Abstract
Nab2, Npl3, and Nab4/Hrp1 are essential RNA binding proteins of the shuttling hnRNP class that are required for the efficient export of mRNA. To characterize the in vivo transcript specificity of these proteins, we identified their mRNA binding partners using a microarray-based assay. Each of the three proteins was coimmunoprecipitated with many different mRNA transcripts. Interestingly, each protein exhibits preferential associations with a distinct set of mRNAs. Notably, some of these appear to denote specific functional classes. For example, the ribosomal protein mRNAs and other highly expressed transcripts significantly favor association with Npl3 over Nab2, and Nab4/Hrp1 is strongly enriched with transcripts required for amino acid metabolism. Significantly, nab4 mutants showed a striking, desensitized growth phenotype when exposed to amino acid stress conditions suggesting a biological consequence to the associations we observed. Supporting the hypothesis that these proteins display transcript specificity, we identified a unique 7-nucleotide sequence overrepresented in the transcripts highly associated with Nab2 and Nab4/Hrp1 using the REDUCE algorithm. Validating our approach, our bioinformatics analysis correctly identified the known binding site for Nab4/Hrp1. These specialized associations of the hnRNP proteins of Saccharomyces cerevisiae suggest the opportunity to regulate the processing of particular transcripts between transcription and translation.
Keywords: hnRNP, protein–RNA interactions, microarray, Nab2, Npl3, Nab4/Hrp1
INTRODUCTION
The biological fate of a transcript is intimately dependent upon the RNA binding proteins with which it is associated. In the nucleus, the predominant form of mRNA is complexed with ribonucleoproteins to form hnRNPs (heterogeneous nuclear ribonucleic acid particles) (Dreyfuss et al. 1993, 2002). An hnRNP protein was classically defined as any protein that was found in association with these nuclear mRNA particles. While many hnRNP proteins share common RNA binding modules, these proteins appear to be functionally diverse and influence many processes, including alternative splicing, 3′ end formation, mRNA export, and RNA stability. In order to better define the function of the hnRNP proteins on specific transcripts, a careful identification of specific hnRNP–RNA interactions is required.
Novel functions of RNA binding proteins have been uncovered by the identification of specific mRNA binding partners on a genome-wide scale in Saccharomyces cerevisiae, Drosophila, and mammalian cells (Takizawa et al. 2000; Tenenbaum et al. 2000; Brown et al. 2001; Herold et al. 2003; Hieronymus and Silver 2003; Shepard et al. 2003; Blanchette et al. 2004; Gerber et al. 2004; Inada and Guthrie 2004; Rehwinkel et al. 2004). The picture that is emerging is one in which RNA binding proteins associate with discrete subsets of mRNAs that share functional attributes. The association with groups of related RNAs by an RNA binding protein can then influence the fate of those transcripts, possibly in a regulated manner. In some cases, well-defined sequence elements have been identified as the protein binding sites.
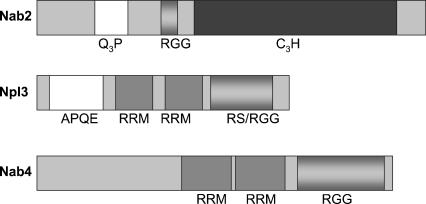
Of the hnRNP proteins, the shuttling, RNA binding proteins are particularly interesting since they maintain the ability to traverse the barrier between the nucleus and the cytoplasm. This shuttling feature may facilitate the export process itself and has also been hypothesized to give the shuttling hnRNPs the capacity to communicate nuclear events to the cytoplasmic machinery (Stutz and Rosbash 1998; Dreyfuss et al. 2002). To date, the best studied of these in S. cerevisiae are the shuttling proteins Nab2, Npl3, and Nab4/Hrp1. In budding yeast, these three are among a small set of proteins that were identified as cross-linked poly(A)-RNA binding proteins (Anderson et al. 1993; Wilson et al. 1994; Henry et al. 1996). While each protein readily associates with RNA, their protein structures suggest that they may do so in different ways (Fig. 1). Nab2 associates with RNA primarily through an extended zinc finger domain (Marfatia et al. 2003). Both Npl3 and Nab4/Hrp1 contain two RRM domains. However, Nab4/Hrp1 has an arginine/glycine-rich domain typical of mammalian hnRNP proteins, while Npl3 has an RGG domain that is interdigitated with arginine/serine dipeptide repeats typical of mammalian SR proteins.
FIGURE 1.
Nab2, Npl3, and Nab4/Hrp1 have shared and unique domains. Schematic representation highlighting the known domains of Nab2, Npl3, and Nab4/Hrp1. (Q3P) glutamine and proline-rich region, (RGG) arginine and glycine-rich region, (C3H) zinc finger region, (APQE) alanine, proline, glutamine, glutamic acid-rich region, (RRM) RNA recognition motif.
Nab2, Npl3, and Nab4/Hrp1 are each functionally nonredundant, as they are each essential. Two possibilities that are not mutually exclusive could explain their essential nature: Either each protein may be involved in the same process but with unique transcripts or each protein may play a unique role in the cell in separate processes. Because conditional mutations in any of these proteins can lead to nuclear mRNA accumulation, the simplest hypothesis implies a function in the mRNA export process (Singleton et al. 1995; Lee et al. 1996; Green et al. 2002; Hector et al. 2002). However, recent data have demonstrated that nuclear events are tightly coupled with one another, such that an export block at the pore can feedback to inhibit at or near the site of transcription (Hilleren et al. 2001; Jensen et al. 2001; Thomsen et al. 2003). Therefore, it is unclear whether Nab2, Npl3, and Nab4/Hrp1 could be involved in directing the export of specific transcripts in parallel export pathways, or if they are involved in earlier RNA processing or remodeling steps that are required to establish export competence.
Despite recent progress in understanding mRNA export in S. cerevisiae, an open question remains whether there is a single export pathway for all transcripts. Notably, the export of Nab2 can be uncoupled from that of the other hnRNP proteins by mutations in TOM1 (Duncan et al. 2000). In the background of tom1 temperature-sensitive mutations, Nab2 is exclusively retained in the nucleus, while other mRNA export factors (Npl3, Nab4/Hrp1, Mex67, Nop1, and Xpo1) continue to display wild-type protein localization (Duncan et al. 2000). It follows that if Nab2 is specifically bound to certain messages, then the export of these messages would be blocked as well. Conversely, the export of those messages not bound to Nab2 would be unaffected. In addition, the selective retention or export of hnRNPs has also been observed during stress conditions (Krebber et al. 1999; Duncan et al. 2000; Henry et al. 2003), arguing against the model of a single, shuttling hnRNP complex. Moreover, a genome-wide study in S. cerevisiae identifying the RNAs associated with two different RNA binding proteins required for export, Yra1 and Mex67, concluded that each factor can associate with disparate transcripts (Hieronymus and Silver 2003).
While the evidence for multiple export pathways seems compelling, the primary function of Nab2, Npl3, and Nab4/Hrp1 may lie in a step upstream of translocation across the nuclear membrane. 3′ end maturation is an essential requirement for export. Notably, the presence of a poly(A) tail is not sufficient for export (Dower and Rosbash 2002) implying that the 3′ end processing reaction deposits a signal or remodels the RNP in a manner that is required for export. Intriguingly, both Nab2 and Nab4/Hrp1 have been implicated in aspects of 3′ end generation. Nab2 has been hypothesized to play a role in poly(A) tail length control and has genetic interactions with the poly(A) binding protein Pab1. Nab4/Hrp1 is part of the Cleavage Factor Complex and may be required for positioning of the cleavage site prior to polyadenylation (Kessler et al. 1997; Minvielle-Sebastia et al. 1997, 1998; Chen and Hyman 1998). Although practically every mRNA has a poly(A) tail, Nab2 and Nab4/Hrp1 may not play a role in the 3′ end processing of every transcript. In contrast, Npl3 may play a role in splicing, which, like 3′ end processing, is required to occur before translocation and greatly enhances the export of properly processed messages. Npl3 is related to the family of SR proteins that, in mammalian cells, are required for constitutive splicing in addition to their widespread involvement in alternative splicing (Graveley 2000; Hastings and Krainer 2001; Caceres and Kornblihtt 2002). Moreover, Npl3 has been identified in a proteomics study in association with U1 snRNPs, an RNP complex that is required for splicing (Gottschalk et al. 1998). Therefore, it is unclear if the essential function of Nab2, Npl3, and Nab4/Hrp1 is at the export step or in an upstream process that is required for export to occur.
In addition to a role in the nucleus and during export, shuttling hnRNPs may act as the communicators of nuclear events to the cytoplasmic machinery. For example, Nab4/Hrp1 is required for nonsense mediated decay (NMD) in S. cerevisiae (Gonzalez et al. 2001). NMD is thought to occur in the cytoplasm to target incorrectly processed messages for degradation. Nonsense-containing messages may arise from errors during replication, transcription, or RNA processing—all nuclear events. Additionally, Npl3 has been found in polysomes and may act as a translational repressor (Windgassen et al. 2004). Mammalian SR proteins have also been found in polysomes but are thought to act as translational enhancers (Sanford et al. 2004).
Nab2, Npl3, and Nab4/Hrp1 share three important features: They all bind RNA, they all shuttle, and they all are essential. However, they also display critical differences, both in their domain structure and their roles in RNA processing. Fundamental to understanding the function of shuttling hnRNP proteins is to define their RNA interactions. Toward this end, we used a microarray-based approach to identify the spectrum of RNAs associated with each of these three proteins on a genomic scale.
RESULTS
Each hnRNP binds to a unique spectrum of transcripts
The genome-wide mRNA binding spectrum of three RNA binding proteins in S. cerevisiae, Nab2, Npl3, and Nab4/Hrp1, were evaluated by RNA-IP microarray analysis. Each protein was tagged with an identical ZZ (tandem IgG binding domains of Protein A) epitope at the carboxy terminus and integrated into its normal locus in the genome to be expressed under the control of its endogenous promoter (Longtine et al. 1998; Preker et al. 2002). Immunoprecipitations were performed in parallel with extracts from tagged and untagged sister strains. The coimmunoprecipitating RNAs were analyzed on an S. cerevisiae DNA microarray after reverse transcription, PCR amplification, and dye incorporation (Takizawa et al. 2000; Shepard et al. 2003). Yeast ORF arrays were employed that contain PCR products for every ORF (DeRisi et al. 1997). For this type of array, the signal from the IP and the mock IP RNA are competitively hybridized. The resulting log2 transformed ratio reflects the relative abundance of that transcript between the two samples. (The data for the microarray and bioinformatics analysis are available at http://www.ucsf.edu/guthrie/kimguisbert2005.htm.)
For the IP versus mock IP experiment, the ratio reveals the enrichment of any given mRNA for the immunoprecipitated protein relative to the mock IP control. As these microarrays simultaneously examined the ratios of >6000 different types of mRNAs, a spectrum of relative enrichment was assembled for each of the proteins. In this analysis, the position of the mRNA in the spectrum relative to other messages is more significant than the actual ratio from the array. By comparing the RNA spectrum for three different hnRNPs, the question of hnRNP specificity can be addressed without the use of an arbitrary cutoff to designate associated versus unassociated messages. Only features that showed clear, strong, and reproducible signal in the microarray in every replicate for every protein were included in the analysis.
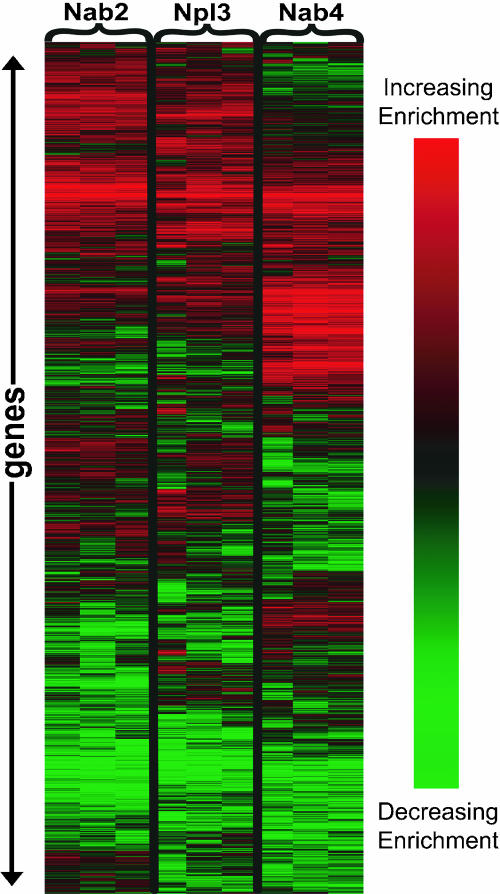
Importantly, each of the proteins displayed a unique RNA binding spectrum (Fig. 2). The RNA association spectrum for each of the proteins displayed an approximately Gaussian distribution (data not shown). The average Pearson correlation value for replicates was 0.729, indicating that the assay is reproducible (Table 1). In contrast, the average Pearson correlation value between the different hnRNP proteins is 0.439, numerically illustrating that the RNA binding spectra of Nab2, Npl3, and Nab4/Hrp1 are not identical (Table 1). Essentially, each protein has a specific “fingerprint” of mRNAs with which it is preferentially associated.
FIGURE 2.
Each protein has a unique RNA binding spectrum. Graphical representation made with Cluster and Treeview (Eisen et al. 1998) of three replicates for each of the three proteins studied, Nab2, Npl3, and Nab4/Hrp1, using the RNA-IP microarray assay. Each line along the Y-axis represents one gene. Only features that gave reliable signal for every replicate for every protein were included. The data were median centered and normalized, then subjected to SOM and average hierarchical clustering in the Cluster program. Brighter red indicates greater association of that transcript for an hnRNP; brighter green indicates less association.
TABLE 1.
Correlation matrix
| vs. | Nab2 | Npl3 | Nab4 |
| Nab2 | 0.778 | 0.577 | 0.350 |
| Npl3 | 0.651 | 0.389 | |
| Nab4 | 0.759 |
Each number represents the average Pearson correlation between experiments in the IP versus mock IP microarray experiment. The average correlation between replicates lies on the diagonal; the average correlation between proteins lies off the diagonal.
Although each protein is associated with a unique spectrum of RNAs, each protein is able to coprecipitate a large number of RNAs. The preferential association of each protein for RNAs could be thought of as a ranked list. Even though this ranked list is unique for each protein, each protein retains the ability to associate with many different RNAs. For example, the GZF3 transcript is highly associated with Nab2 as the 99th percentile ranked transcript, but is not absent from the Npl3 and Nab4/Hrp1 arrays; rather the GZF3 transcript ranks in the 46th and 81st percentile for Npl3 and Nab4/Hrp1, respectively.
A small fraction of genes (<2% of all genes) were found highly enriched for all three proteins. These may represent messages that are simultaneously bound by multiple shuttling hnRNP proteins. Alternatively, these could indicate RNAs that are bound nonspecifically to all proteins. Interestingly, a significant portion of these genes lie near the telomeric or centromeric region and include the abundant Ty and Y′ elements.
Preferential association of mRNA for hnRNPs is segregated by functional classification
A list of transcripts reproducibly associated with each protein was identified by subjecting the entire data set to the statistical analysis of microarrays (SAM) analysis (Tusher et al. 2001). The data sets were analyzed in pairwise combinations in order to easily identify the messages that exhibited the most drastic difference in association between proteins. SAM analysis uses multiple gene-specific t-tests to define a list of genes with a given false discovery rate (FDR). For all the analyses provided here, the FDR was set at a maximum of 5%.
The purpose of the SAM analysis is not to delineate associated versus unassociated messages, but to identify reproducibly, preferentially associated messages. This list of genes was then analyzed to determine if any functional classifications were statistically overrepresented. To determine the functional classifications, the GO annotations (March 2003 annotation) from the Saccharomyces Genome Database (SGD, available at http://yeastgenome.org) were utilized (Ashburner et al. 2000). Classifications that were over-represented with a p-value of <5.0e-4 are displayed in Table 2. Npl3 is enriched over Nab2 with transcripts encoding for ribosomal protein genes (RPGs). Nab4/Hrp1 is enriched over both Nab2 and Npl3 in pathways responding to metabolism and stress. Nab2 is enriched over Nab4/Hrp1 for proteins required for DNA-dependent transcription. Therefore, each hnRNP protein displays a propensity to preferentially associate with specific messages and this specificity correlates with functional classifications.
TABLE 2.
Function classes are overrepresented in association with each hnRNP
| Functional categories associated | ||
| Nab2 | Npl3 | Nab4 |
| transcription | ribosomal subunits | alcohol metabolism, energy pathways, ergosterol metabolism, branched chain amino acid, carboxylic acid metabolism |
Transcripts that were overrepresented in association with one hnRNP relative to another hnRNP were identified by using SAM analysis with pairwise comparisons of the mean IP versus mock IP microarray data. This should identify only those messages that show a significant and reproducible difference in association between the proteins studied. This list was then subjected to the GO termfinder available at SGD (http://yeastgenome.org) to identify functional classes that are overrepresented in association with each hnRNP. Above are the functional classes that displayed a p-value of less than 5e-4.
Differences between proteins are highlighted when compared directly against each other
In the above IP versus mock IP analysis, the Nab2 RNA binding spectra and the Npl3 RNA binding spectra were the most similar to one another. If these two proteins indeed bind similar messages to the equivalent extent, then the profiles of these spectra when compared directly against each other would be predicted to look uniform. Conversely, differences between the RNA binding profiles would be enhanced. Therefore, to increase the precision of identifying differentially associated messages between Nab2 and Npl3, the Nab2 associated RNAs were directly hybridized against the Npl3 associated RNAs on the microarray (i.e., IP vs. IP). This type of analysis has several advantages. First, the data will directly reveal any binding preferences for RNAs between the two proteins. Second, since Nab2 and Npl3 both reside primarily in the nucleus, it is reasonable to assume that they are exposed to a similar milieu of RNAs with which to bind. In this manner, the direct comparison may be a more accurate reflection of the binding differences between these two proteins, as a mock IP presumably is more reflective of the total RNA population than the nuclear RNA population. Lastly, in the case of Nab2 and Npl3, sufficient quantities of RNA coprecipitated to forgo amplification, thereby removing any bias introduced during the amplification step.
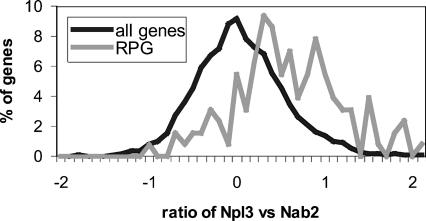
Strikingly, several distinct and reproducible differences were uncovered using this IP versus IP approach. In agreement with the first set of experiments (IP vs. mock IP), the mRNAs encoding the ribosomal protein genes (RPGs) as a class were biased for association with Npl3 and against association with Nab2 (Fig. 3). This bias cannot be explained by differences in gene expression between the two tagged strains as the control total RNA microarrays showed no significant deviations in expression and displayed a uniform distribution of RPGs (data not shown). Therefore, the bias of the RPGs for Npl3 is not due to differences in the strain or in the extract preparation. Within the distribution of the RPGs, Npl3 showed no further preference for intron-containing or essential RPGs.
FIGURE 3.
RPGs are skewed towards association with Npl3 over Nab2. Histogram displaying the distribution of transcripts in the Nab2 versus Npl3 direct IP versus IP microarray experiment relative to the transcript’s ratio on the array. Greater distance to the right of the histograms indicates greater association with Npl3 and vice versa. The distribution of the RPGs are normalized and displayed in gray. (RPGs) ribosomal protein genes.
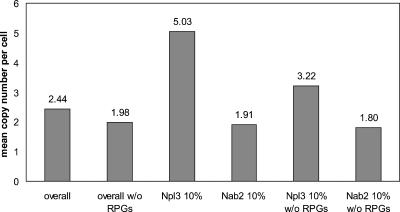
RPGs are among the most highly transcribed genes in the genome. Interestingly, Npl3 shows a preferential association with other highly transcribed genes (Fig. 4). Using the transcriptome information, the median expression values were calculated for the top 10% of messages most highly enriched on either side of our binding spectrum (Fig. 4; Holstege et al. 1998; Wang et al. 2002). After subtracting the abundant RPG messages, the median expression level of messages associated with Npl3 is still much higher than the median expression of all mRNAs without the RPGs. Therefore, Npl3 is preferentially associated over Nab2 with messages that are abundantly expressed in the cell.
FIGURE 4.
Mean abundance of Nab2 and Npl3 enriched targets. The average expression estimates (in copies/cell) based upon the Nab2 IP versus Npl3 IP microarray experiments. Estimates were drawn from Wang et al. (2002). (RPGs) Ribosomal protein genes.
Identification of RNA motifs correlated with hnRNP association
The analysis thus far has shown that hnRNPs can display preferential associations for particular classes of transcripts. In order to identify potential sequence binding motifs, the data set was subjected to the motif analysis algorithm REDUCE (Regulatory Element Detection Using Correlation with Expression) (Bussemaker et al. 2001). This algorithm performs an unbiased search on a single microarray experiment for every possible motif (oligonucleotide sequence) where the presence of the motif correlates with the transcript’s enrichment with the target protein (Bussemaker et al. 2001). Up to 7-nt-long motifs were searched, as longer motifs occur rarely and cannot yield sensible statistics. Although this algorithm was developed to identify DNA motifs involved in transcriptional regulation, the principle is conceptually identical to finding motifs associated between RNAs and proteins. The sense strand was searched from 600 nt before the start codon to 300 nt downstream of the stop codon. This region includes the 3′ UTR for ~98% of the genome (Graber et al. 2002) and is estimated to include nearly all of the transcription start sites (Spellman et al. 1998). The REDUCE algorithm identified unique seven-letter motifs overrepresented in the transcripts associated with each of the three immunoprecipitated proteins. Once all the overrepresented motifs were identified, REDUCE created a model motif that best fit the microarray data. Model motifs that were identified in at least two independent microarray-REDUCE analyses are shown in Table 3 with the highest p-value for the given motif.
TABLE 3.
A unique motif is associated with Nab2 and Nab4/Hrp1
| Motifs | p-value (neg. log) |
| Nab2 | |
| AAAAAG | 7.50 |
| Nab4 | |
| TATATAA | 8.26 |
| ACATACA | 3.60 |
Motifs greater than 5 nt long that were identified in more than one independent REDUCE analysis are shown with the highest p-value calculated for that motif. (REDUCE) Regulatory Element Detection Using Correlation with Expression, an algorithm described by Bussemaker et al. (2001).
Validating our approach, the REDUCE algorithm identified the UAUAUA motif overrepresented in transcripts that are associated with Nab4/Hrp1. Importantly, this element is identical to the core efficiency element (EE) of UA repeats that Nab4/Hrp1 associates with in in vitro binding experiments (Kessler et al. 1997; Chen and Hyman 1998). Nab4/Hrp1, in association with the Efficiency Element, has been hypothesized to play a role in the cleavage event that precedes polyadenylation during 3′ end formation (Minvielle-Sebastia et al. 1998).
An A-rich sequence was identified that strongly correlates with association with Nab2 (Table 3). Notably, Nab2 was shown to associate with high affinity (~7.5 nM) with a 25-mer of poly(A) RNA (Hector et al. 2002). Although REDUCE identified C-rich motifs enriched with Npl3, none of the motifs was >5 nt long, a requirement for inclusion in our data set. The failure to identify a larger motif for Npl3 could indicate that Npl3 binds to a more degenerate motif (REDUCE analysis does not allow for any wobble positions), that Npl3 has less specificity for a single RNA sequence, or that Npl3 does not require a 5-nt-long sequence to associate with RNA. To our knowledge, no work has been published that has examined the sequence specificity or affinity of Npl3.
Since the REDUCE algorithm was used to identify words up to seven letters, the regions flanking the motif were subsequently analyzed to determine if a larger context could be revealed. All occurrences of the top ranking motif were extracted from the SGD along with flanking sequences upstream and downstream of the motif (Cherry et al. 1997). This list of sequences was then ranked according to their ratio from the RNA-IP microarray experiment. Approximately 200 of the highest scoring occurrences of the word by RNA-IP microarray analysis were subjected to MEME local area alignment (available at http://meme.sdsc.edu/meme/website/) (Bailey and Elkan 1994). By this means, an expanded motif for both Nab2 and Nab4/Hrp1 was identified. A graphical summary of the alignment is shown in Figure 5 using Weblogo, a tool developed by Crooks et al. (2004) based upon the sequence logos created by Schneider and Stephens (1990) (Weblogo is available at http://weblogo.berkeley.edu). To avoid the identification of common promoter elements, the 5′ UTR was not searched for expanded motifs.
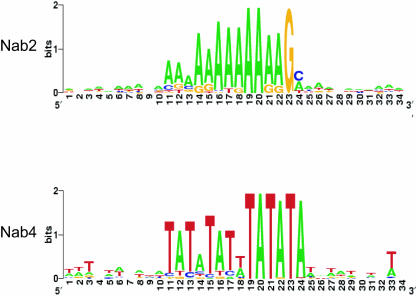
FIGURE 5.
Extended motifs identified for Nab2 and Nab4/Hrp1. Sequence logo display of the extended motifs identified by MEME analysis of the highest scoring messages containing the motif generated using the Weblogo tool. The height of the letters indicates the relative frequency of that letter at that position and the overall height of the stack indicates the sequence conservation in terms of information content in bits.
This analysis extended the A-rich sequence found with Nab2 to 12 nt, AAAAAAAAAAAG. Interestingly, the MEME analysis identified a T-rich element in a subset of genes that lies directly upstream or downstream of the A-rich element (data not shown). The T-rich element may have been too degenerate to detect with the REDUCE analysis. The identification of a close complement sequence to the central A-rich motif may suggest an RNA stem structure for some Nab2 associated transcripts.
The extended motif for Nab4/Hrp1 is nearly a duplicated occurrence of the Nab4/Hrp1 motif, from TATATAA to TATATATTTATATA. Intriguingly, the Nab2 and Nab4/Hrp1 expanded motifs are only visible when the 3′ UTR occurrences of the motif were aligned. This may indicate that the occurrences of the motif in the coding region are false positives or that the shorter nucleotide motif is sufficient for binding.
Growth phenotype correlates with association in the RNA-IP microarray assay
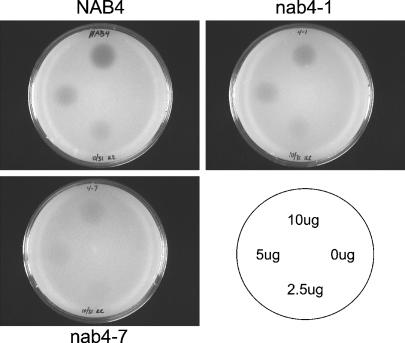
To evaluate the biological significance of the preferential hnRNP-mRNA associations we observed, we focused on the strong association of Nab4/Hrp1 with transcripts required for amino acid metabolism. In particular, transcripts required for branched-chain amino acid metabolism were highly enriched. To test the hypothesis that mutant nab4 protein may have an effect on cell growth when Nab4/Hrp1 associated messages are required for cell viability, halo assays were performed with the nab4 mutants, nab4-1 and nab4-7 (Minvielle-Sebastia et al. 1998) to test for sensitivity to sulfometuron methyl (SMM) (Fig. 6). SMM is a specific inhibitor of acetolactate synthase, which causes a depletion of branched-chain amino acids in the cell (Jia et al. 2000). Surprisingly, the nab4 mutants showed a growth advantage in comparison to wild-type cells when exposed to SMM. Strikingly, the nab4-7 mutant strain is almost completely resistant. This growth advantage seems to be specific for amino acid starvation as other stresses tested, including salt stress, hypo-osmolarity stress, and nonfermentable media, showed no discernable difference between wild-type and the nab4-1 mutant strain (data not shown).
FIGURE 6.
nab4 mutant strains are resistant to branched-chain amino acid stress. Growth assay where a lawn of cells are treated with drops of SMM at the quantities indicated in the lower right panel. SMM specifically inhibits acetolactate synthase that is required for the production of branched-chain amino acids. The zone of growth inhibition (halo) is proportional to the resistance of the strain to the drug. (SMM) sulfometuron methyl.
DISCUSSION
To explore the function of the shuttling hnRNP proteins in S. cerevisiae, we compared the genome-wide spectrum of RNAs that are associated with three essential hnRNP proteins, Nab2, Npl3, and Nab4/Hrp1. We discovered that not only were each of these proteins reproducibly associated with a different spectrum of mRNAs, but these associations can be divided into distinct functional categories. This work confirms that hnRNPs can preferentially recognize a subset of the transcriptome and provides predictions as to how this specificity may be utilized by the cell.
We have preserved the entire distribution of preferential associations to describe the RNA binding spectrum for each hnRNP as the association for any given transcript relative to the remainder of the transcriptome. For example, in Figure 3, the distribution of the ribosomal protein genes is displayed against the distribution of all genes to illustrate their preferential association as a class of genes toward Npl3 and against Nab2. Likewise, the RNA binding spectra of the three hnRNPs can be contrasted to display the relative association of any given transcript between the three proteins.
When interpreting the RNA binding spectra for the hnRNP proteins, the limitations of our assays must also be considered. Since the hnRNPs are predominantly nuclear proteins, we have assumed our RNA binding spectra reflect the nuclear RNA pool. However, the immunoprecipitations were performed on cell extracts and some nuclear protein–RNA interactions may have been disturbed during extract preparation (Mili and Steitz 2004). Moreover, in our interpretation of our results, we choose the strict standard that our assay had to produce a signal in every replicate for every protein. Therefore, a low ranking placement or absence from the RNA binding spectra may not necessarily reflect an inability to interact.
Complex networks of hnRNP–RNA interactions
The comparison of the genome-wide RNA binding profile reveals that each protein exhibits preferential binding to specific mRNAs and also illustrates that each protein can associate with a large number of transcripts. Therefore, each of the hnRNPs studied must be able to associate, to some degree, with the same transcripts. However, it is not possible to distinguish whether the proteins are bound simultaneously or sequentially to the same molecule.
If the hnRNPs are simultaneously bound, then the hnRNPs may be used in a combinatorial manner to dictate transcript fate. Such a concept has been advanced as post-transcriptional eukaryotic operons (Keene and Tenenbaum 2002). In this hypothesis, RNA binding proteins can exert a regulatory role on their associated messages well after transcription has taken place. In this manner, messages that are required for a particular response can be coordinately controlled as to localization, stability, etc. In the case of the shuttling hnRNPs studied here, it is possible that in order to establish the proper RNP structure for nuclear export, each transcript must have a specific set of hnRNPs; perhaps a specific “code” of hnRNPs is required to signal the formation of an export competent transcript.
Alternatively, each hnRNP could be required to associate with the given transcript in a sequential fashion. In this view, different mRNA processing events are associated with a hand-off event shifting the mRNA from one hnRNP to another. Such hand-off events between hnRNPs could facilitate the coupling of nuclear events that has recently been shown.
The presence of a unique motif correlates with hnRNP association
Although each hnRNP can associate with a large number of mRNAs, each hnRNP displayed a unique preferential association toward different mRNAs. This apparent specificity may be derived from unique sequence elements embedded in the message itself. In agreement with this hypothesis, our motif analysis using the REDUCE algorithm revealed unique motifs statistically overrepresented in the messages that are associated with each of the three proteins. In the simplest model, the identified motif represents a sequence-specific binding site for a particular hnRNP protein. Notably, the biochemically identified specific binding motif for Nab4/Hrp1 (Kessler et al. 1997; Minvielle-Sebastia et al. 1997; Chen and Hyman 1998) was correctly identified in our bioinformatics analysis. Additionally, REDUCE identified an A-rich sequence with Nab2. Nab2 has been shown to associate with high affinity (~7.5 nM) with a 25-mer of poly(A) RNA (Hector et al. 2002). Although the Hector et al. study did not examine the specificity of Nab2, the correlation between a high affinity interaction of Nab2 with a poly(A) oligo and our identification of an A-rich sequence in association with Nab2 is striking. These data support the hypothesis that transcript specificity can be driven by a sequence-specific interaction between the RNA and the transcript.
Consequences of hnRNP–RNA specificity
What are the consequences of hnRNP–RNA specificity for the cell? By identifying the RNAs associated with Nab4/Hrp1, we have discovered a previously unappreciated role of Nab4/Hrp1 during amino acid starvation. Interestingly, strains with mutant versions of Nab4/Hrp1 grow much better than wild-type cells when exposed to branched-chain amino acid stress. We are currently testing whether Nab4/Hrp1 has a negative regulatory role in this stress response. Similar to this discovery, the RNA specificity of Nab2 and Npl3 may also be used to discover new roles for the hnRNPs in other processes. Although these biological data serve to validate our microarray analysis, the precise function of these proteins on the partner mRNAs remains unknown. Two notable predictions for hnRNP function are described below.
Consequences: Multiple routes of mRNA export?
In light of the evidence that export of hnRNPs can be differentially controlled (in mutants or during stress), the apparent mRNA specificity demonstrated here implies that the export of mRNAs could also be differentially controlled (Krebber et al. 1999; Duncan et al. 2000; Henry et al. 2003). Although Nab2 and Npl3 had the most similar binding profiles in the RNA-IP microarray experiments, we observed numerous distinctions between the two RNA binding spectra. Notably the large class of RPGs, as well as other abundant messages, was significantly biased toward association with Npl3. Many of these abundant messages are required for rapid growth during logarithmic phase, but are not highly expressed during stress responses (Gasch et al. 2000). One intriguing hypothesis is that Npl3, the most abundant of the nuclear shuttling hnRNPs, is responsible for the export of the bulk of the mRNAs during rapid growth. Given that Npl3 is known to transiently localize to the cytoplasm during stress (heat shock, salt stress, ethanol stress) (Krebber et al. 1999) and that many of these abundant messages are transiently down-regulated during the stress response (Gasch et al. 2000), the absence of Npl3 from the nucleus during this period could further suppress the expression of the abundant messages with which Npl3 is normally associated. Interestingly, Npl3 is no longer preferentially associated with RPGs over Nab2 in stationary phase cells (data not shown), a condition where the expression of the RPGs is repressed relative to logarithmically growing cells.
Consequences: Specialized 3′ end processing?
In addition to a role in export, both Nab2 and Nab4/Hrp1 are involved in reactions that must presumably occur on nearly every mRNA transcript, namely 3′ end processing. Nab4/Hrp1 is both an hnRNP protein and a component of the Cleavage Factor I complex (Kessler et al. 1997) and has been hypothesized to direct cleavage at the correct site prior to polyadenylation (Minvielle-Sebastia et al. 1998). Nab2 has genetic interactions with Pab1, the poly(A) binding protein, and has been hypothesized to play a role in poly(A) tail length control (Hector et al. 2002). Although 3′ end processing is required of all mRNAs, Nab2 and Nab4/Hrp1 may not be required for the proper processing of all transcripts. In light of the recent evidence reporting that Nab4/Hrp1 is rapidly translocated to the cytoplasm during hypo-osmotic stress, this seems a likely hypothesis (Henry et al. 2003). During this stress, Nab4/Hrp1 is largely depleted from the nucleus and therefore presumably unable to perform its nuclear role of 3′ end processing. Either the minimal Nab4/Hrp1 remaining in the nucleus is sufficient for function or Nab4/Hrp1 is not required for the processing of messages during this stress response. Moreover, if Nab2 or Nab4/Hrp1 were exclusively associated with RNA via the poly(A) tail, then the prediction would be that they are equivalently associated with all mRNAs. The RNA specificity of Nab2 and Nab4/Hrp1 reported here indicates the possibility of much more gene-specific control during 3′ end processing than is currently appreciated.
Since they were first discovered more than 30 years ago, a revolution has occurred in the way hnRNP proteins are thought to function in the cell. No longer are they thought of as the histones of mRNA; they now appear to play vital roles in various stages of mRNA processing. Our work here confirms that hnRNP proteins, like many other RNA binding proteins, possess transcript specificity. Moreover, in a theme that has been discovered in other genome-wide searches, the specificity often extends to families of RNAs that are related by the functional category of the proteins that they encode. The specificity discovered here implicates the hnRNPs in previously unknown roles in the cell (i.e., Nab4/Hrp1 in amino acid stress) and also redefines their known functions to include RNA specificity.
MATERIALS AND METHODS
Strain preparation
Yeast manipulations were executed according to Guthrie and Fink (1981). Strains were prepared with ZZ (tandem IgG-binding domains of Protein A) epitope tags for each of the desired proteins according to the Longtine et al. (1998) method as described previously in Preker et al. (2002). ZZ tags were created in a diploid strain. (Primer sequences are available upon request.) The diploid transformants were then sporulated to generate two tagged strains with two untagged sister strains. The strains were confirmed to have no growth phenotype, normal cell morphology, wild-type localization of the target protein, and wild-type localization of poly(A) RNA (data not shown). In addition, total RNA microarrays were performed hybridizing the tagged strain against the untagged sister strain to confirm that no significant abnormalities in total RNA expression existed (data not shown).
The nab4 mutant strains were generously provided by the Swan-son Laboratory and were first described by Minvielle-Sebastia et al. (1998). Each strain has the endogenouse NAB4 gene deletion covered by CEN plasmid carrying either a wild-type or mutant version of NAB4.
Immunoprecipitation and RNA preparation
Immunoprecipitations were performed essentially according to Takizawa et al. (2000) with minor modifications. To elute the associated RNA, the beads were incubated with ProteinaseK (RNase-free) for 2 h at 37°C followed by a Hot Phenol RNA extraction of the supernatant. The purified RNA was then precipitated and quantified by OD260.
Probe preparation and microarray hybridization
For microarrays requiring amplification, coimmunoprecipitated RNA were reverse transcribed using Primer A (GTTTCCCAGT CACGATCNNNNNNNNN). After reverse transcription, the reactions were then PCR amplified with Primer B (GTTTCCCAGT CACGATC) and Primer A. Reactions were desalted by repeated dialysis using Microcon-30 membranes (Amicon). For microarrays without amplification, the RNA was reverse transcribed using dT19V (dTTTTTTTTTTTTTTTTTTTV) oligomer. The reactions were purified as described above.
The cDNA, from samples both with and without amplification, were coupled to either Cy3 or Cy5 dye (Amersham) and prepared for microarray hybridization essentially as described by DeRisi et al. (1997). The labeled probe was applied to a standard yeast ORF array (also see http://microarrays.org for more information on microarray construction and probe preparation). The hybridized arrays were washed in two successive solutions of 0.5× SSC, 0.25%SDS, and 0.05× SSC. The washed arrays were then scanned on either a 4000A or 4000B array scanner from Axon.
The data for all the microarrays were filtered to include only features that had no apparent physical deformity, correlation >0.5, median signal >500 in either channel. Independent immunoprecipitations were performed at least in triplicate. The data presented for Nab2 IP versus Npl3 IP microarray represents the mean ratio of seven independent experiments. For inclusion in this data set, the feature must be present in at least four of the seven replicates. Also, the Nab2 IP versus Npl3 IP microarrays were performed in sister strains of the opposite mating type; therefore the mating specific genes were deleted from the data set prior to data analysis. For the IP versus mock microarray analysis for the three hnRNPs, the feature had to be present in all nine of the independent experiments. Across three independent arrays for each protein, for a total of nine microarrays, the stringent criterion reduced the number of genes for further analysis to 4645 genes. Data from experiments that were dye flipped were transformed by multiplying the log2 transformed ratios by −1. Hierarchical clustering and Tree-view display of the microarray data was performed using the software described in Eisen et al. (1998).
Statistical analysis of microarray data
SAM analysis was developed by Tusher et al. (2001). In brief, SAM analysis utilizes a series of t-tests in order to define a list of genes with a given false discovery rate (FDR), which is indirectly modulated by the user. This FDR estimates the number of genes that are false positives within a set of experiments. To establish significant differences between pairwise comparisons using SAM, only data that were in every single replicate were chosen to be evaluated. The FDR for the IP versus mock IP experiments was set to a maximum of 5%. The FDR for the Nab2 IP versus Npl3 IP experiments was set to a maximum of 1%.
Halo assays
Halo assays were performed essentially as described by Hoffmann et al. (2002). Briefly, 0.5% agar was heated, cooled to 50°C, and then 4 mL were aliquoted into prewarmed 15 mL conical tubes. Ten microliters of saturated cultures were gently mixed with the agar, then poured onto a standard YPD plate. Serial dilutions of sulfometuron methyl (SMM) were applied to the plates as 5 μL drops in DMSO. Yeast were allowed to grow at room temperature for 2 d, then photographed.
Acknowledgments
We thank M. Swanson for generously providing the nab4 mutant strains. We thank H. Madhani, J. DeRisi, and C. Kung for their invaluable assistance in creating the microarrays. We thank J. De-Risi, H. Madhani, L. Blackburn, and E. O’Shea for critical reading of the manuscript. We thank E. Guisbert and all the members of the Guthrie Laboratory, particularly P. Preker, for critical reading of the manuscript as well as for advice and support. This work has been supported by a National Institute of Health Grant (GM21119). K.S.K.G. is an HHMI Predoctoral Fellow. C.G. is an American Cancer Society Research Professor of Molecular Genetics.
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.7234205.
REFERENCES
- Anderson, J.T., Wilson, S.M., Datar, K.V., and Swanson, M.S. 1993. NAB2: A yeast nuclear polyadenylated RNA-binding protein essential for cell viability. Mol. Cell. Biol. 13: 2730–2741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburner, M., Ball, C.A., Blake, J.A., Botstein, D., Butler, H., Cherry, J.M., Davis, A.P., Dolinski, K., Dwight, S.S., Eppig, J.T., et al. 2000. Gene ontology: Tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 25: 25–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey, T.L. and Elkan, C. 1994. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc. Int. Conf. Intell. Syst. Mol. Biol. 2: 28–36. [PubMed] [Google Scholar]
- Blanchette, M., Labourier, E., Green, R.E., Brenner, S.E., and Rio, D.C. 2004. Genome-wide analysis reveals an unexpected function for the Drosophila splicing factor U2AF(50) in the nuclear export of intronless mRNAs. Mol. Cell 14: 775–786. [DOI] [PubMed] [Google Scholar]
- Brown, V., Jin, P., Ceman, S., Darnell, J.C., O’Donnell, W.T., Tenenbaum, S.A., Jin, X., Feng, Y., Wilkinson, K.D., Keene, J.D., et al. 2001. Microarray identification of FMRP-associated brain mRNAs and altered mRNA translational profiles in fragile X syndrome. Cell 107: 477–487. [DOI] [PubMed] [Google Scholar]
- Bussemaker, H.J., Li, H., and Siggia, E.D. 2001. Regulatory element detection using correlation with expression. Nat. Genet. 27: 167–171. [DOI] [PubMed] [Google Scholar]
- Caceres, J.F. and Kornblihtt, A.R. 2002. Alternative splicing: Multiple control mechanisms and involvement in human disease. Trends Genet. 18: 186–193. [DOI] [PubMed] [Google Scholar]
- Chen, S. and Hyman, L.E. 1998. A specific RNA-protein interaction at yeast polyadenylation efficiency elements. Nucleic Acids Res. 26: 4965–4974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherry, J.M., Ball, C., Weng, S., Juvik, G., Schmidt, R., Adler, C., Dunn, B., Dwight, S., Riles, L., Mortimer, R.K., et al. 1997. Genetic and physical maps of Saccharomyces cerevisiae. Nature 387: 67–73. [PMC free article] [PubMed] [Google Scholar]
- Crooks, G.E., Hon, G., Chandonia, J.M., and Brenner, S.E. 2004. WebLogo: A sequence logo generator. Genome Res. 14: 1188–1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeRisi, J.L., Iyer, V.R., and Brown, P.O. 1997. Exploring the metabolic and genetic control of gene expression on a genomic scale. Science 278: 680–686. [DOI] [PubMed] [Google Scholar]
- Dower, K. and Rosbash, M. 2002. T7 RNA polymerase-directed transcripts are processed in yeast and link 3′ end formation to mRNA nuclear export. RNA 8: 686–697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreyfuss, G., Matunis, M.J., Pinol-Roma, S., and Burd, C.G. 1993. hnRNP proteins and the biogenesis of mRNA. Annu. Rev. Biochem. 62: 289–321. [DOI] [PubMed] [Google Scholar]
- Dreyfuss, G., Kim, V.N., and Kataoka, N. 2002. Messenger-RNA-binding proteins and the messages they carry. Nat. Rev. Mol. Cell Biol. 3: 195–205. [DOI] [PubMed] [Google Scholar]
- Duncan, K., Umen, J.G., and Guthrie, C. 2000. A putative ubiquitin ligase required for efficient mRNA export differentially affects hnRNP transport. Curr. Biol. 10: 687–696. [DOI] [PubMed] [Google Scholar]
- Eisen, M.B., Spellman, P.T., Brown, P.O., and Botstein, D. 1998. Cluster analysis and display of genome-wide expression patterns. Proc. Natl. Acad. Sci. 95: 14863–14868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasch, A.P., Spellman, P.T., Kao, C.M., Carmel-Harel, O., Eisen, M.B., Storz, G., Botstein, D., and Brown, P.O. 2000. Genomic expression programs in the response of yeast cells to environmental changes. Mol. Biol. Cell 11: 4241–4257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerber, A.P., Herschlag, D., and Brown, P.O. 2004. Extensive association of functionally and cytotopically related mRNAs with Puf family RNA-binding proteins in yeast. PLoS Biol. 2: E79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez, C.I., Bhattacharya, A., Wang, W., and Peltz, S.W. 2001. Nonsense-mediated mRNA decay in Saccharomyces cerevisiae. Gene 274: 15–25. [DOI] [PubMed] [Google Scholar]
- Gottschalk, A., Tang, J., Puig, O., Salgado, J., Neubauer, G., Colot, H.V., Mann, M., Seraphin, B., Rosbash, M., Luhrmann, R., et al. 1998. A comprehensive biochemical and genetic analysis of the yeast U1 snRNP reveals five novel proteins. RNA 4: 374–393. [PMC free article] [PubMed] [Google Scholar]
- Graber, J.H., McAllister, G.D., and Smith, T.F. 2002. Probabilistic prediction of Saccharomyces cerevisiae mRNA 3′-processing sites. Nucleic Acids Res. 30: 1851–1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graveley, B.R. 2000. Sorting out the complexity of SR protein functions. RNA 6: 1197–1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green, D.M., Marfatia, K.A., Crafton, E.B., Zhang, X., Cheng, X., and Corbett, A.H. 2002. Nab2p is required for poly(A) RNA export in Saccharomyces cerevisiae and is regulated by arginine methylation via Hmt1p. J. Biol. Chem. 277: 7752–7760. [DOI] [PubMed] [Google Scholar]
- Guthrie, C. and Fink, G.R. 1981. Guide to yeast genetics and molecular biology. Academic Press, San Diego, CA.
- Hastings, M.L. and Krainer, A.R. 2001. Pre-mRNA splicing in the new millennium. Curr. Opin. Cell Biol. 13: 302–309. [DOI] [PubMed] [Google Scholar]
- Hector, R.E., Nykamp, K.R., Dheur, S., Anderson, J.T., Non, P.J., Urbinati, C.R., Wilson, S.M., Minvielle-Sebastia, L., and Swanson, M.S. 2002. Dual requirement for yeast hnRNP Nab2p in mRNA poly(A) tail length control and nuclear export. EMBO J. 21: 1800–1810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry, M., Borland, C.Z., Bossie, M., and Silver, P.A. 1996. Potential RNA binding proteins in Saccharomyces cerevisiae identified as suppressors of temperature-sensitive mutations in NPL3. Genetics 142: 103–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry, M.F., Mandel, D., Routson, V., and Henry, P.A. 2003. The yeast hnRNP-like protein Hrp1/Nab4 sccumulates in the cytoplasm after hyperosmotic stress: A novel Fps1-dependent response. Mol. Biol. Cell 14: 3929–3941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herold, A., Teixeira, L., and Izaurralde, E. 2003. Genome-wide analysis of nuclear mRNA export pathways in Drosophila. EMBO J. 22: 2472–2483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hieronymus, H. and Silver, P.A. 2003. Genome-wide analysis of RNA–protein interactions illustrates specificity of the mRNA export machinery. Nat. Genet. 33: 155–161. [DOI] [PubMed] [Google Scholar]
- Hilleren, P., McCarthy, T., Rosbash, M., Parker, R., and Jensen, T.H. 2001. Quality control of mRNA 3′-end processing is linked to the nuclear exosome. Nature 413: 538–542. [DOI] [PubMed] [Google Scholar]
- Hoffman, G.A., Garrison, T.R., and Dohlman, H.G. 2002. Analysis of RGS proteins in Saccharomyces cerevisiae. Methods Enzymol. 344: 617–631. [DOI] [PubMed] [Google Scholar]
- Holstege, F.C., Jennings, E.G., Wyrick, J.J., Lee, T.I., Hengartner, C.J., Green, M.R., Golub, T.R., Lander, E.S., and Young, R.A. 1998. Dissecting the regulatory circuitry of a eukaryotic genome. Cell 95: 717–728. [DOI] [PubMed] [Google Scholar]
- Inada, M. and Guthrie, C. 2004. Identification of Lhp1p-associated RNAs by microarray analysis in Saccharomyces cerevisiae reveals association with coding and noncoding RNAs. Proc. Natl. Acad. Sci. 101: 434–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen, T.H., Patricio, K., McCarthy, T., and Rosbash, M. 2001. A block to mRNA nuclear export in S. cerevisiae leads to hyperadenylation of transcripts that accumulate at the site of transcription. Mol. Cell 7: 887–898. [DOI] [PubMed] [Google Scholar]
- Jia, M.H., Larossa, R.A., Lee, J.M., Rafalski, A., Derose, E., Gonye, G., and Xue, Z. 2000. Global expression profiling of yeast treated with an inhibitor of amino acid biosynthesis, sulfometuron methyl. Physiol. Genomics 3: 83–92.11015603 [Google Scholar]
- Keene, J.D. and Tenenbaum, S.A. 2002. Eukaryotic mRNPs may represent posttranscriptional operons. Mol. Cell 9: 1161–1167. [DOI] [PubMed] [Google Scholar]
- Kessler, M.M., Henry, M.F., Shen, E., Zhao, J., Gross, S., Silver, P.A., and Moore, C.L. 1997. Hrp1, a sequence-specific RNA-binding protein that shuttles between the nucleus and the cytoplasm, is required for mRNA 3′-end formation in yeast. Genes & Dev. 11: 2545–2556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krebber, H., Taura, T., Lee, M.S., and Silver, P.A. 1999. Uncoupling of the hnRNP Npl3p from mRNAs during the stress-induced block in mRNA export. Genes & Dev. 13: 1994–2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, M.S., Henry, M., and Silver, P.A. 1996. A protein that shuttles between the nucleus and the cytoplasm is an important mediator of RNA export. Genes & Dev. 10: 1233–1246. [DOI] [PubMed] [Google Scholar]
- Longtine, M.S., McKenzie 3rd, A., Demarini, D.J., Shah, N.G., Wach, A., Brachat, A., Philippsen, P., and Pringle, J.R. 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14: 953–961. [DOI] [PubMed] [Google Scholar]
- Marfatia, K.A., Crafton, E.B., Green, D.M., and Corbett, A.H. 2003. Domain analysis of the Saccharomyces cerevisiae heterogeneous nuclear ribonucleoprotein, Nab2p. Dissecting the requirements for Nab2p-facilitated poly(A) RNA export. J. Biol. Chem. 278: 6731–6740. [DOI] [PubMed] [Google Scholar]
- Mili, S. and Steitz, J.A. 2004. Evidence for reassociation of RNA-binding proteins after cell lysis: Implications for the interpretation of immunoprecipitation analysis. RNA 10: 1692–1694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minvielle-Sebastia, L., Preker, P.J., Wiederkehr, T., Strahm, Y., and Keller, W. 1997. The major yeast poly(A)-binding protein is associated with cleavage factor IA and functions in premessenger RNA 3′-end formation. Proc. Natl. Acad. Sci. 94: 7897–7902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minvielle-Sebastia, L., Beyer, K., Krecic, A.M., Hector, R.E., Swanson, M.S., and Keller, W. 1998. Control of cleavage site selection during mRNA 3′ end formation by a yeast hnRNP. EMBO J. 17: 7454–7468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preker, P.J., Kim, K.S., and Guthrie, C. 2002. Expression of the essential mRNA export factor Yra1p is autoregulated by a splicing-dependent mechanism. RNA 8: 969–980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehwinkel, J., Herold, A., Gari, K., Kocher, T., Rode, M., Ciccarelli, F.L., Wilm, M., and Izaurralde, E. 2004. Genome-wide analysis of mRNAs regulated by the THO complex in Drosophila melanogaster. Nat. Struct. Mol. Biol. 11: 558–566. [DOI] [PubMed] [Google Scholar]
- Sanford, J.R., Gray, N.K., Beckmann, K., and Caceres, J.F. 2004. A novel role for shuttling SR proteins in mRNA translation. Genes & Dev. 15: 1771–1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider, T.D. and Stephens, R.M. 1990. Sequence logos: A new way to display consensus sequences. Nucleic Acids Res. 18: 6097–6100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shepard, K.A., Gerber, A.P., Jambhekar, A., Takizawa, P.A., Brown, P.O., Herschlag, D., DeRisi, J.L., and Vale, R.D. 2003. Widespread cytoplasmic mRNA transport in yeast: Identification of 22 bud-localized transcripts using DNA microarray analysis. Proc. Natl. Acad. Sci. 100: 11429–11434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singleton, D.R., Chen, S., Hitomi, M., Kumagai, C., and Tartakoff, A.M. 1995. A yeast protein that bidirectionally affects nucleocytoplasmic transport. J. Cell Sci. 108: 265–272. [DOI] [PubMed] [Google Scholar]
- Spellman, P.T., Sherlock, G., Zhang, M.Q., Iyer, V.R., Anders, K., Eisen, M.B., Brown, P.O., Botstein, D., and Futcher, B. 1998. Comprehensive identification of cell cycle-regulated genes of the yeast Saccharomyces cerevisiae by microarray hybridization. Mol. Biol. Cell 9: 3273–3297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stutz, F. and Rosbash, M. 1998. Nuclear RNA export. Genes & Dev. 12: 3303–3319. [DOI] [PubMed] [Google Scholar]
- Takizawa, P.A., DeRisi, J.L., Wilhelm, J.E., and Vale, R.D. 2000. Plasma membrane compartmentalization in yeast by messenger RNA transport and a septin diffusion barrier. Science 290: 341–344. [DOI] [PubMed] [Google Scholar]
- Tenenbaum, S.A., Carson, C.C., Lager, P.J., and Keene, J.D. 2000. Identifying mRNA subsets in messenger ribonucleoprotein complexes by using cDNA arrays. Proc. Natl. Acad. Sci. 97: 14085–14090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomsen, R., Libri, D., Boulay, J., Rosbash, M., and Jensen, T.H. 2003. Localization of nuclear retained mRNAs in Saccharomyces cerevisiae. RNA 9: 1049–1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tusher, V.G., Tibshirani, R., and Chu, G. 2001. Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl. Acad. Sci. 98: 5116–5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Y., Liu, C.L., Storey, J.D., Tibshirani, R.J., Herschlag, D., and Brown, P.O. 2002. Precision and functional specificity in mRNA decay. Proc. Natl. Acad. Sci. 99: 5860–5865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson, S.M., Datar, K.V., Paddy, M.R., Swedlow, J.R., and Swanson, M.S. 1994. Characterization of nuclear polyadenylated RNA-binding proteins in Saccharomyces cerevisiae. J. Cell Biol. 127: 1173–1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Windgassen, M., Strum, D., Cajigas, I.J., Gonzalez, C.I., Seedorf, M., Bastians, H., and Krebber, H. 2004. Yeast shuttling SR proteins Npl3p, Gbp2p, and Hrb1p are part of the translating mRNPs, and Npl3 can function as a translational repressor. Mol. Cell. Biol. 24: 10479–10491. [DOI] [PMC free article] [PubMed] [Google Scholar]