Abstract
The family of adenosine deaminases acting on RNA (ADARs) targets adenosines in RNA that is mainly double stranded. Some substrates are promiscuously deaminated whereas others, such as the mammalian glutamate receptor B (gluR-B) pre-mRNA, are more selectively deaminated. Many DNA/RNA-base modification enzymes use a base flipping mechanism to be able to reach their target base and it is believed that ADARs function in a similar way. In this study we used molecular dynamics (MD) simulations to describe two sites on the gluR-B pre-mRNA, the selectively targeted R/G site and the nontargeted 46 site, in an attempt to explain the substrate specificity. We used regular MD and also a forced base flipping method with umbrella sampling to calculate the free energy of base opening. Spontaneous opening of the mismatched adenosine was observed for the R/G site but not for the 46 site.
Keywords: base opening, editing enzymes, umbrella sampling, potential of mean force, mismatch
INTRODUCTION
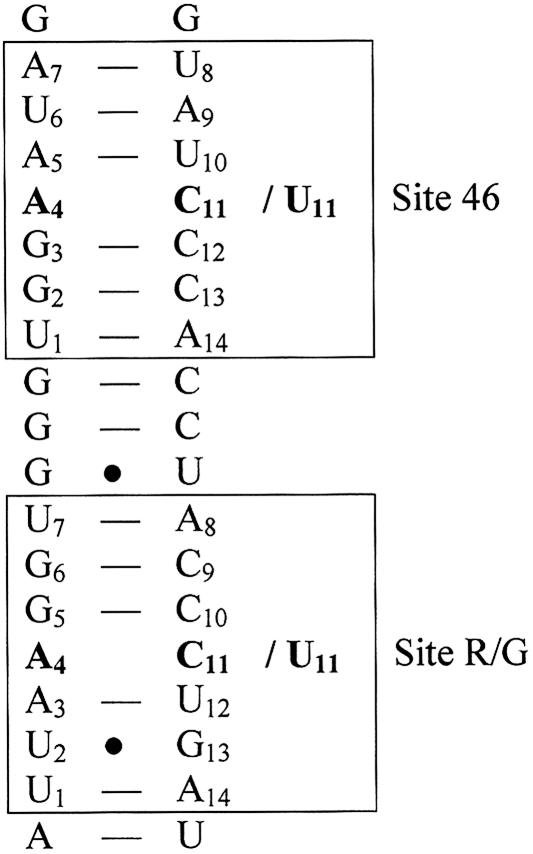
The adenosine deaminase that acts on RNA (ADAR) proteins convert adenosines to inosines within RNAs that are largely double stranded (dsRNA). A number of substrates coding for genes involved in neurotransmission have been found in the mammalian brain (Sommer et al. 1991; Lomeli et al. 1994; Burns et al. 1997; Bhalla et al. 2004). Since inosine is recognized as guanosine at translation, editing within a coding sequence will often lead to a change in codon usage. Evidence has been found that ADARs form homodimers on dsRNA, a necessary event for editing activity (Jaikaran et al. 2002; Cho et al. 2003; Gallo et al. 2003). ADAR enzymes deaminate adenosines nonselectively within completely base-paired dsRNA that is longer than 50 bp (Nishikura et al. 1991; Polson and Bass 1994). More selective deamination occurs in unbranched helices that are disrupted by loops, mismatches, or bulges (for review, see Bass 2002). ADARs have a preference for deaminating adenosines in a certain sequential context. Human ADAR2 has a 5′ neighbor preference (U ≈ A > C = G) and a 3′ neighbor preference (U = G > C = A) (Lehmann and Bass 2000). However, when ADARs deaminate selectively there are adenosines that do not get edited even though they appear to be in the right context. One substrate that is only deaminated at certain adenosines is the mammalian glutamate receptor subunit B (gluR-B) pre-mRNA. The R/G site of gluR-B is efficiently edited by ADAR2 in vivo (Higuchi et al. 2000). Editing at this site results in a change from arginine to glycine in the subunit. The adenosine at the R/G site is mismatched with a cytidine, and the conclusion of a recent study on selectivity was that the mismatch directs the enzyme to the specific site of interaction (Kallman et al. 2003). Further, an A-C mismatch has been shown to be preferred over A-A or A-G mismatches or A-U base pairs for efficient editing in most cases (Wong et al. 2001). A few base pairs downstream of the R/G editing site there is another adenosine–cytidine mismatch, hereafter referred to as the 46 site, but this adenosine has not been proven to be deaminated in vivo (Fig. 1 ▶). The nearest neighbors for the R/G editing site are as follows, with the deaminated adenosine in bold: 5′-AAG-3′ and for the 46 site 5′-GAA-3′. This is in favor of the R/G site according to the general nearest neighbor preferences for ADAR2. Another difference between the two sites is the existence of a G•U wobble pair, 2 bp away from the mismatched R/G site.
FIGURE 1.
Part of the sequence for glutamate receptor B pre-mRNA. The boxed areas represent the simulated heptamers; the two sites have been simulated both with the mismatched base pair A-C (46-wt and R/G-wt) and with a Watson–Crick base pair A-U (46-mut and R/G-mut), shown in bold.
Little is known about substrate activation and the mechanism of binding and deamination. There are sequence similarities between ADARs and DNA methyltransferases that use a base flipping mechanism (Hough and Bass 1997). It has been proposed that the adenosine flips out of the helix before deamination for several reasons. If a 2-aminopurine replaces the adenosine at the R/G site, ADAR2 induces an increase in fluorescence, which suggests base flipping before A to I editing occurs (Stephens et al. 2000). The proposed movement of an attacking water for hydrolytic deamination of adenosine makes it likely that it is a base flipping reaction where the reactive nucleotide is flipped out of the duplex (Yi-Brunozzi et al. 2001).
Opening of bases within double-stranded DNA and RNA is a common event in the cell. Enzymes that would not otherwise have access to the bases can after opening perform modifications and alterations. Base-pair opening has been studied experimentally through NMR and crystallography (for review, see Cheng and Roberts 2001), but since it occurs on a millisecond time scale (Leroy et al. 1988; Folta-Stogniew and Russu 1994; Snoussi and Leroy 2001), the pathway is not altogether understood. A theoretical study showed that after about a 50° rotation of the bases thymine and guanine the imino protons become accessible (Giudice and Lavery 2003). For a base to achieve a fully flipped state with 180° rotation the time scale becomes much longer (Spies and Schowen 2002).
The first crystal structure of a protein bound to DNA with a flipped base was of Hha I methyltransferase (Klimasauskas et al. 1994). The protein binds to the major groove of the DNA, suggesting a minor groove pathway for the opening event. Soon after, another crystal structure was solved, uracil DNA glycosylase, where the protein binds to the minor groove, presenting the idea of a major groove flipping (Slupphaug et al. 1996). Base-pair opening has been studied theoretically in nucleic acids for some time using molecular dynamics simulations (Cubero et al. 1999). The main objective of recent calculations has been to determine the pathway and the free energy changes involved when a Watson–Crick base pair is being broken (Giudice et al. 2001; Banavali and MacKerell 2002; Varnai and Lavery 2002; Giudice and Lavery 2003; Pan and MacKerell 2003). In these studies the major groove pathway seems to be slightly more favorable although both pathways are feasible. Most studies have been performed on DNA Watson–Crick base pairs and only a few recent studies have been performed on the opening of RNA bases (Giudice and Lavery 2003) or a DNA wobble pair (Varnai et al. 2004). A comparison between dsDNA and dsRNA flexibility (Pan and MacKerell 2003) indicates that the base pairs in dsRNA undergo spontaneous opening whereas this could not be seen for DNA.
We have chosen to study the mechanism of base flipping in an ADAR editing substrate by molecular dynamics (MD) simulation, both with the adenosine free to start flipping out spontaneously and using umbrella sampling to force the adenosine to flip out, to study differences in free energy for base flipping in different sequences. We have compared the R/G editing site, both the wild-type A-C pair and a mutated A-U pair, with the 46 site, also with the wild-type sequence and a mutated A-U pair, in dsRNA free in solution. The aim of this investigation was to better understand the site selectivity of ADARs. It is conceivable that ADAR2 recognizes an already flipped out adenosine, in analogy with the suggestion that uracil DNA glycosylase uses the spontaneous opening of DNA base pairs to search for uracil (Cao et al. 2004), or that it recognizes bases near the mismatch at the R/G site that differ from the bases near the 46 site.
It is possible that the selectivity of ADAR2 is not dependent on how easily the adenosine flips out. However, in the absence of a structure of ADAR2, structural differences between an editing substrate and a nonsubstrate can give us further knowledge about the mechanism of ADAR substrate recognition.
RESULTS
Free molecular dynamics simulations
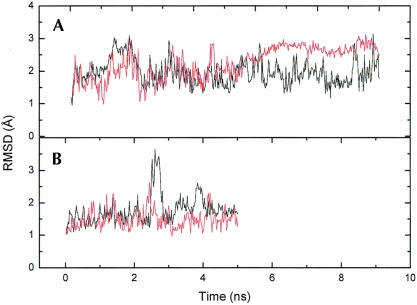
The selectively edited R/G site and the nonedited 46 site are both contained in the helix of the hairpin recognized by ADAR2. Five sequences were used in the MD simulations, four heptamers and a longer one with 17 bp. The four RNA heptamers are the R/G site, with the (a) wild-type A-C base pairing (R/G-wt) and the (b) mutated A-U base pair (R/G-mut), the 46 site, with the (c) wild-type A-C pair (46-wt) and the (d) mutated A-U pair (46-mut) (Fig. 1 ▶). The longer sequence includes both the R/G-wt and 46-wt sequence (with the wild-type A-C pair) and also the 3 bp in between, a total of 17 bp, and was simulated in three independent runs of 6 ns each. The two shorter wt-RNA sequences were simulated for 10 ns while the mutated ones, with the correct Watson–Crick pair, were simulated for 5 ns. The fluctuations in the root-mean-square deviation (RMSD) for the 46-wt coordinates against starting coordinates were significantly reduced after ~5 ns, and we therefore only used the first 5 ns of this simulation for the analysis (Fig. 2 ▶).
FIGURE 2.
Root mean square deviation for the coordinates during the MD simulations of the heptamers. (A) the wt R/G (black) and 46 (red) sequences, and (B) the mut R/G (black) and 46 (red) sequences.
The main focus is on the heptamer simulations. The full-length RNA molecule exhibits the same pattern of behavior as the heptamers and will only be mentioned when there are significant differences.
Overall structure and stability
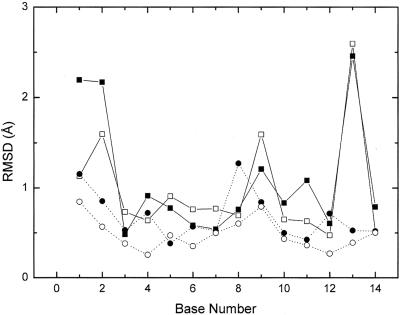
The evolution of the RMSD with time (Fig. 2 ▶) shows that the simulations are stable, but there are some local structural changes. At ~2.7 ns R/G-mut shows a rapid increase in RMSD, which is due to a stretching of the entire molecule, but shortly after, the molecule relaxes back to the original structure. This stretching was also seen in one of the simulations of the longer sequence (data not shown). For both wild-type sequences the average RMSD is around 1.9 Å while for R/G-mut it is 1.7 Å and for 46-mut it is 1.5 Å, including all atoms. From the RMSD per base between the average simulated structures and the starting structures it can be seen that the two 46 sequences have a lower RMSD compared to the R/G sequences with the exception of nucleotide 8, which is positioned at the end of 46-wt. (Fig. 3 ▶). It is also notable that the R/G sequences have a large RMSD for U2, C9, and G13 due to an interaction between C9 and the wobble pair U2•G13 (see below). R/G-wt also has a large RMSD for U1, again a nucleotide at the end of the helix.
FIGURE 3.
Root mean square deviation on the heptamer average structures of the bases. Filled squares with solid lines are R/G-wt, open squares with solid lines are R/G-mut, filled triangles with dotted lines are 46-wt, and open circles with dotted lines are 46-mut.
Base mobility
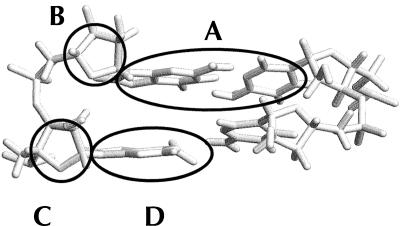
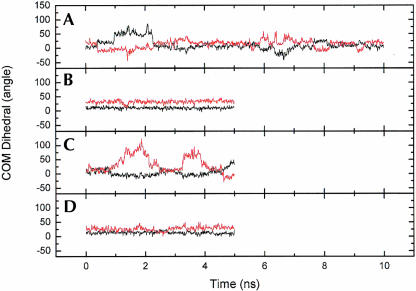
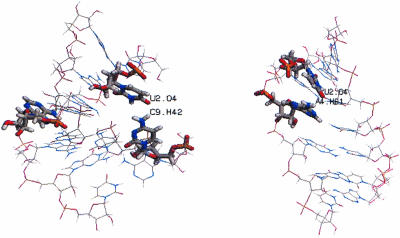
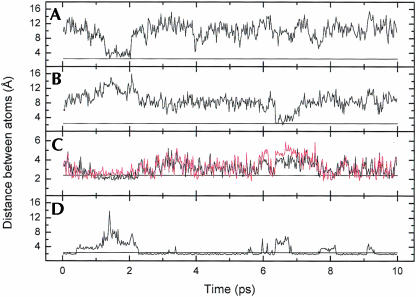
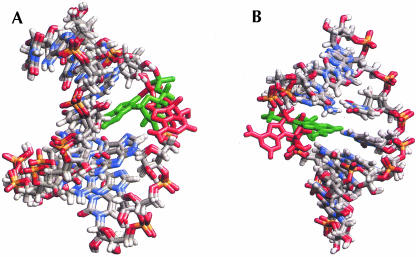
Flipping out motions of the bases at the R/G and 46 sites were analyzed using a pseudo-dihedral angle (Banavali and MacKerell 2002) defined by the centers of mass of the following four units (Fig. 4 ▶): (A) the base pair 3′ to the flipping base, (B) the sugar attached to the 3′ base, (C) the sugar attached to the flipping base, and (D) the flipping base. The same angle (COM dihedral) is also used as the reaction coordinate in the free energy calculations below. For A4 the COM dihedral is positive for minor groove flipping and negative for major groove flipping while for C11/U11, on the facing strand, the definition of the dihedral is reversed, causing a positive dihedral to correspond to flipping through the major groove and a negative dihedral to flipping through the minor groove. This means that opposite signs of A4 and C11 indicate that both bases have moved out into the same groove. In the COM dihedral analysis we can see that in R/G-wt A4 moves out from the stack twice, after ~1 ns and 6.5 ns, respectively (Fig. 5A ▶). The snapshots in Figure 6 ▶ show in the left picture, the first event of flipping, that C9 turns toward U2 and in the right picture, in the second event, that A4 turns toward U2. When C9 turned toward U2, the helix became more bent than before and therefore gave A4 an opportunity to start flipping out through the minor groove (COM angle of 75°), whereas when A4 moved out toward U2 it moved through the major groove (COM angle of −25°). The curvature of the helices was analyzed with the program CURVES (Lavery 1988) and it showed that when A4 moved toward the minor groove the shortening of the helix was ~40% compared to the starting structure, indicating a major bend in the helix. When A4 moved through the major groove the bending was less severe, with a helix shortening of ~23%.
FIGURE 4.
The COM dihedral is defined by the centers of mass of (A) the base pair 3′ to the flipping base, (B) the sugar attached to the 3′ base, (C) the sugar attached to the flipping base, and (D) the flipping base.
FIGURE 5.
COM dihedral for A4 (black) and C11/U11 (red) during the heptamer MD simulations. (A) R/G-wt, (B) R/G-mut, (C) 46-wt, and (D) 46-mut.
FIGURE 6.
R/G-wt sequence at two different times when A4 is out of the helix. Snapshots at 2 ns (left) and at 6.5 ns (right).
The base flipping motions of A4 and C11 are clearly correlated in R/G-wt and 46-wt and occur over a wider range of the COM dihedral for A4 in R/G-wt (Fig. 7 ▶). It can also be seen that for 46-wt C11 is much more flexible than A4. There is a correlation of movement though between the two bases into the same groove: When the cytidine moves from its original position into the major groove, the adenosine also moves to the major groove but it only changes the COM angle by a 10th compared to the cytidine, as can be the curvature only increases by ~7% and there is no apparent change in the overall structure. For both mutated sequences the COM angle is very stable, indicating no movement of the bases.
FIGURE 7.
Correlation plots of the COM dihedral for A4 and C/U11 in (A) R/G-wt (the solid line is the linear regression line with correlation coefficient R = −0.72 and slope = −0.53), (B) 46-wt (R = −0.73, slope = −1.85), (C) R/G-mut, and (D) 46-mut. Movement toward the minor groove corresponds to COM > 0 for the adenine base and to COM < 0 for the C or U base on the opposite strand.
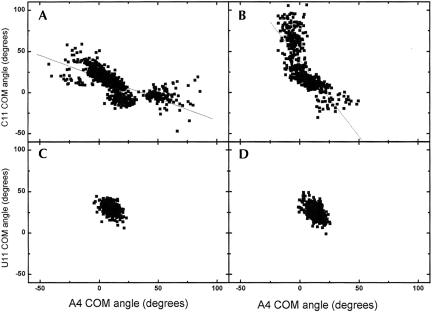
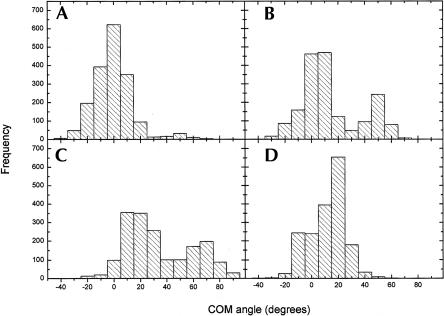
The flexibility of the mismatched bases seen for the heptamers was also seen in the three 17-bp simulations. A histogram with the three simulations combined was constructed from sampling the COM angles every 10 ps of the simulations. The highest frequencies for A4 in the 46 site are around 0°, which indicates that the base is mostly in the stack (Fig. 8A ▶). A4 in the R/G site on the other hand has a tendency to move toward the minor groove with a high frequency at 50° (Fig. 8B ▶). As seen for the heptamers, C11 in the 46 site moves out through the major groove and is more flexible than C11 in the R/G site (Fig. 8C,D ▶).
FIGURE 8.
Combined histogram for the three 17-bp simulations showing the frequencies for a mismatched base in a certain COM angle. (A) A4 at the 46 site, (B) A4 at the R/G site, (C) C11 at the 46 site, and (D) C11 at the R/G site.
Hydrogen bonds
During the 5-ns trajectory there is a hydrogen bond between A4 N1 and C11 H41 in 46-wt for ~50% of the time (Fig. 9C ▶). This bond is weakened significantly at the two occasions C11 is moving from the helix. In Figure 9B ▶ it can be seen that this event is highly correlated with the forming of the new hydrogen bonds C11 H2′–C12 O5′ and C11 H2′–C12 O4′. The distance between H2′ and either O5′ or O4′ is about the same throughout the trajectory, indicating that H2′ alternates binding to O5′ and O4′. It has been proposed that hydrogen bonding to nearby base pairs as a base moves out along a groove constitutes a novel mechanism of sequence dependence for dynamical processes involving DNA (Banavali and MacKerell 2002), and that for RNA the 2′OH(n)-O4′(n + 1) bond is responsible for the stabilization of an already open state rather than for the opening of the base pair (Pan and MacKerell 2003). The hydrogen bond between A4 N1 and C11 H41 in the R/G-wt sequence was also weakened twice in the simulation, at the same time as the spontaneous flipping occurred (Fig. 10 ▶). Like for the 46-wt, atoms in the sugar or backbone hydrogen bond to the nearest 3′ or 5′ sugar or backbone. In the R/G-wt simulation, the first motion of A4 occurred after ~1 ns and lasted for ~1 ns. When studying the hydrogen bonds of the involved bases between 1 and 2 ns it was found that A4 lost the hydrogen bond to the mismatched C11. However, a new bond between A4 H61 and U12 O2 was present for ~40% during this time, stabilizing the helix. U2 was out of the helix but still had contact with its wobble-pairing partner G13. C9 moved out of the helix and its H42 hydrogen bonded with U2 O4. On the other occasion when A4 was out of the helix A4 turned toward U2, making a hydrogen bond between A4 H61 and U2 O4.
FIGURE 9.
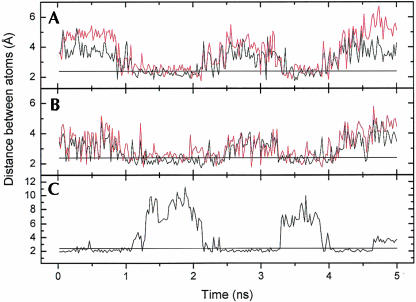
Distances between atoms during the simulations in the 46-wt sequence. The straight line is at 2.4 Å to indicate the maximum distance for hydrogen bonds to occur. (A) C11 H2′–C12 O5′ (black) and C11 H2′–C12 O4′ (red), (B) A4 H2′–A5 O5′ (black) and A4 H2′–A5 O4′ (red), and (C) A4 N1–C11 H41.
FIGURE 10.
Distances between atoms during the simulation in the R/G-wt sequence. The straight line is at 2.4 Å to show where there are hydrogen bonds present. (A) U2 O4–C9 H42, (B) A4 H61–U2 O4, (C) A3 H2′–A4 O5′ (black) and A3 H2′–A4 O4′ (red), and (D) A4 N1–C11 H41.
Forced base flipping
Potential of mean force (PMF)
The free energy calculations were performed according to a protocol that has been shown to work for flipping out one of the bases in a G-C base pair in DNA (Banavali and MacKerell 2002; Huang et al. 2003; Horton et al. 2004); this also allows a direct comparison to be made between our RNA data and the published DNA results. Starting structures for each of the umbrella sampling windows were prepared without any disturbance to the rest of the structure (Fig. 11 ▶).
FIGURE 11.
Superposition of the R/G-wt initial model structure on the umbrella sampling starting structures with the adenosine base (red) flipped out through (A) the minor groove (+120° COM) and (B) the major groove (−120° COM). The position of the adenosine in the initial model is shown in green.
The A4 base was moved out of the stack due to a force on the COM dihedral, through both the major and the minor grooves. If ADAR2 in fact uses a base flipping mechanism in order to perform the deamination, the assumption was that the adenosine at the most commonly deaminated site, the R/G site, would have a lower energy barrier than the 46 site. We also predicted that the mutated sequences would have a higher energy barrier than the wild-type because of the second hydrogen bond between the bases. The A4 base started at a COM dihedral of 0°, and was forced to move to the fully flipped states at +180° (minor groove) and −180° (major groove). Recent studies show that the preferred pathway for base flipping is via the major groove (Giudice et al. 2003; Huang et al. 2003), although the free energy difference between the pathways is small. The wild-type structures have the most favorable state at a COM angle of ~5° whereas the mutated sequences have an energy minimum at ~10° (Fig. 12 ▶).
FIGURE 12.
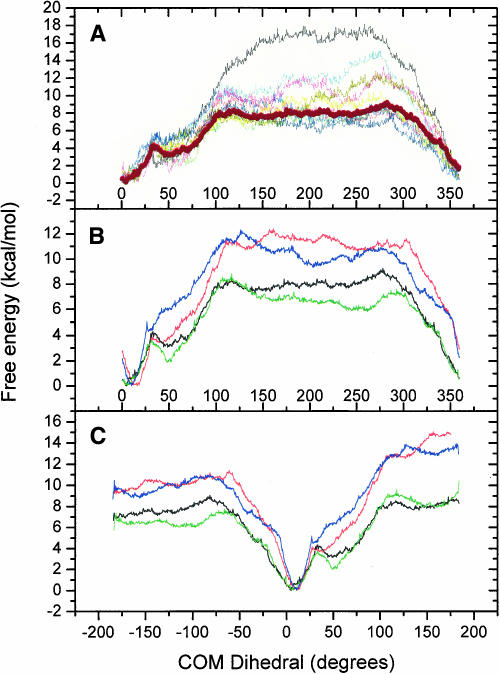
The potential of mean force for the four heptamers (A) the 46-wt sequence in 20-ps intervals for the 200-ps production run. 0–20 ps (black), 20–40 ps (cyan), 40–60 ps (magenta), 60–80 ps (yellow), 80–100 ps (dark yellow), 100–120 ps (red), 120–140 ps (green), 140–160 ps (navy), 160–180 ps (purple), 180–200 ps (blue). The thick wine colored line is 40–200 ps used for analysis. (B) Periodic calculations and (C) nonperiodic calculations. For B and C the black curve is 46-wt, red is 46-mut, green is R/G-wt, and blue is R/G-mut.
The first 40 ps of each simulation window were considered equilibration and the following 160 ps were used for analysis. Figure 12A ▶ shows the 46-wt simulation divided into 20-ps intervals where the thick line corresponds to the data used in Figure 12, B and C ▶. The free energy was calculated with enforced periodicity (Fig. 12B ▶) to allow comparison with a previous study (Banavali and MacKerell 2002) and without periodicity (Fig. 12C ▶). COM dihedral angle values of 180° and −180° formally represent the same state along this reaction coordinate, but by visual inspection it can clearly be seen that the structures are very different. Therefore the system cannot be treated as if the base has flipped 360° but rather as if it has started to flip out through either the major or the minor groove and the following results are based on the calculations without the enforced periodicity. However, the overall appearance of the curves is very similar and the general conclusions can be drawn from either Figure 12B or 12C ▶.
The shape of the PMF curve for flipping the base through the minor groove is different from the major groove, especially for the wild-type sequences (Fig. 12C ▶). In the major groove pathway the energy quickly rises for all sequences and flattens out at ~−60° while for the minor groove the curves have a gentler slope, with a significant local minimum at ~50° for the wild-type sequences before flattening out at ~100°. The plateaus are reached when there are no direct or water mediated hydrogen bonds left between A4 and the opposing base. Within the R/G sequences and the 46 sequences the ones with a mismatched A4 flipped out more easily than the ones with A-U base pairs, which is as expected because of the second hydrogen bond. The second hydrogen bond is also responsible for the steeper slope in the minor groove when compared to the wild-type simulations. The different stages in the PMF curves correspond to direct or water mediated hydrogen bonds between A4 and U/C11. For example, the rise in energy after the local minimum in the minor groove pathway for the wild-type sequences is due to the presence of a new hydrogen bond between A4 H61 and C11 N3 that needs to be broken.
When the base is moving out from the stack it acquires new hydrogen bonds to nearby bases as could also be seen in the regular MD, starting from ~20° with a maximum occurrence at ~50°, which corresponds to the local minimum. At this stage non-Watson–Crick hydrogen bonds are formed between A4 H61 and C/U11 O2 for R/G, A4 H61 and C11 N3 for 46-wt, and A4 H62 and U11 O4 for 46-mut. Additional strong stabilizing hydrogen bonds occur between A4 and the nearest 5′ base, an event described previously, for all four heptamers (described earlier for the 46 site, 3′ described for R/G). U2 in the wobble pair turns toward C9 at 60°, 110°, and 140° in a way similar to that during the regular MD. Despite the increased flexibility in the R/G site molecule compared to the 46 site, the same amount of free energy is needed for the base to flip the full 180°. Two differences between R/G-wt and 46-wt are worth mentioning though: (1) the local minimum at 50° is a little more than 1 kcal/mol higher for 46-wt and (2) 46-wt requires more energy than R/G-wt to flip from −50° to −180°.
An initial set of umbrella sampling simulations with added positional harmonic restraints on the ends of the helices (between U2-G13 and U6-A9 in addition to the outermost base pairs as described in Materials and Methods) was also performed (data not shown). Since the free energy curves were essentially the same as the ones described here, indicating a stable method, we have only presented the analysis of the less restrained systems.
DISCUSSION
To better understand site selective recognition by the ADAR enzymes we have in this study investigated four different 7-bp sequences in two ways: molecular dynamics simulations and free energy calculations with umbrella sampling. The two sites, R/G and 46, are in contexts of similar nucleotide composition, with the same number of G-C base pairs and one more A-U base pair in the 46 sequence. One major difference between the two sequences is a wobble pair situated 2 bp away from the mismatched R/G site. Since the R/G site is deaminated by ADAR2 in vivo and the 46 site is not, the hypothesis was that we would see a different behavior of the adenosine base between the sequences and the possible role of the wobble pair in the structural differences. In a recent study where damaged DNA (a G•U wobble pair) was simulated it was suggested that the damaged DNA has increased flexibility, which would lower the energy barrier for flipping (Fuxreiter et al. 2002). The wobble pair in our simulations creates even more flexibility in the strand and helps to slightly bend the molecule in the free MD simulations. However, in the free energy calculations the difference between the mismatched sequences and the Watson–Crick base paired sequences was not as great as in the aforementioned study. During the simulation of the wild-type R/G the adenosine in the mismatched base pair flipped out of the stack spontaneously on two occasions, flipping through both the minor and major grooves. When A4 turned toward U2 in the wobble pair it was less accessible compared to when U2 turned toward C9 and opened a way for the adenosine to flip out. This spontaneous flipping could not be seen in any of the other sequences. On the other hand, in the 46-wt simulation the cytidine in the mismatched pair moves in and out of the stack. The cyti-dine is surrounded by pyrimidines whereas the adenosine is surrounded by purines, which should allow more flexibility for the cytidine due to the weaker stacking interactions in the pyrimidine stack (Norberg and Nilsson 1995). This argument is also true for the R/G site but here we could see a greater flexibility in the adenosine, possibly facilitated by the wobble pair interactions.
The simulations of the longer sequence including both the R/G and the 46 sites confirmed the results found in the heptamer simulations that the adenosine at the R/G site flips more easily than the cytidine, whereas for the 46 site it is the opposite. Spontaneous flipping/breathing of bases in dsRNA on a nanosecond time scale was reported, even for bases in a Watson–Crick base pair (Pan and MacKerell 2003). However, in our simulations spontaneous flipping/ breathing was only found for the bases in the A-C mismatches.
Looking at the free energy curves it can be concluded that the most favorable pathway for the start of the flipping is through the minor groove, due to the local minimum at ~50°. The deepest minimum is found for the R/G-wt, which corresponds well with the histogram for the 17-bp sequences in which there was a high frequency at 50° for R/G-wt. For the enzyme to have enough access to the base for the deamination process it might be sufficient if the base is only flipped out ~50°. Base flipping more than 50° is not a spontaneous event in these simulations and the base might need the enzyme for this kind of movement. For all sequences there seems to be a slightly higher barrier to reach a stable fully flipped out state going through the minor groove, with an ~3 kcal/mol difference for the mutated sequences and ~1 kcal/mol difference for the wild type compared to the major groove. Since the starting structures of the fully flipped state, ±180°, do not have the same conformation it might be reasonable to assume that the pathways are not the same and therefore the free energy curves should also differ. Overall, the free energies obtained here are comparable to studies using the same protocol for DNA (Banavali and MacKerell 2002) or different protocol and force-field (Giudice and Lavery 2003). Comparing our results with the previous study using the same procedure (Banavali and MacKerell 2002) it could be concluded that flipping a Watson–Crick paired adenosine requires a few kilocalories per mole less than either a cytidine or guanosine, consistent with the smaller number of hydrogen bonds in the A-U pair or in a DNA G-T wobble pair (Varnai et al. 2004).
Since there is no determined structure of the simulated sequences the arrangement of the hydrogen bonds within the mismatch and in the wobble pair is unknown. When the bases were mutated to generate the mismatch and the wobble pair there were no hydrogen donors and acceptors sufficiently close to cause formation of hydrogen bonds. The hydrogen bonds found after minimization represent only one of several possible arrangements; the way the hydrogen bonds are arranged in the mismatch could in principle affect how easily the adenosine moves out, with the behavior in the R/G-mut and 46-mut sequences as limiting cases. We chose to use these structures instead of forcing the bases into some other hydrogen bonding conformation, and in the free simulations the systems did return to this arrangement after all the base excursions that we observed.
Conclusions
A4 in the R/G sequence is the base that is most prone to move out from the stack, mainly populating a conformation around 50° into the minor groove, where it is stabilized by a non-Watson–Crick hydrogen bond between A4 and C11 and hydrogen bonds between atoms in the sugar ring to the nearest 5′ base. This amount of movement from the stack might be enough for the enzymatic reaction to take place.
The wobble pair is the most significant difference between the sequences around the R/G editing site and the 46 site. It is involved in the two occasions when the target for ADAR2, adenosine 4, is moving out of the helix and also creates a bent molecule during the forced base flipping. However, the wobble pair alone is probably not the cause of base flipping. When ADAR2 binds to the dsRNA it is most likely that it induces some structural change that creates access to the adenosine to be deaminated. One possibility is that the first bound ADAR2 monomer induces the flipping whereas the second monomer performs the actual deamination.
MATERIALS AND METHODS
Free molecular dynamics simulation
The RNA helices were created in a double helix A RNA conformation (Arnott et al. 1972). Initially all bases were generated as Watson–Crick (WC) base pairs. The wobble pair was generated as C-G, the C was then mutated to a U, and the mismatch was generated as A-U followed by mutation of U to a C. Hydrogens were added as previously described (Brünger and Karplus 1988). There were no hydrogen bonds present between the bases in the mismatch and the wobble pair when the sequences were generated. After the minimization (see below) the following hydrogen bonds were found in the wobble pair: U2 O2–G13 H1 and U2 H3–G13 O6; and in the mismatch: A4 N1–C11 H41. All calculations were performed with the program CHARMM (Brooks et al. 1983) using the CHARMM27 nucleic acid force field (Foloppe and MacKerell 2000; MacKerell and Banavali 2000). The heptamers were centered in boxes of TIP3P (Jorgensen et al. 1983) minimized water with dimensions 46 × 36 × 36 Å3. The longer sequence was centered in a truncated octahedron, of the size that it fits in a cube of side length 75 Å. The whole system was then minimized with 100 steps of steepest descent plus 100 steps of adopted basis set Newton–Raphson. Water molecules within 1.8 Å of the RNA were deleted. Water molecules were replaced by sodium ions to create a neutral system, 12 for the heptamers and 32 for the larger molecule. The final system for the heptamers consisted of 5865 atoms, including the 12 sodium ions and 1986 water molecules. The larger system consisted of a total of 19,987 atoms, 32 sodium atoms and 6322 water molecules included. Periodic boundary conditions were applied and SHAKE (Ryckaert et al. 1977) was used on all bonds involving hydrogens. Electrostatic interaction energies and forces were smoothly shifted to zero at 12 Å. The nonbonded list was constructed using a 14-Å cutoff and was updated every time an atom moved >1 Å since the last update. During the simulation the hydrogen bonds of the 2 bp at the ends of the helix (1 and 7 in the short helices, 1 and 17 in the long helix) were assigned a noe-restraining potential with force constant 10 kcal/mol/Å2 to keep the helix ends from fraying. The system was heated from 50 to 300 K for 10 ps. The heptamers were simulated for 5 ns (mutated strands) and for 10 ns (wild-type strands). The 17-bp sequence was simulated in three independent runs with different starting velocities for 6 ns, resulting in a total of 18 ns. During the simulation the temperature was checked to make sure it stayed within ±10 K. A time step of 0.002 ps was used for integrating Newton’s equations of motion. Coordinates were stored every 1.0 ps for analysis.
Forced base flipping of adenosine in the helix
The method used has been described previously (Banavali and MacKerell 2002) and will only briefly be described here. The same system setup as described above used in the MD simulation was also used in the forced base flipping. The COM dihedral that was used as the reaction coordinate (Fig. 4 ▶) was also used in the base mobility analysis. A harmonic potential, wi(x) = ki(x − xi)2, was used for the umbrella sampling along the COM dihedral (x), with the reference value (xi) in window i going from 0° to ±180° in 5° increments/decrements. First a set of initial structures was generated by 0.5-ps simulation for each window with a force constant ki of 10,000 kcal/mol/rad2. The final structure in each window was used to start the simulation for the next window.
The production run for each window was started with a 5-ps equilibration step and 200-ps simulation. A total of 72 windows were simulated resulting in 14.4 ns of production simulation for the 0–360° of flipping. During the production the harmonic force constant was reduced to 1000 kcal/mol/rad2. All simulations were performed at 300 K. The potential of mean force was constructed from the COM dihedrals in all windows using the Weighted Histogram Analysis Method (Kumar et al. 1992; Boczko and Brooks 1993). Of the 200 ps of simulation in each window the first 40 ps were considered equilibration time and the last 160 ps were used for analysis (Fig. 12A ▶).
Methods and parameters used for analyses
Hydrogen bonds were analyzed from the trajectories using the criterion that the hydrogen donor–acceptor distance is <2.4 Å, and a water bridge was considered to be present between two groups that simultaneously formed hydrogen bonds to the same water molecule. The occupancy of a hydrogen bond during a portion of the simulation was calculated as the fraction of snapshots taken at 2-ps intervals that the bond was present during that part of the simulation. The RMSD from the starting structure were calculated after superimposing all heavy atoms on the starting structures for each coordinate set in the trajectories.
Acknowledgments
We are grateful to the Swedish Research Council for financial support.
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.7147805.
REFERENCES
- Arnott, S., Smith, P.J.C., and Chandrasekharan, R. 1972. CRC handbook of biochemistry and molecular biology: Nucleic acids. CRC Press, Cleveland, OH.
- Bass, B.L. 2002. RNA editing by adenosine deaminases that act on RNA. Annu. Rev. Biochem. 71: 817–846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhalla, T., Rosenthal, J.J.C., Holmgren, M., and Reenan, R. 2004. Control of human potassium channel inactivation by editing of a small mRNA hairpin. Nat. Struct. Biol. 11: 950–956. [DOI] [PubMed] [Google Scholar]
- Boczko, E.M. and Brooks, C.L. 1993. Constant-temperature free-energy surfaces for physical and chemical processes. J. Phys. Chem. 97: 4509–4513. [Google Scholar]
- Brooks, B.R., Bruccoleri, R.E., Olafson, B.D., States, D.J., Swaminathan, S., and Karplus, M. 1983. CHARMM: A program for macromolecular energy minimization and dynamics calculations. J. Comp. Chem. 4: 187–217. [Google Scholar]
- Brünger, A.T. and Karplus, M. 1988. Polar hydrogen positions in proteins: Empirical energy placement and neutron diffraction comparison. Proteins 4: 148–156. [DOI] [PubMed] [Google Scholar]
- Burns, C.M., Chu, H., Reuter, S.M., Hutchinson, L.K., Canton, H., Sanders-Bush, E., and Emeson, R.B. 1997. Regulation of serotonin-2C receptor G-protein coupling by RNA editing. Nature 387: 303–308. [DOI] [PubMed] [Google Scholar]
- Cao, C., Jiang, Y.L., Stivers, J.T., and Song, F. 2004. Dynamic opening of DNA during the enzymatic search for a damaged base. Nat. Struct. Mol. Biol. 11: 1230–1236. [DOI] [PubMed] [Google Scholar]
- Cheng, X. and Roberts, R.J. 2001. AdoMet-dependent methylation, DNA methyltransferases and base flipping. Nucleic Acids Res. 29: 3784–3795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho, D.S., Yang, W., Lee, J.T., Shiekhattar, R., Murray, J.M., and Nishikura, K. 2003. Requirement of dimerization for RNA editing activity of adenosine deaminases acting on RNA. J. Biol. Chem. 278: 17093–17102. [DOI] [PubMed] [Google Scholar]
- Cubero, E., Sherer, E.C., Luque, F.J., Orozco, M., and Laughton, C.A. 1999. Observation of spontaneous base pair breathing events in the molecular dynamics simulation of a diflourotoluene-containing DNA oligonucleotide. J. Am. Chem. Soc. 121: 8653–8654. [Google Scholar]
- Foloppe, N. and MacKerell, A.D. 2000. All-atom empirical force field for nucleic acids: I. Parameter optimization based on small molecule and condensed phase macromolecular target data. J. Comp. Chem. 21: 86–104. [Google Scholar]
- Folta-Stogniew, E. and Russu, I.M. 1994. Sequence dependence of base-pair opening in a DNA dodecamer containing the CACA/ GTGT sequence motif. Biochemistry 33: 11016–11024. [DOI] [PubMed] [Google Scholar]
- Fuxreiter, M., Luo, N., Jedlovszky, P., Simon, I., and Osman, R. 2002. Role of base flipping in specific recognition of damaged DNA by repair enzymes. J. Mol. Biol. 323: 823–834. [DOI] [PubMed] [Google Scholar]
- Gallo, A., Keegan, L.P., Ring, G.M., and O’Connell, M.A. 2003. An ADAR that edits transcripts encoding ion channel subunits functions as a dimer. EMBO J. 22: 3421–3430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giudice, E. and Lavery, R. 2003. Nucleic acid base pair dynamics: The impact of sequence and structure using free-energy calculations. J. Am. Chem. Soc. 125: 4998–4999. [DOI] [PubMed] [Google Scholar]
- Giudice, E., Varnai, P., and Lavery, R. 2001. Energetic and conformational aspects of A : T base-pair opening within the DNA double helix. Chem. Phys. Chem. 2: 673–677. [DOI] [PubMed] [Google Scholar]
- ———. 2003. Base pair opening within B-DNA: Free energy pathways for GC and AT pairs from umbrella sampling simulations. Nucleic Acids Res. 31: 1434–1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higuchi, M., Maas, S., Single, F.N., Hartner, J., Rozov, A., Burnashev, N., Feldmeyer, D., Sprengel, R., and Seeburg P.H. 2000. Point mutation in an AMPA receptor gene rescues lethality in mice deficient in the RNA-editing enzyme ADAR2. Nature 406: 78–81. [DOI] [PubMed] [Google Scholar]
- Horton, J.R., Ratner, G., Banavali, N.K., Huang, N., Choi, Y., Maier, M.A., Marquez, V.E., MacKerell Jr., A.D., and Cheng X. 2004. Caught in the act: Visualization of an intermediate in the DNA base-flipping pathway induced by HhaI methyltransferase. Nucleic Acids Res. 32: 3877–3886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hough, R.F. and Bass, B.L. 1997. Analysis of Xenopus dsRNA adenosine deaminase cDNAs reveals similarities to DNA methyltransferases. RNA 3: 356–370. [PMC free article] [PubMed] [Google Scholar]
- Huang, N., Banavali, N.K., and MacKerell, A.D. 2003. Protein-facilitated base flipping in DNA by cytosine-5-methyltransferase. Proc. Natl. Acad. Sci. 100: 68–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaikaran, D.C., Collins, C.H., and MacMillan, A.M. 2002. Adenosine to inosine editing by ADAR2 requires formation of a ternary complex on the GluR-B R/G site. J. Biol. Chem. 277: 37624–37629. [DOI] [PubMed] [Google Scholar]
- Jorgensen, W.L., Chandrasekar, J., Madura, J.D., Impey, R.W., and Klein, M.L. 1983. Comparison of simple potential functions for simulating water. J. Chem. Phys. 79: 926–935. [Google Scholar]
- Kallman, A.M., Sahlin, M., and Ohman, M. 2003. ADAR2 A → I editing: Site selectivity and editing efficiency are separate events. Nucleic Acids Res. 31: 4874–4881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klimasauskas, S., Kumar, S., Roberts, R.J., and Cheng, X. 1994. HhaI methyltransferase flips its target base out of the DNA helix. Cell 76: 357–369. [DOI] [PubMed] [Google Scholar]
- Kumar, S., Bouzida, D., Swendsen, R.H., Kollman, P.A., and Rosenberg, J.M. 1992. The weighted histogram analysis method for free-energy calculations on biomolecules. 1. The method. J. Comp. Chem. 13: 1011–1021. [Google Scholar]
- Lavery, R.S.H. 1988. The definition of generalized helicoidal parameters and of axis of curvature for irregular nucleic acids. J. Biomol. Struct. Dyn. 6: 63–91. [DOI] [PubMed] [Google Scholar]
- Lehmann, K.A. and Bass, B.L. 2000. Double-stranded RNA adenosine deaminases ADAR1 and ADAR2 have overlapping specificities. Biochemistry 39: 12875–12884. [DOI] [PubMed] [Google Scholar]
- Leroy, J.L., Charretier, E., Kochoyan, M., and Gueron, M. 1988. Evidence from base-pair kinetics for two types of adenine tract structures in solution: Their relation to DNA curvature. Biochemistry 27: 8894–8898. [DOI] [PubMed] [Google Scholar]
- Lomeli, H., Mosbacher, J., Melcher, T., Hoger, T., Geiger, J.R., Kuner, T., Monyer, H., Higuchi, M., Bach, A., and Seeburg, P.H. 1994. Control of kinetic properties of AMPA receptor channels by nuclear RNA editing. Science 266: 1709–1713. [DOI] [PubMed] [Google Scholar]
- Nishikura, K., Yoo, C., Kim, U., Murray, J.M., Estes, P.A., Cash, F.E., and Liebhaber, S.A. 1991. Substrate specificity of the dsRNA unwinding/modifying activity. EMBO J. 10: 3523–3532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norberg, J. and Nilsson, L. 1995. Stacking free-energy profiles for all 16 natural ribodinucleoside monophosphates in aqueous-solution. J. Am. Chem. Soc. 117: 10832–10840. [Google Scholar]
- Pan, Y. and MacKerell Jr., A.D. 2003. Altered structural fluctuations in duplex RNA versus DNA: A conformational switch involving base pair opening. Nucleic Acids Res. 31: 7131–7140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polson, A.G. and Bass, B.L. 1994. Preferential selection of adenosines for modification by double-stranded RNA adenosine deaminase. EMBO J. 13: 5701–5711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryckaert, J.P., Ciccotti, G., and Berendsen, H.J.C. 1977. Numerical integration of the Cartesian equations of motion of a system with constraints: Molecular dynamics of n-alkanes. J. Comp. Chem. 23: 327–341. [Google Scholar]
- Slupphaug, G., Mol, C.D., Kavli, B., Arvai, A.S., Krokan, H.E., and Tainer, J.A. 1996. A nucleotide-flipping mechanism from the structure of human uracil-DNA glycosylase bound to DNA. Nature 384: 87–92. [DOI] [PubMed] [Google Scholar]
- Snoussi, K. and Leroy, J.L. 2001. Imino proton exchange and base-pair kinetics in RNA duplexes. Biochemistry 40: 8898–8904. [DOI] [PubMed] [Google Scholar]
- Sommer, B., Kohler, M., Sprengel, R., and Seeburg, P.H. 1991. RNA editing in brain controls a deteminant of ion flow in the glutamate-gated channels. Cell 67: 11–19. [DOI] [PubMed] [Google Scholar]
- Spies, M.A. and Schowen, R.L. 2002. The trapping of a spontaneously “flipped-out” base from double helical nucleic acids by host–guest complexation with beta-cyclodextrin: The intrinsic base-flipping rate constant for DNA and RNA. J. Am. Chem. Soc. 124: 14049–14053. [DOI] [PubMed] [Google Scholar]
- Stephens, O.M., Yi-Brunozzi, H.Y., and Beal, P.A. 2000. Analysis of the RNA-editing reaction of ADAR2 with structural and fluorescent analogues of the GluR-B R/G editing site. Biochemistry 39: 12243–12251. [DOI] [PubMed] [Google Scholar]
- Varnai, P. and Lavery, R. 2002. Base flipping in DNA: Pathways and energetics studied with molecular dynamic simulations. J. Am. Chem. Soc. 124: 7272–7273. [DOI] [PubMed] [Google Scholar]
- Varnai, P., Canalia, M., and Leroy, J.L. 2004. Opening mechanism of G.T/U pairs in DNA and RNA duplexes: A combined study of imino proton exchange and molecular dynamics simulation. J. Am. Chem. Soc. 126: 14659–14667. [DOI] [PubMed] [Google Scholar]
- Wong, S.K., Sato, S., and Lazinski, D.W. 2001. Substrate recognition by ADAR1 and ADAR2. RNA 7: 846–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi-Brunozzi, H.Y., Stephens, O.M., and Beal, P.A. 2001. Conformational changes that occur during an RNA-editing adenosine deamination reaction. J. Biol. Chem. 276: 37827–37833. [DOI] [PubMed] [Google Scholar]