probing results for the wild-type and A3243G samples increase in tRNA concentration from left to right. (B) Bar graphs extracted from PAGE analysis shown in A. Results represent cleavage intensities at each individual position. The solid line to the right of the gel shown in A represents the analyzed area. Results are normalized according to the total signal along the sample lane. Bars represent the results of terbium cleavage for the mutant A3243G (top) and wild-type (bottom) with [tRNA] = 75 nM (black) and 600 nM (gray). Multiple trials (> 3) of all probing experiments were performed to ensure the reproducibility of the trends shown.
Comparison of the A3243G hs mt tRNALeu(UUR) dimer with other RNA dimers
The diabetes and MELAS-related A3243G hs mt tRNALeu(UUR) forms a dimeric complex that is significantly more stable than other tRNA aggregates reported (Loehr and Keller 1968; Yang et al. 1972; Kholod 1999). It is observed at physiological temperatures, and importantly, its formation is more efficient at 37°C than at 25°C. The hs mt tRNA dimer is also magnesium-dependent, which presents another contrast to other tRNA complexes that require incubation under low ionic strength conditions (Madore et al. 1999).
The RNA dimers formed by viral genomes represent a more analogous system to the hs mt tRNA structure reported here. Dimerization of the HIV-1 viral genome occurs through hybridization of a palindromic hexamer very similar to the sequence present in this tRNA (Skripkin et al. 1994). Dimerization of HIV-1 is also highly dependent on temperature and RNA concentration, with an optimal temperature for dimerization close to 37°C (Marquet et al. 1991). The association constant for the HIV dimer is 4.10 × 106 M−1 (Kd = 250 nM) (Marquet et al. 1991). These values represent striking parallels to the A3243G hs mt tRNALeu(UUR) dimer, which has the same temperature dependence and a very similar Kd.
Other intriguing similarities between the hs mt tRNA and HIV-1 RNA complexes are the magnesium-binding sites that may exist within the dimeric interface (Ennifar et al. 2001). The HIV-1 RNA dimer is formed between two identical looped regions of the viral RNA with the sequence 5′-GUGCAC-3′. In the structure, a magnesium cation lies in the major groove of the interface and is bound to the RNA dimer through coordination with water to the Hoogsteen face of two guanine residues and to the O4 of two uridines. It is intriguing that a metal-ion binding site was detected in the hs mt tRNA dimer at the second guanine of the 5′-GGGCCC-3′ dimerization interface. This finding indicates that the tRNA and HIV RNA dimers may use metal ions at analogous positions for structural stabilization.
Structural abnormalities in the A3243G hs mt tRNALeu(UUR) mutant are significant because this single base substitution has been associated with a variety of physiological disorders and many different biochemical defects. The observation that a nonnative structure is formed may explain the deleterious effect of the mutation on aminoacylation, processing, and translation, as the formation of a high-affinity dimeric complex would interfere with many of the processes required for tRNA function. The finding that the complex is formed under physiological conditions in vitro presents the possibility that it may exist in vivo.
MATERIALS AND METHODS
Preparation of tRNA constructs
Plasmid DNA templates were prepared for in vitro transcription as described (Wittenhagen and Kelley 2002, 2003). Plasmids were harvested from Escherichia coli DH5α in milligram quantities and digested with Mva1 (Ambion) to generate the 3′CCA end. The digested DNA was then phenol/chloroform-extracted (pH 8.0), ethanol-precipitated, and resuspended in distilled H2O. The DNA was further purified using G-25 columns (Amersham Pharmacia). Transcription reactions were performed using template DNA (200–400 μg in 1 mL), T7 RNA polymerase (overexpressed in E. coli), RNasin (400 units/mL, Promega), 40 mM Tris-HCl, pH 8.0, 10 mM NaCl, 2 mM spermidine, 20 mM MgCl2, 4 mM NTPs, and 5 mM dithiothreitol. Samples were incubated at 37°C for 6 h, with the addition of a second aliquot of polymerase after 3 h. The DNA template was then digested with DNase I (60 units/mL, Takara). RNA products were extracted with 5:1 phenol (pH 4.7)/chloroform and ethanol-precipitated. Transcription products were further purified by 12% denaturing PAGE using a 0.5X TBE buffer (45 mM Tris base/45 mM boric acid/1 mM EDTA) on a 26 cm × 16.5 cm × 3 mm gel for 4 h. Purified transcripts were recovered by electroelution and then ethanol-precipitated. tRNA was resuspended in 0.5X TE (5 mM Tris-HCl, pH 8.0, 0.5 mM EDTA). All solutions were prepared with diethyl pyrocarbonate (DEPC)-treated water.
Absorbance at 260 nm was quantified in order to determine the concentrations of tRNA in solution. Values were obtained by applying an extinction coefficient of 895,000 M−1 (mononucleotide) cm−1 (http://wwwscripps.edu/mb/gottesfeld/ExtCoeff.html). tRNA samples were annealed with incubation at 70°C for 5 min in 0.5X TE followed by addition of MgCl2 (10 mM) and immediate cooling on ice.
Native gel electrophoresis
All samples were annealed at 70°C (as described above) with or without Mg2+ and mixed with native loading buffer to obtain a tRNA concentration of 1 μM. For the temperature-dependence assays, samples were annealed without Mg2+ and incubated at either 25°C or 37°C. The time course was initiated with the addition of 10 mM Mg2+. Aliquots were removed after 0, 10, 20, 30, 60, 90, 120, and 180 min and chilled on ice to halt the reaction. To determine the Mg2+ dependence, samples were annealed with 0–40 mM Mg2+ and immediately placed on ice. Samples were analyzed by 12% native PAGE in 0.5X TB (45 mM Tris, pH 7.5, 45 mM boric acid). Gels were stained with ethidium bromide and visualized with an Epi Chemi II Darkroom (UVP Imaging).
Preparation of 5′-32P-labeled hs mt tRNALeu(UUR) transcripts
Purified transcripts (500 pmol) were 5′-dephosphorylated using calf intestine alkaline phosphatase (2.5 U, Takara). Next, 100 pmol of each tRNA were quantitatively phosphorylated using γ-32P-ATP (MP Biomedicals, 7000 Ci/mmole) and 100 units of T4 poly-nucleotide kinase (New England Biolabs) in a total volume of 50 μL containing 70 mM Tris-HCl (pH 7.6), 10 mM MgCl2, and 5 mM dithiothreitol. The labeling reactions were incubated at 37°C for 30 min and then purified using G-25 columns. The labeled samples were further purified using 12% denaturing PAGE with 0.5X TBE buffer. Gel-purified samples were recovered by electro-elution and ethanol-precipitated. Samples were resuspended in 0.5X TE buffer.
Enzymatic probing of transcripts
Probing experiments were performed on both the wild-type and mutant A3243G hs mt tRNALeu(UUR) transcripts using 25 and 6 × 10−2 U of nuclease S1 (Fermentas) and RNase T2 (Sigma), respectively. Reactions were performed under nondenaturing conditions in a total reaction volume of 20 μL containing 40 mM Tris-HCl, pH 7.5, 40 mM NaCl, 10 mM MgCl2, and 1 mM spermine (1 mM ZnCl2 was present in nuclease S1 probing samples). Prior to adding reaction mixtures, the radiolabeled tRNA (2 × 104 CPM) supplemented with cold tRNA (0.05 μg/μL final concentration) was annealed in 40 mM Tris-HCl buffer by heating to 70°C for 5 min. After the addition of NaCl, MgCl2, spermine, and ZnCl2 (when applicable), samples were cooled on ice for 20 min. Probing enzymes were then added to the reaction mixtures, which were then incubated at 25°C for 5 min. The reactions were stopped by the addition of 5 μL of denaturing PAGE loading buffer (8 M Urea/45 mM Tris base/45 mM boric acid/1 mM EDTA). Multiple trials (> 3) of all probing experiments were performed to ensure the reproducibility of the trends observed.
Sequencing reactions
A guanine ladder was prepared using ribonuclease T1 (Fermentas). Radiolabeled wild-type hs mt tRNALeu(UUR) (2 × 104 CPM) supplemented with cold tRNA to a final concentration of 0.1 μg/ μL in 50 mM Tris base/2 mM EDTA (5 μL total volume) was heated to 70°C for 5 min and then cooled on ice. RNase T1 (0.02 U) was added, and the mixture was incubated at 37°C for 20 min. To stop the reaction, the sample was diluted with 5 μL denaturing PAGE loading buffer.
An alkaline ladder was also prepared using radiolabeled wild-type hs mt tRNALeu(UUR) (2 × 104 CPM). tRNA was diluted in 5 μL 10 mM NaHCO3/2 mM EDTA and heated at 90°C for 8 min. The reaction was stopped by the addition of 5 μL denaturing PAGE loading buffer.
Terbium cleavage reactions
Samples were prepared using the same conditions as in the nuclease probing experiments. Reaction volumes were 20 μL consisting of 40 mM Tris-HCl, pH 7.5, 40 mM NaCl, 10 mM MgCl2, and 1 mM spermine. Radiolabeled tRNA transcripts (2 × 104 CPM) were supplemented with unlabeled tRNA to create sample solutions of 75, 150, 300, and 600 nM tRNA. Mixtures were annealed in buffer by heating to 70°C for 5 min, followed by addition of NaCl, MgCl2, and spermine. Samples were then cooled on ice for 20 min. Next, TbCl3 was added to a final concentration of 200 μM. Reaction mixtures were incubated at 25°C for 30 min, and were quenched by adding 5 μL denaturing PAGE loading buffer. Multiple trials (> 3) of all terbium probing experiments were performed to ensure the reproducibility of the trends observed.
PAGE analysis of probing reactions
Reactions were analyzed using 40 cm (h) × 21 cm (w) × 0.4 mm (d) 20% denaturing polyacrylamide gels (Bio-Rad sequencing gel electrophoresis apparatus) buffered with 0.5X TBE. Gels were typically run at 25 mA for 80 min. Gels were visualized by exposure to a Kodak K-plate for 2 h, and imaging was achieved with a Bio-Rad Molecular Imager FX Pro-Plus phosphorimager. Histograms were extracted from individual gel lanes and normalized according to the amount of radioactivity present in the sample.
Acknowledgments
Financial support of this project was provided by the National Institutes of Health (R01 GM063890-01A2). We also acknowledge Eric Westhof for helpful comments on this work.
Article and publication are at http://www.rnajournal.org/cgi/doi/10.1261/rna.7143305.
REFERENCES
- Behlen, L.S., Sampson, J.R., DiRenzo, A.B., and Uhlenbeck, O.C. 1990. Lead-catalyzed cleavage of yeast tRNAPhe mutants. Biochemistry 29: 2515–2523. [DOI] [PubMed] [Google Scholar]
- Borner, G.V., Zeviani, M., Tiranti, V., Carrara, F., Hoffmann, S., Gerbitz, K.D., Lochmuller, H., Pongratz, D., Klopstock, T., Melberg, A., et al. 2000. Decreased aminoacylation of mutant tRNAs in MELAS but not in MERRF patients. Hum. Mol. Genet. 9: 467–475. [DOI] [PubMed] [Google Scholar]
- Chomyn, A., Martinuzzi, A., Yoneda, M., Daga, A., Hurko, O., Johns, D., Lai, S.T., Nonaka, I., Angelini, C., and Attardi, G. 1992. MELAS mutation in mtDNA binding site for transcription termination factor causes defects in protein synthesis and in respiration but no change in levels of upstream and downstream mature transcripts. Proc. Natl. Acad. Sci. 89: 4221–4225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chomyn, A., Enriquez, J.A., Micol, V., Fernandez-Silva, P., and Attardi, G. 2000. The mitochondrial myopathy, encephalopathy, lactic acidosis, and stroke-like episode syndrome-associated human mitochondrial tRNALeu(UUR) mutation causes aminoacylation deficiency and concomitant reduced association of mRNA with ribosomes. J. Biol. Chem. 275: 19198–19209. [DOI] [PubMed] [Google Scholar]
- El Meziane, A., Lehtinen, S.K., Holt, I.J., and Jacobs, H.T. 1998. Mitochondrial tRNALeu isoforms in lung carcinoma cybrid cells containing the np 3243 mtDNA mutation. Hum. Mol. Genet. 7: 2141–2147. [DOI] [PubMed] [Google Scholar]
- Ennifar, E., Walter, P., Ehresmann, B., Ehresmann, C., and Dumas, P. 2001. Crystal structures of coaxially stacked kissing complexes of the HIV-1 RNA dimerization initiation site. Nat. Struct. Biol. 8: 1064–1068. [DOI] [PubMed] [Google Scholar]
- Flierl, A., Reichmann, H., and Seibel, P. 1997. Pathophysiology of the MELAS 3243 transition mutation. J. Biol. Chem. 272: 27189–27196. [DOI] [PubMed] [Google Scholar]
- Florentz, C. 2002. Molecular investigations on tRNAs involved in human mitochondrial disorders. Biosci. Rep. 22: 81–98. [DOI] [PubMed] [Google Scholar]
- Fosse, P., Motte, N., Roumier, A., Gabus, C., Muriaux, D., Darlix, J.L., and Paoletti, J. 1996. A short autocomplementary sequence plays an essential role in avian sarcoma-leukosis virus RNA dimerization. Biochemistry 35: 16601–16609. [DOI] [PubMed] [Google Scholar]
- Goto, Y., Nonaka, I., and Horai, S. 1990. A mutation in the tRNA(Leu)(UUR) gene associated with the MELAS subgroup of mitochondrial encephalomyopathies. Nature 348: 651–653. [DOI] [PubMed] [Google Scholar]
- Hampel, A., Cherayil, J., and Bock, R. 1971. Specific aggregation of yeast glycine tRNA. Biochim. Biophys. Acta 228: 482–491. [DOI] [PubMed] [Google Scholar]
- Hargittai, M.R.S. and Musier-Forsyth, K. 2000. Use of terbium as a probe of tRNA tertiary structure and folding. RNA 6: 1672–1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helm, M., Florentz, C., Chomyn, A., and Attardi, G. 1999. Search for differences in post-transcriptional modification patterns of mitochondrial DNA-encoded wild-type and mutant human tRNALys and tRNALeu(UUR). Nucleic Acids Res. 27: 756–763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helm, M., Brule, H., Friede, D., Giegé, R., Putz, D., and Florentz, C. 2000. Search for characteristic structural features of mammalian mitochondrial tRNAs. RNA 6: 1356–1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs, H.T. and Holt, I.J. 2000. The np 3243 MELAS mutation: Damned if you aminoacylate, damned if you don’t. Hum. Mol. Genet. 9: 463–465. [DOI] [PubMed] [Google Scholar]
- Janssen, G.M., Maassen, J.A., and van Den Ouweland, J.M. 1999. The diabetes-associated 3243 mutation in the mitochondrial tRNALeu(UUR) gene causes severe mitochondrial dysfunction without a strong decrease in protein synthesis rate. J. Biol. Chem. 274: 29744–29748. [DOI] [PubMed] [Google Scholar]
- Kadowaki, T., Kadowaki, H., Mori, Y., Tobe, K., Sakuta, R., Suzuki, Y., Tanabe, Y., Sakura, H., Awata, T., Goto, Y., et al. 1994. A subtype of diabetes mellitus associated with a mutation of mitochondrial DNA. N. Engl. J. Med. 330: 962–968. [DOI] [PubMed] [Google Scholar]
- Kholod, N. 1999. Dimer formation by tRNAs. Biochemistry (Moscow) 64: 298–306. [PubMed] [Google Scholar]
- King, M.P., Koga, Y., Davidson, M., and Schon, E.A. 1992. Defects in mitochondrial protein synthesis and respiratory chain activity segregate with the tRNALeu(UUR) mutation associated with mitochondrial myopathy, encephalopathy, lactic acidosis, and strokelike episodes. Mol. Cell. Biol. 12: 480–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koga, A., Koga, Y., Akita, Y., Fukiyama, R., Ueki, I., Yatsuga, S., and Matsuishi, T. 2003. Increased mitochondrial processing intermediates associated with three tRNA(Leu(UUR)) gene mutations. Neuromuscul. Disord. 13: 259–262. [DOI] [PubMed] [Google Scholar]
- Laing, L., Gluick, T., and Draper, D. 1994. Stabilization of RNA structure by Mg ions. J. Mol. Biol. 237: 577–587. [DOI] [PubMed] [Google Scholar]
- Laughrea, M. and Jette, L. 1996. Kissing-loop model of HIV-1 genome dimerization. Biochemistry 35: 1589–1598. [DOI] [PubMed] [Google Scholar]
- Loehr, J.S. and Keller, E.B. 1968. Dimers of alanine transfer RNA with acceptor activity. Proc. Natl. Acad. Sci. 61: 1115–1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maassen, J.A. 2002. Mitochondrial diabetes: Pathophysiology, clinical presentation, and genetic analysis. Am. J. Med. Genet. 115: 66–70. [DOI] [PubMed] [Google Scholar]
- Madore, E., Florentz, C., Giege, R., and Lapointe, J. 1999. Magnesium-dependent alternative foldings of active and inactive Escherichia coli tRNA(Glu) revealed by chemical probing. Nucleic Acids Res. 27: 3583–3588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maechler, P. and Wollheim, C.B. 2001. Mitochondrial function in normal and diabetic β-cells. Nature 414: 807–812. [DOI] [PubMed] [Google Scholar]
- Marquet, R., Baudin, F., Gabus, C., Darlix, J.L., Mougel, M., Ehresmann, C., and Ehresmann, B. 1991. Dimerization of human immunodeficiency virus (type 1) RNA: Stimulation by cations and possible mechanism. Nucleic Acids Res. 19: 2349–2357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masucci, J.P. and Schon, E.A. 1996. tRNA processing in human mitochondrial disorders. Mol. Biol. Rep. 22: 187–193. [DOI] [PubMed] [Google Scholar]
- Matsumura, K. and Komiyama, M. 1997. Enormously fast RNA hydrolysis by lanthanide (III) ions underphysiological conditions: Eminent candidates for novel tools of biotechnology. J. Biochem. 122: 387–394. [DOI] [PubMed] [Google Scholar]
- Muriaux, D., Fosse, P., and Paoletti, J. 1996. A kissing complex together with a stable dimer is involved in the HIV-1 RNA dimerization process in vitro. Biochemistry 35: 5075–5082. [DOI] [PubMed] [Google Scholar]
- Paillart, J.C., Marquet, R., Skripkin, E., Ehresmann, B., and Ehresmann, C. 1994. Mutational analysis of the bipartite dimer linkage structure of human immunodeficiency virus type 1 genomic RNA. J. Biol. Chem. 269: 27486–27493. [PubMed] [Google Scholar]
- Panganiban, A.T. and Fiore, D. 1988. Ordered interstrand and intra-strand DNA transfer during reverse transcription. Science 241: 1064–1069. [DOI] [PubMed] [Google Scholar]
- Park, H., Davidson, E., and King, M.P. 2003. The pathogenic A3243G mutation in human mitochondrial tRNALeu(UUR) decreases the efficiency of aminoacylation. Biochemistry 42: 958–964. [DOI] [PubMed] [Google Scholar]
- Rossmanith, W. and Karwan, R.M. 1998. Impairment of tRNA processing by point mutations in mitochondrial tRNALeu(UUR) associated with mitochondrial diseases. FEBS Lett. 433: 269–274. [DOI] [PubMed] [Google Scholar]
- Roy, M.D., Wittenhagen, L.M., Vozzella, B.E., and Kelley, S.O. 2004. Interdomain communication between weak structural elements within a disease-related human tRNA. Biochemistry 43: 384–392. [DOI] [PubMed] [Google Scholar]
- Schleich, T. and Goldstein, J. 1964. Gel filtration properties of CCD-prepared E. coli B sRNA. Proc. Natl. Acad. Sci. 52: 774–749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schon, E.A., Koga, Y., Davidson, M., Moraes, C.T., and King, M.P. 1992. The mitochondrial tRNALeu(UUR) mutation in MELAS: A model for pathogenesis. Biochim. Biophys. Acta. 1101: 206–209. [PubMed] [Google Scholar]
- Schon, E.A., Bonilla, E., and DiMauro, S. 1997. Mitochondrial DNA mutations and pathogenesis. J. Bioenerg. Biomem. 29: 131–149. [DOI] [PubMed] [Google Scholar]
- Skripkin, E., Paillart, J.C., Marquet, R., Ehresmann, B., and Ehresmann, C. 1994. Identification of the primary site of the human immunodeficiency virus type 1 RNA dimerization in vitro. Proc. Natl. Acad. Sci. 91: 4945–4949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohm, B., Frugier, M., Brule, H., Olszak, K., Przykorska, A., and Florentz, C. 2003. Towards understanding human mitochondrial leucine aminoacylation identity. J. Mol. Biol. 328: 995–1010. [DOI] [PubMed] [Google Scholar]
- Sprinzl, M., Horn, C., Brown, M., Ioudovitch, A., and Steinberg, S. 1998. Compilation of tRNA sequences and sequences of tRNA genes. Nucleic Acids Res. 26: 148–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Ouweland, J.M., Lemkes, H.H., Ruitenbeek, W., Sandkuijl, L.A., de Vijlder, M.F., Struyvenberg, P.A., van de Kamp, J.J., and Maassen, J.A. 1992. Mutation in mitochondrial tRNA(Leu)(UUR) gene in a large pedigree with maternally transmitted type II diabetes mellitus and deafness. Nat. Genet. 1: 368–371. [DOI] [PubMed] [Google Scholar]
- Wittenhagen, L.M. and Kelley, S.O. 2002. Dimerization of a pathogenic human mitochondrial tRNA. Nat. Struct. Biol. 9: 586–590. [DOI] [PubMed] [Google Scholar]
- ———. 2003. Impact of disease-related mitochondrial mutations on tRNA structure and function. Trends Biochem. Sci. 28: 605–611. [DOI] [PubMed] [Google Scholar]
- Wittenhagen, L.M., Roy, M.D., and Kelley, S.O. 2003. The pathogenic U3271C human mitochondrial tRNALeu(UUR) mutation disrupts a fragile anticodon stem. Nucleic Acids Res. 31: 596–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, S.K., Söll, D.G., and Crothers, D.M. 1972. Properties of a dimer of tRNATyr. Biochemistry 11: 2311–2320. [DOI] [PubMed] [Google Scholar]
- Yasukawa, T., Suzuki, T., Ueda, T., Ohta, S., and Watanabe, K. 2000. Modification defect at anticodon wobble nucleotide of mitochondrial tRNAsLeu(UUR) with pathogenic mutations of mitochondrial myopathy, encephalopathy, lactic acidosis, and stroke-like episodes. J. Biol. Chem. 275: 4251–4257. [DOI] [PubMed] [Google Scholar]
- Zachau, H.G. 1968. Serine specific transfer ribonucleic acids. 16. Aggregation of serine specific transfer ribonucleic acids. Eur. J. Biochem. 5: 559–566. [DOI] [PubMed] [Google Scholar]