Abstract
RNA editing and alternative splicing are two processes that increase protein diversity. The relationship between the two processes is not well understood. There are a few examples of correlations between editing and alternative splicing, but these are all nearby effects. A search for alternative splicing among 16 edited genes in Drosophila reveals two novel instances of alternative splicing. In one example where alternative splicing occurs downstream of editing, a strong correlation between editing efficiency and splice site selection is observed. In contrast, when editing occurs downstream of alternative splicing, no correlation is seen. These results suggest some models for the coupling of editing and splicing processes.
Keywords: editing, alternative splicing, ADAR
INTRODUCTION
Editing of exonic codons by adenosine deaminases that act on RNA (ADARs) and alternative splicing are two processes that increase protein diversity. The relative importance of both ADAR editing and alternative splicing increases with organismal complexity. In invertebrates, ADAR knockouts are viable with behavioral defects (Palladino et al. 2000; Tonkin et al. 2002). In mammals, however, ADAR editing of exons appears to be essential for viability (Brusa et al. 1995; Higuchi et al. 2000). Likewise, the extent to which an organism relies on alternative splicing for its protein repertoire increases from worms to humans (Kim et al. 2004b). In agreement with this association, there is a positive correlation between inosine content (Paul and Bass 1998) and the amount of alternative splicing (Yeo et al. 2004) where both editing and alternative splicing are most to least abundant in the brain, lung, and muscle, respectively. Given this concordance, and the reliance of exonic editing on intron-encoded exonic complementary sequences (ECS) (Higuchi et al. 1993), one might expect that the efficiency of the two processes would, in some cases, be coupled in vivo. The recent discovery of widespread editing of transcribed Alu elements, and their association with alternative splicing, is consistent with that expectation (Sorek et al. 2002; Athanasiadis et al. 2004; Kim et al. 2004a).
There are a few examples of an association between alternative splicing and editing at (Rueter et al. 1999) or near (Maas et al. 2001; Flomen et al. 2004) splice sites and some suggestions that this may be a common phenomenon (Reenan et al. 2000; Bass 2002). The ADAR2 protein regulates its own synthesis by creating an alternative splice site that leads to an out-of-frame product (Rueter et al. 1999). Previous experiments have also shown changes in splice site selection of the exonically edited 5-HT2C receptor in response to changes in ADAR2 activity (Maas et al. 2001) or in vitro A-to-G substitutions at the editing sites (Flomen et al. 2004). In this case alternative splicing removes the edited region of the receptor mRNA, thereby preventing detection of an association between editing efficiency and alternative splicing in vivo. Editing of the glutmate receptor subunit B pre-mRNA was found to inhibit splicing in vitro, but in vivo both processes can occur without interference (Bratt and Ohman 2003). This indicates a mechanism in vivo for coordinating the two processes and the discrepancy between the in vitro and in vivo results underscores the importance of obtaining in vivo data about the correlation of the two events.
In each of the previous examples the editing site and the affected splice site are in close proximity to one another. To determine if there might be correlations in the two processes that occur over long distances we studied two Drosophila genes that are both edited (Hoopengardner et al. 2003) and show alternative splicing. In one case the alternative splice sites are located far upstream (5′) of the editing site, and in the other case the alternative splices sites are located far downstream (3′) of the editing site. We find no difference in editing efficiency for the two upstream alternative products, but the two downstream products show dramatic differences in their editing efficiencies.
RESULTS AND DISCUSSION
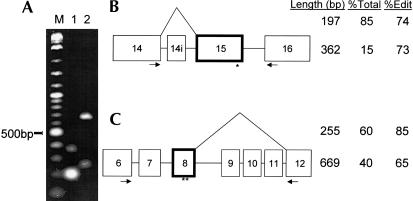
To search for alternatively spliced products, we performed RT-PCR on each of the 16 recently discovered Drosophila edited genes (Hoopengardner et al. 2003) using primers for exons on either side of edited exons. For two of the genes, we discovered unreported alternative splice products that could not be attributed to intron retention events (Fig. 1A ▶). Sequencing of the two products revealed their exon–intron structures (Fig. 1B,C ▶). CG15899, which encodes a calcium channel protein, contains an additional exon of 165 nt within the intron between exons 14 and 15 of the reported gene product (The Flybase Consortium, 2003). CG32975, which encodes a nicotinic acetylcholine receptor, has a product that skips exons 9–11 of the reported form (Grauso et al. 2002). Whereas the intron between exons 8 and 9 is ~4 kb in length, the longer intron between exons 8 and 12 is ~5 kb in length. The relative abundance of each splice product was estimated from their band intensities in an ethidium bromide stained agarose gel (Fig. 1 ▶).
FIGURE 1.
Alternative splicing of CG15899 and CG32975. (A) Agarose gel electrophoresis of alternative splice products. (Lanes 1,2) RT-PCR products from amplified regions of CG15899 and CG32975, respectively. (Lane M) A 2-log (100 bp - 10 kb) DNA base-pair ladder. (B,C) Schematic representation of the CG15899 (B) and CG32975 (C) alternative splice products relative to GenBank entries CG15899RB and CG32975RB, respectively. Edited exons are indicated (bold) with approximate editing site location highlighted (asterisk). Primer locations are depicted with closed arrows. Exons are numbered as in the associated GenBank entry. Exon “14i” of CG15899 comes from within the intron of the reference gene. Three columns (Length, %Total, %Edit) represent the size (in base pairs) of the PCR products, the percentage of the total population that the spliceform represents and the percentage of the spliceform population that is edited, respectively.
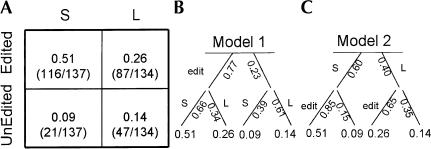
Using spliceform-specific primers we then amplified each spliceform separately and cloned and sequenced a large number of each product to determine the fraction of each that were edited (Fig. 1B,C ▶). For CG15899, with the alternative exon 5′ of the edited exon and the edited nucleotide at the 3′ end of the edited exon, we see no difference in the editing efficiency between the alternatively spliced products. In both products the ratio of edited to unedited is ~3/1. For CG32975, with the alternative splice sites 3′ of the edited exon, we find a large difference in the editing efficiencies of the two products (Fig. 2A ▶). The ratio of edited to unedited is >5/1 (116/21) for the short product while for the long product the same ratio is <2/1 (87/47). Combining the ratios of edited to unedited for each spliceform with the relative abundance of each the two spliceforms (Fig. 1 ▶), we can calculate the fraction of the total gene product in each of the four states, short or long and edited or unedited (Fig. 2 ▶). In the edited population the ratio of short to long product is 2/1 (0.51 vs. 0.26) while in the unedited population the short product is less abundant than the long product at a ratio of 1/1.5 (0.09 vs. 0.14).
FIGURE 2.
Abundance of edited and unedited CG32975 spliceforms. (A) Matrix elements are the fraction of the total that occur in each combination of spliceform and edited state. These numbers were obtained as the product of the percentage total and percent edited from Figure 1 ▶. Numbers in parentheses are raw counts of edited and unedited clones for each spliceform. S and L represent the short (255 bp) and long (669 bp) spliceforms, respectively. The fraction of the total mRNA in each spliceform (60% S and 40% L) were determined in Figure 1 ▶. (B,C) Splicing and editing decision trees for the proposed models. Model 1 (B) represents splicing decisions (lower branches) made as a result of prior editing decisions (upper branches). Model 2 (C) represents editing decisions (lower branches) made as a result of prior splice site choice (upper branches). Numbers on each branch represent the probability of that decision. Branch labels S and L correspond to selection of the splice site for the short (255 bp) or long (669 bp) product, respectively. Branches labeled edit or unlabeled represent the presence or absence of RNA editing, respectively. Numbers at the leaves represent the total frequency with which the path leading to that leaf is traversed and correspond to the observed values from A.
These results show a correlation between editing efficiency and the choice of alternative splice sites. We propose two general models to explain the results.
The editing event strengthens a splicing enhancer signal within exon 8, allowing it to be bound by an enhancer protein (model 1, Fig. 2B ▶). The enhanced splice site is now more efficient at splicing out the longer intron, leading to the shorter product. It is not exactly clear how an enhacer within this exon would alter the choice of a distant acceptor site, but there are previous studies showing that longer introns tend to be flanked by stronger splice sites (Clark and Thanaraj 2002). In this case the enhancement for the longer intron by the edited exon would be about threefold, altering the ratio from the longer product being more abundant to the shorter product predominating.
This model presumes no role for editing in directing splicing (model 2, Fig. 2C ▶). Here the more distant splice acceptor site is preferentially chosen by a ratio of ~1.5/1. Because editing requires an ECS within the intron we still expect that editing will occur before splicing is completed, but we imagine that the choice of splice site affects the efficiency of the editing event. For example, a helicase (Reenan et al. 2000) is required to unwind the structure of the intervening intron as the spliceo-some travels from the splice acceptor site toward the donor site. The greater distance traveled from the short form acceptor site allows more time for the editing machinery to operate and a greater amount of editing to occur.
The two models could be distinguished by analyzing the splicing pattern in the absence of RNA editing. Model 2 predicts that the 1.5/1 ratio of short to long products does not depend on editing. Model 1 predicts that the shorter product would be more highly prevalent, to a ratio of ~2/1, if the editing site were changed to match the edited sequence, i.e., if it were pre-edited. On the other hand, if editing were prevented, for instance, by modifying the ECS or in an ADAR mutant, then we would expect the longer form to predominate by a ratio of ~1.5/1. The two models are not mutually exclusive, and it is possible some combination of each leads to the observed result. It is also possible that some more complicated process is involved.
In previous studies that have demonstrated an association between splicing and editing, the two sites were very close together (Rueter et al. 1999; Maas et al. 2001; Bratt and Ohman 2003; Flomen et al. 2004). Those examples could be explained by the editing sites altering the splice signals themselves or mRNA secondary structures that interfere with splicing.
RNA editing is known to be important in many metazoan organisms. Invertebrates that lack editing activity show behavioral phenotypes (Palladino et al. 2000; Tonkin et al. 2002), whereas in mammals ADAR editing is essential for viability (Brusa et al. 1995; Higuchi et al. 2000). There is strong evidence that there are many editing sites yet to be discovered (Paul and Bass 1998) but experimental detection remains a difficult problem. Editing can alter a protein sequence and thereby affect its function (Bass 2002). It can affect the splicing of pre-mRNA, as shown in this report and in previous work. It can also alter gene expression, such as in the autoregulation of ADAR2 (Rueter et al. 1999) and by affecting mRNA degradation (Tonkin and Bass 2003). It is clear that a comprehensive analysis of gene functions and of gene expression patterns must take into account editing events and their correlations with other cellular processes.
MATERIALS AND METHODS
RNA isolation and reverse-transcriptase (RT)-PCR
Total RNA was extracted from Canton-S fly heads with Trizol Reagent (Invitrogen) and treated with DNAse (DNA-Free, Ambion). Total RNA was used to synthesize cDNA using Superscript II reverse transcriptase (Invitrogen) and random hexamer primers. Amplification of CG15899 used primers 5′-CGCTCGTCACAGT GTTCCAG-3′ and 5′-GCGCTCGCGACGTTCATTTC-3′. Gene CG32975 was amplified using primers 5′-CACTCTGCCGCCA GATTCG-3′ and 5′-GCACTCGTCATCTTTACGTAGC-3′.
CG15899 splice product identification
Amplified products from CG15899 were T/A cloned into pGEM-TEasy Vector (Promega) and clones were screened for appropriately sized bands upon PCR amplification with M13F (−41) and M13R (−48) primers. Appropriately sized PCR products were cleaned by Exonuclease I/Shrimp Alkaline Phosphatase Treatment (USB) and sequenced by dideoxy sequencing.
CG32975 splice product identification
Amplified products from CG32975 represented a previously identified product (669 bp) and a smaller product that was larger than the CG32975RA spliceform. This product was characterized using a single primer specific to the expected CG32975RA spliceform coupled to one of the original primers used for amplification. The 3′ spliceform-specific primer produced a PCR product of the appropriate size, suggesting the product represented retention of CG32975RB exon 7 and deletion of exons 9, 10, and 11.
Amplification of specific spliceforms
Each splice product was amplified separately from bulk cDNA using a single spliceform-specific primer and a shared primer. For CG15899 the spliceform-specific primers were 5′-CAGCAT ACTCGGCATGTATCTG-3′ for spliceform 362 and 5′-CACAGTG TTCCAGATACTGACGCAAG-3′ for spliceform 197. These were coupled to the three prime primer 5′-GCGCTCGCGACGTTCAT TTC-3′. For CG32975 the spliceform-specific primers were 5′-CGTCATCTTTACGTAGCCATTCGGACATTTCG-3′ for spliceform 255 and 5′-CTTTACGTAGCTGATCAGTTATAAAG-3′ for spliceform 669. These were coupled to a shared forward primer 5′-TGCCTCCATGGCCTTGCTCGG-3′. All products were then Exo/Sap treated and sequenced.
Quantification of RNA editing
Splice products were amplified and cloned into the BamHI/HindIII site of the pUC19 Vector. Primers used for amplification of CG32975 splice products were forward: 5′-GCCGGATCCGGTA CATATTTCAATTGCATAATGTTTATGG-3′ and reverse: 5′-GC GAAGCTTCCATTCGGACATTTCGTGCG-3′. Primers used for amplification of CG15899 were forward: 5′-GGCGGATCCATAC TGACGCAAGAGGATTGG-3′ (form 362), 5′-GGCGGATCC GATACTGACGCAAGAGGATTGG-3′ (form 197) and reverse 5′-GTCAAGCTTGCTCGCGACGTTCATTTCGC-3′. Relative A-to-G abundance was determined by sequencing individual clones with plasmids containing appropriately sized inserts.
Quantification of spliceform abundance
Populations of mixed spliceforms obtained from RT-PCR of Drosophila cDNA were subjected to gel electrophoresis and stained with ethidium bromide. The stained gel was scanned on a Typhoon Scanner (Amersham) and the relative abundance of each band was measured with the image analysis program Imagequant (Amersham). Values were normalized against background readings. Final values were obtained as the average of three readings. Relative molar abundances were obtained by correcting for differences in product size.
Acknowledgments
We thank Barak Cohen, Rob Mitra, and the members of their laboratories for many helpful comments. We thank Larry Schriefer and Yiing Lin for technical assistance. G.D.S. was supported by NIH grant HG00249 and R.A. was supported by NIH training grant GM08802.
Article and publication are at http://www.rnajournal.org/cgi/doi/10.1261/rna.7280605.
REFERENCES
- Athanasiadis, A., Rich, A., and Maas, S. 2004. Widespread A-to-I RNA editing of Alu-containing mRNAs in the human transcriptome. PLoS Biol. 2: e391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bass, B.L. 2002. RNA editing by adenosine deaminases that act on RNA. Annu. Rev. Biochem. 71: 817–846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bratt, E. and Ohman, M. 2003. Coordination of editing and splicing of glutamate receptor pre-mRNA. RNA 9: 309–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brusa, R., Zimmermann, F., Koh, D.S., Feldmeyer, D., Gass, P., Seeburg, P.H., and Sprengel, R. 1995. Early-onset epilepsy and post-natal lethality associated with an editing-deficient GluR-B allele in mice. Science 270: 1677–1680. [DOI] [PubMed] [Google Scholar]
- Clark, F. and Thanaraj, T.A. 2002. Categorization and characterization of transcript-confirmed constitutively and alternatively spliced introns and exons from human. Hum. Mol. Genet. 11: 451–464. [DOI] [PubMed] [Google Scholar]
- Flomen, R., Knight, J., Sham, P., Kerwin, R., and Makoff, A. 2004. Evidence that RNA editing modulates splice site selection in the 5-HT2C receptor gene. Nucleic Acids Res. 32: 2113–2122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Flybase Consortium. 2003. The flybase database of the Drosophila genome projects and community literature. Nucleic Acids Res. 31: 172–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grauso, M., Reenan, R.A., Culetto, E., and Satelle, D.B. 2002. Novel putative nicotinic acetylcholine receptor subunit genes, Dα5, Dα6 and Dα7, in Drosophila melanogaster identify a new and highly conserved target of adenosine deaminase acting on RNA-mediated A-to-I pre-mRNA editing. Genetics 160: 1519–1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higuchi, M., Single, F.N., Kohler, M., Sommer, B., Sprengel, R., and Seeburg P.H. 1993. RNA editing of AMPA receptor subunit GluR-B: A base-paired intron–exon structure determines position and efficiency. Cell 75: 1361–1370. [DOI] [PubMed] [Google Scholar]
- Higuchi, M., Maas, S., Single, F.N., Hartner, J., Rozov, A., Burnashev, N., Feldmeyer, D., Sprengel, R., and Seeburg, P.H. 2000. Point mutation in an AMPA receptor gene rescues lethality in mice deficient in RNA-editing enzyme ADAR2. Nature 406: 78–81. [DOI] [PubMed] [Google Scholar]
- Hoopengardner, B., Bhalla, T., Staber, C., and Reenan R. 2003. Nervous system targets of RNA editing identified by comparative genomics. Science 301: 832–836. [DOI] [PubMed] [Google Scholar]
- Kim, D.D., Kim, T.T., Walsh, T., Kobayashi, Y., Matise, T.C., Buyske, S., and Gabriel, A. 2004a. Widespread RNA editing of embedded alu elements in the human transcriptome. Genome Res. 14: 1719–1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, H., Klein, R., Majewski, J., and Ott, J. 2004b. Estimating rates of alternative splicing in mammals and invertebrates. Nat. Genet. 36: 915–916. [DOI] [PubMed] [Google Scholar]
- Maas, S., Patt, S., Schrey, M., and Rich, A. 2001. Underediting of glutamate receptor GluR-B mRNA in malignant gliomas. Proc. Natl. Acad. Sci. 98: 14687–14692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palladino, M.J., Keegan, L.P., O’Connell, M.A., and Reenan, R.A. 2000. A-to-I pre-mRNA editing in Drosophila is primarily involved in adult nervous system function and integrity. Cell 102: 437–439. [DOI] [PubMed] [Google Scholar]
- Paul, M.S. and Bass, B.L. 1998. Inosine exists in mRNA at tissue specific levels and is most abundant in brain mRNA. EMBO J. 17: 1120–1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reenan, R.A., Hanrahan, C.J., and Ganetzky, B. 2000. The mlenapts RNA helicase mutation in Drosophila results in a splicing catastrophe of the para Na+ channel transcript in a region of RNA editing. Neuron 25: 139–149. [DOI] [PubMed] [Google Scholar]
- Rueter, S.M., Dawson, T.R., and Emeson, R.B. 1999. Regulation of alternative splicing by RNA editing. Nature 399: 75–80. [DOI] [PubMed] [Google Scholar]
- Sorek, R., Ast, G., and Graur, D. 2002. Alu-containing exons are alternatively spliced. Genome Res. 12: 1060–1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tonkin, L.A. and Bass, B.L. 2003. Mutations in RNAi rescue aberrant chemotaxis of ADAR mutants. Science 302: 1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tonkin, L.A., Saccomanno, L., Morse, D.P., Brodigan, T., Krause, M., and Bass, B.L. 2002. RNA editing by ADARs is important for normal behavior in Caenorhabditis elegans. EMBO J. 21: 6025–6035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeo, G., Holste, D., Kreiman, G., and Burge, C.B. 2004. Variation in alternative splicing across human tissues. Genome Biol. 5: R74. [DOI] [PMC free article] [PubMed] [Google Scholar]