Abstract
In bacteria, translation of mRNAs lacking stop codons produces truncated polypeptides and traps ribosomes in unproductive complexes. Potentially harmful truncated proteins are tagged with short peptides encoded by the mRNA-like domain of tmRNA and targeted for digestion by housekeeping proteases. We show that altered Escherichia coli transfer-messenger RNAs (tmRNAs) produce in vivo fusion proteins with peptide tags that extend far beyond the conventional termination signal of the wild-type tmRNA. Regions of tmRNA capable of serving as templates for protein synthesis include helix 5, as well as pseudoknots 2, 3, and 4. The removal of all six in-frame stop codons negatively affects tmRNA processing, thereby preventing translation of the 3′ portion of the tRNA-like domain. These findings provide evidence that trans-translation can be accompanied by the unfolding of significant portions of the tmRNA molecule. Many of these conformational changes are likely to be required during trans-translation to maintain the ribosomal subunits in close proximity to the tmRNA for monitoring its transit.
Keywords: tmRNA, trans-translation, protein tagging, ribosome rescue, RNA structure
INTRODUCTION
Translation of mRNAs without stop codons stalls ribosomes and produces truncated proteins. To recycle ribosomes and remove the defective proteins, bacteria have developed trans-translation, a surveillance and quality control mechanism which tags the C-termini of partially translated proteins with short peptides to yield fusion proteins that are readily degraded by housekeeping proteases. The peptide tag is encoded within a stable RNA molecule called transfer-messenger RNA (tmRNA) which is transcribed from the ssrA gene (Tu et al. 1995; Keiler et al. 1996).
Genes for tmRNA have been identified in all bacterial as well as certain plastid, mitochondrial, and bacteriophage genomes (Zwieb et al. 2003). The tmRNAs are synthesized as precursors that are processed by RNases BN, D, E, III, P, PH, and T (Makarov and Apirion 1992; Li et al. 1998; Lin-Chao et al. 1999). Comparative sequence analysis (Williams and Bartel 1996; Zwieb et al. 1999b) and chemical probing of Escherichia coli tmRNA (Felden et al. 1997) revealed that the folded tmRNA is composed of three domains. The 3′ and 5′ terminal segments form the tRNA-like domain (TLD). An adenosine-rich segment and a short open reading frame (ORF), demarcated by resume and stop codons, form the mRNA-like domain (MLD). The TLD and ORF are connected by a pseudoknot-rich domain (PKD). Four pseudoknots (designated pk1–pk4 in Fig. 1A ▶) are present in most tmRNAs, but fewer pseudoknots may be contained within the tmRNAs of plastids and endosymbionts (Gueneau de Novoa and Williams 2004).
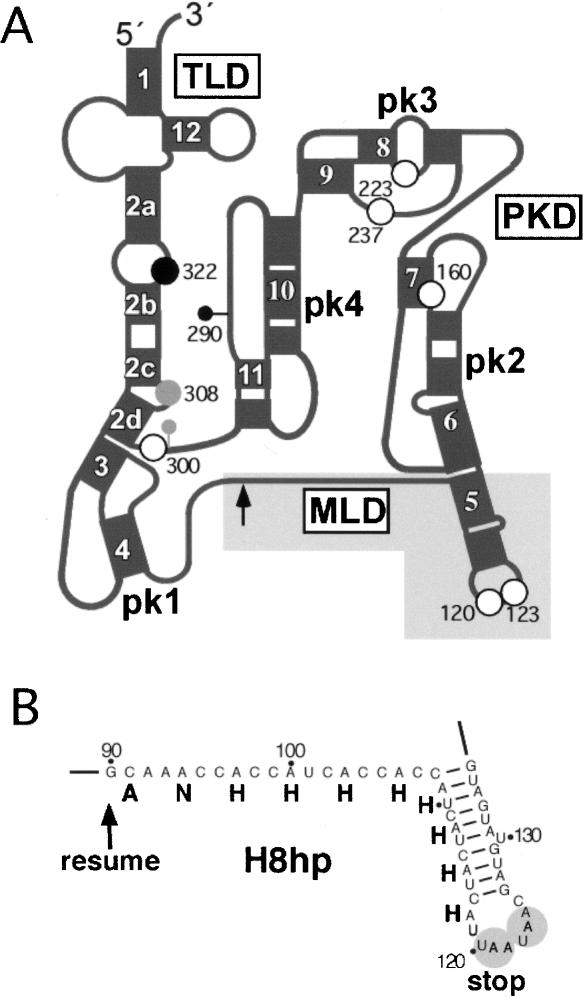
FIGURE 1.
Structural features of E. coli tmRNA. (A) Schematic representation of tmRNA secondary structure. The mRNA-, tRNA-, and pseudoknot-rich domains are denoted as MLD, TLD, and PKD, respectively. Helices and pseudoknots are numbered 1–12 and pk1-pk4, respectively. The position of the resume codon at residue 90 is indicated by an arrow. White dots indicate the six in-frame stop codons at positions 120, 123, 161, 222, 237, and 300. The gray dot indicates a stop generated by the insertion of one nucleotide between positions 299 and 300 (gray lollipop). The black dot indicates a stop generated by the insertion of two adenosines between 289 and 290 (black lollipop). (B) Detailed view of the MLD of tmRNA(H8hp) highlighted in gray in panel A. The ANHHHHHHHH sequence denotes protease-resistant, histidine-rich peptide tag. The tandem stop codons 120-UAA-122 and 123-UAA-125 are highlighted.
Numerous studies in E. coli show that trans-translation is initiated by the formation of a ternary complex between the TLD of aminoacylated tmRNA, elongation factor Tu (EF-Tu), and GTP (Rudinger-Thirion et al. 1999; Barends et al. 2000, 2001). This ternary complex binds to the stalled ribosome in a process that is facilitated by protein SmpB and ribosomal protein S1 (Karzai et al. 1999; Wower et al. 2000; Hallier et al. 2004; McGinness and Sauer 2004). Translation resumes and proceeds through elongation to add a short peptide tag to the C-terminus of the defective protein. Upon termination of translation at the tmRNA-provided tandem stop codons located within a loop formed by helix 5 (Fig. 1 ▶), the tagged protein and deacylated tmRNA are released. Cryo-electron microscopy of ribosome-bound tmRNA in the presence and absence of ribosomal protein S1 at an early stage of trans-translation indicates that the TLD, MLD, and pk1 associate intimately with the ribosome, whereas pk2 to pk4 surround the beak of the 30S ribosomal subunit (Valle et al. 2003). How tmRNA is accommodated on the ribosome during the subsequent steps of trans-translation remains unresolved.
Recently, we have developed a very sensitive in vivo assay for investigating the functions of tmRNA in trans-translation (Wower et al. 2004). In this assay, pETrpmA-At-3, a derivative of the pET23a plasmid (Novagen), directs co-expression of tmRNA(H8hp), protein SmpB, and truncated ribosomal protein L27 in the ssrA− E. coli strain IW363. Synthesis of truncated L27 is induced by IPTG and, because tmRNA(H8hp) encodes a peptide tag which is resistant to proteolysis and contains eight histidines, the tagged protein is stable and easily detected in fractionated E. coli lysates, either by staining with Coomassie Blue or the use of anti-His-tag antibodies. Here we have used this strategy to investigate the role of the stop codons and to reveal the mechanism which governs the transit of tmRNA through the ribosome.
RESULTS
Mutations of the tandem stop codons at positions 120 and 123
The tag-encoding ORF in E. coli tmRNA ends with 120-UAA-122 and 123-UAA-125 (Fig. 1A ▶). To examine the contribution of the tandem stop codons to tmRNA-mediated protein tagging, nucleotide changes were introduced into tmRNA(H8hp), an E. coli tmRNA derivative with an altered helix 5 sequence and a modified MLD designed to direct the synthesis of a protease-resistant His-tagged fusion protein (Fig. 1B ▶). Mutant pETrpmA-At-3 plasmids were transformed into the ssrA− E. coli strain IW363. Synthesis of tagged truncated ribosomal protein L27 was induced by treating the cells with IPTG (Wower et al. 2004). Alterations of residues at positions 120, 121, 122, and 123 in tmRNA(H8hp) produced tagged polypeptides with molecular masses that were consistent with the predicted addition of 11 or 12 amino acid tags (Table 1 ▶). Similar high expression levels were observed for all mutant tmRNAs and the unmutated tmRNA(H8hp) (Fig. 2A ▶, lanes 1–7). A maximal tagging efficiency of ∼50% was reached in all investigated constructs.
TABLE 1.
Activities of mutant tmRNAs
| Changes in tmRNA(H8hp) | Processing | Stop | AA in taga | Predicted MW | Tagging |
| none | +++ | U120 | 11 | 12,473 | +++ |
| U120C | +++ | U123 | 12 | 12,601 | +++ |
| A121G | +++ | U120 | 11 | 12,473 | +++ |
| A122C | +++ | U123 | 12 | 12,636 | +++ |
| A122G | +++ | U120 | 11 | 12,473 | +++ |
| U123C | +++ | U120 | 11 | 12,473 | +++ |
| A122G/A125G | +++ | U120 | 11 | 12,473 | +++ |
| U120C/U123C | +++ | U222 | 45 | 16,214 | +++ |
| h5-tetraloop | +++ | U161 | 23 | 13,845 | +++ |
| A301U | ++ | U120 | 11 | 12,473 | +++ |
| A302C | +++ | U120 | 11 | 12,473 | +++ |
| A302G | ++ | U120 | 11 | 12,473 | +++ |
| insAA(289:290) | ++ | U120 | 11 | 12,473 | +++ |
| wsb | − | none | – | – | − |
| osc | ++ | U300 | 71 | 18,967 | +++ |
| os-A302C | + | none | – | – | − |
| os-U301G | + | U300 | 71 | 18,967 | − |
| os-U301A/A302G | ++ | U300 | 71 | 18,967 | − |
| os-insAA(289:290) | + | U322 | – | – | − |
| os-insU(299:300) | + | U308 | 74 | 19,298 | + |
| os-insA(299:300) | + | U308 | 74 | 19,298 | + |
| os-insC(299:300) | + | U308 | 74 | 19,298 | + |
| os-insG(299:300) | + | U308 | 74 | 19,310 | + |
aAA in tag = number of amino acids in peptide tag.
bws denotes tmRNA(H8hp) without stop codons.
cos is tmRNA(H8hp) with a single stop codon at position 300.
Processing of precursor tmRNA: +++ (>50%); ++ (∼50%); + (<50%); − (<5%).
Tagging: +++ (∼50%); + (undetectable with Coomassie Blue but detectable by Western); − (undetectable even by Western).
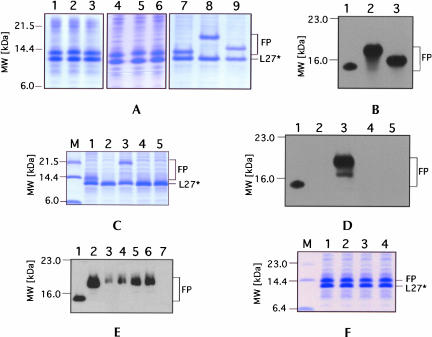
FIGURE 2.
In vivo tagging of truncated protein L27 by tmRNA(H8hp) and its derivatives bearing mutations in and around the tandem stop codons 120-UAA-UAA-125 (A,B) and the triplet 300-UAA-302 (C–E). Cell lysates from IPTG-induced E. coli IW363 cells co-expressing truncated protein L27, protein SmpB, and mutant tmRNAs were fractionated on a 12.5% SDS-polyacrylamide gel. Tagged proteins were detected either by staining gel with Coomassie Blue (A,C,F) or by Western blot analysis using anti-His-tag antibodies (B,D,E). TmRNA mutants tested for tagging are presented as follows: (A) 1, H8hp; 2, U120C; 3, A121G; 4, A122C; 5, A122G; 6, U123C; 7, A122G/A125G; 8, U120C/U123C; 9, h5-tetraloop. (B) 1, H8hp; 2, U120C/U123C; 3, h5-tetraloop. (C) 1, H8hp; 2, ws; 3, os; 4, os-U301G; 5, os-U301A/A302G. (D) 1, H8hp; 2, ws; 3, os; 4, os-U301G; 5, os-U301A/A302G. (E) 1, H8hp; 2, os; 3, os-insU(299:300); 4, os-insA(299:300); 5, os-insC(299:300); 6, os-insG(299:300); 7, os-insAA(289:290). (F) 1, H8hp; 2, A301U; 3, A301G; 4, A302C. To detect tagging by tmRNA derivatives with single-and double-nucleotide insertions, the equivalent of 20 tagging reactions was analyzed in individual lanes on the gel (E, lanes 3–7); otherwise equivalent portions of tagging reactions were used for the analysis. Lane M in panel C shows molecular weight markers. L27* and FP denote truncated protein L27 and fusion proteins, respectively.
To test whether removal of the tandem stop codons would allow tagging, we generated a mutant tmRNA(H8hp) in which the uridines at positions 120 and 123 were changed to cytosines. In yet another construct, the naturally occurring nanonucleotide CUUAAUAAC loop of helix 5 was replaced with a GAGA tetraloop in order to determine whether the larger loop was required. None of these changes affected the ability of tmRNA(H8hp) mutants to tag truncated L27. However, polypeptides of considerably increased molecular mass were observed (Fig. 2A ▶, lanes 8,9).
Mass spectroscopic analysis demonstrated that tmRNA(H8hp) with the U120C/U123C mutation produced a fusion protein of 16,039 Da (Fig. 2A ▶, lane 8; 2B ▶, lane 2). This measured molecular mass agreed well with the predicted size of a 16,214-Da polypeptide that would be synthesized if tagging was terminated in pk3 at the in-frame stop codon 222-UGA-224. The tmRNA with the helix 5-tetraloop mutation produced a fusion protein of 13,700 Da (Fig. 2A ▶, lane 9; 2B ▶, lane 3). This mass spectroscopic measurement was close to the predicted value of 13,834 Da for a polypeptide for which synthesis would terminate at 161-UAG-163 in pk2. These results demonstrated not only that a single stop codon—UAA, UGA, or UAG—was sufficient to effectively terminate trans-translation, but also that the ORF in tmRNA could be extended to include pk2 and pk3.
To investigate whether even larger tagged proteins could be synthesized, the in-frame stop-codons 222-UGA-224, 237-UAA-239, and 246-UAG-248 were altered by introducing G223U, A238U, A247U, and A301U changes into the U120C/U123C mutant tmRNA(H8hp), thus generating a tmRNA without stop codons (abbreviated as ws-tmRNA). When the activity of the ws-tmRNA was tested in vivo, not even minute levels of tagging were observed (see Fig. 2C,D ▶, lane 2). Similarly, no tagging was observed when the 300-UAA-302 triplet in os-tmRNA was changed to either UGA or UAG (Fig. 2C,D ▶, lanes 4,5). However, efficient tagging was restored and large amounts of a polypeptide migrating at 21.5 kDa were produced (Fig. 2C,D ▶, lane 3) by reverting the uridine at position 301 of the ws-tmRNA to an adenosine to form a mutant tmRNA containing one stop codon at 300-UAA-302 (designated os-tmRNA). Mass spectroscopic analysis determined the mass of this polypeptide to be 18,827 Da, consistent with a predicted molecular mass of 18,967 Da and the assumption that trans-translation terminated at 300-UAA-302. These results demonstrated that the ORF of tmRNA could be extended beyond pk4 but not into the TLD.
Mutations in and around 300-UAA-302
Because a properly folded TLD is required for tagging (Rudinger-Thirion et al. 1999; Barends et al. 2001), we suspected that removal of all in-frame stop codons in ws-tmRNA would disrupt the TLD and lead to the observed tagging incompetence documented in Figure 2C,D ▶. Aiming to exclude the possibility that ws-tmRNA was somehow impaired in its processing ability, E. coli lysates were subjected to Northern analysis as described in Materials and Methods. Surprisingly, processing of ws-tmRNA was significantly impaired in both uninduced and induced cells (Fig. 3B ▶, right panel, lane 4). In contrast, only small amounts (<1%) of precursors were observed with tmRNA(H8hp), U120C/U123C, and the helix 5-tetraloop mutant RNAs (Fig. 3A ▶, lanes 1–3). Changing A302 to C or G, as well as introducing a U301G mutation in the ws-tmRNA background, yielded processing levels between 20% and 40% (Fig. 3B ▶, right panel, lanes 3–5). Similar results were obtained when a single U, A, C, or G residue was inserted between C299 and U300, or when two adenosine residues were inserted between U289 and A290 (Fig. 3C ▶). These insertions shifted the 300-UAA-302 stop codon out-of-frame and were expected to produce new stop codons between helical sections 2c and 2d, or in the loop between helical sections 2a and 2b (see Fig. 1A ▶). When tested in vivo, the mutant os-tmRNAs with single nucleotide insertions displayed detectable tagging activities that were however 100- to 500-fold lower than the tagging supported by the os-tmRNA (Fig. 2E ▶, lanes 3–6). In contrast, no traces of tagging could be detected when os-tmRNAs with two nucleotide insertions were tested (Fig. 2E ▶, lane 7). These results indicated that, unlike the pk2-pk4 segment, helix 2 could not serve as template for the synthesis of peptide tags.
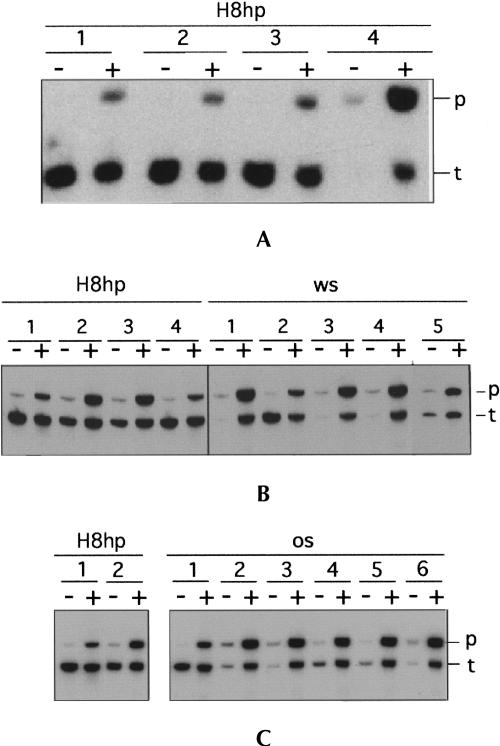
FIGURE 3.
Maturation of tmRNA(H8hp), ws-tmRNA, os-tmRNA, and their derivatives. Northern blot analysis: Aliquots of 1 μg total RNA extracted from un-induced and IPTG-induced E. coli IW363 cells were separated on a 5% Tris-buffered EDTA-urea polyacrylamide gel and blotted to a Zeta-probe membrane. [32P]-labeled oligonucleotide TM4 was hybridized to both precursor and mature tmRNAs. Tested tmRNA mutants are presented as follows: (A) 1, H8hp; 2, U120C/U123C; 3, h5-tetraloop; 4, ws. (B) Left panel: 1, H8hp; 2, A301U; 3, A302G; 4, A302C. Right panel: 1, ws; 2, os; 3, U301G; 4, A302C; 5, A302G. (C) Left panel: 1, H8hp; 2, os-insAA(289:290). Right panel: 1, H8hp; 2, os-insU(299:300); 3, os-insA(299:300); 4, os-insC (299:300); 5, os-insG(299:300); 6; os-insG(299:300). The tmRNA species used to produce each group of mutants are indicated above each panel. Precursor and mature tmRNA are denoted as p and t, respectively.
Mutations eliminating the 300-UAA-302 triplet, or shifting it into an out-of-frame position, inhibited expression of ws- and os-tmRNA (Fig. 3A ▶, lane 4; 3B ▶, right panel, lanes 3–5; 3C, right panel, lanes 3–6) in the absence of truncated protein L27. In contrast, when the synthesis of protein L27 was induced by IPTG, ample amounts of defective tmRNAs were produced. The cause for this unusual pattern of expression of the ssrA gene, which is controlled by a σ70-type promoter (Chauhan and Apirion 1989), is under investigation.
DISCUSSION
To complete the translation of truncated proteins and facilitate recycling of stalled ribosomes, tmRNA provides missing stop codons (Keiler et al. 1996). In E. coli tmRNA, a tandem pair of UAA stops, apparently directing the addition of 10 amino acids to the truncated protein, is situated within a loop of helix 5 (Felden et al. 1996; Williams and Bartel 1996). We demonstrate that a single stop codon is sufficient to terminate trans-translation in vivo, even under experimental conditions where numerous truncated L27 mRNA molecules are engaged with translation-arrested ribosomes. Why the tmRNAs of several bacterial species contain clusters of tandem stop codons within the terminal loop of helix 5 remains to be determined (Zwieb et al. 1999a,b; see also Major et al. 2002); however, one can speculate that their function comes into play when cells are exposed to stress. Interestingly, the context in which tandem stop codons were placed (nonaloop vs. tetraloop) did not affect tagging efficiency. This indicated, in agreement with the observed translation of pseudoknotted regions, that helix 5 unfolds and thereby obliterates any potential influences caused by the loop configuration.
Changing the tandem stop codons to sense codons did not affect tagging, but produced tagged peptides that were considerably longer than those synthesized by wild-type E. coli tmRNA. In exploring the limits for extending the ORF in tmRNA, we converted all in-frame stops into sense codons. These changes produced the tagging-incompetent ws-tmRNA. Restoring the 300-UAA-302 stop codon converted the ws-tmRNA into the tagging-competent os-tmRNA. Mutating the 300-UAA-302 triplet in wild-type tmRNA had no affect on its tagging activity (see Fig. 2F ▶), and attempts to extend the ORF beyond stop codon 300-UAA-302 produced tmRNA mutants that tagged poorly or not at all (see Fig. 2E ▶). In agreement with phylogenetic data, which show that 300-UA-301 is highly conserved whereas the nucleotide at position 302 can be A, U, or C (Zwieb et al. 1999b), the A301U, A301G, and A302G changes were particularly detrimental to tmRNA-mediated tagging. Northern analyses revealed that the tagging defects had their roots, at least in part, in the defective maturation of tmRNA precursors. As shown in Figure 3 ▶, up to 40% of the tmRNA precursors bearing mutations in or close to the 300-UAA-302 triplet could be trimmed to mature but tagging-incompetent tmRNAs. How trimming of the tmRNA termini by RNases BN, D, E, III, P, PH, and T (Makarov and Apirion 1992; Li et al. 1998; Lin-Chao et al. 1999) could be influenced by the 300-UAA-302 sequence remains to be determined. The strong conservation of 300-UA-301 suggests an involvement of these residues with other segments of tmRNA that may be important for the structural integrity near the resume codon (Zwieb et al. 1999a).
Our finding that the ORF in tmRNA can be extended to include pk2 through pk4 supports a mechanistic model for trans-translation which explains how the rather bulky free tmRNA molecule changes its conformation during its transit through the ribosome (Fig. 4 ▶). At the resumption step, the TLD approaches the ribosomal A site which accommodates aminoacyl-tRNAs in canonical protein synthesis. At that stage of trans-translation, helix 5 and the pk2–pk4 segment of tmRNA appear to form a loop around the beak on the head of the 30S ribosomal subunit (Valle et al. 2003). As the peptide tag is being extended during elongation, it is likely that helix 5, pk2, and maybe even pk3 and pk4 need to unfold to enter the mRNA channel of the 30S ribosomal subunit. This model is supported by earlier observations indicating that the ribosome protects a 30-nucleotide segment in the linear mRNA encompassing residues at positions −15 and +16 (Steitz 1969; Yusupova et al. 2001). Although the single-stranded part of the MLD in E. coli tmRNA is 30 nucleotides long, stretching of this segment of the tmRNA molecule in the mRNA channel is difficult to imagine, because of the circular arrangement of the MLD and pk2-pk4. More likely, the TLD is positioned outside of the ribosome while tagging is terminated, consistent with our finding that the 3′ part of helix 2 and the TLD are not suitable to serve as a template (see Fig. 2E ▶). Our proposal is reminiscent of the recent demonstration that the 50-nt bypassed region of the T4 gene 60 mRNA is also unfolded (Gallant et al. 2003).
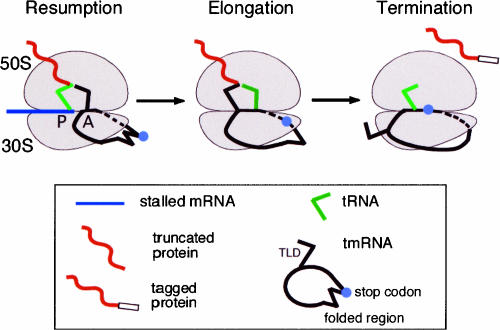
FIGURE 4.
Transit of tmRNA through the ribosome during trans-translation. Resumption: The ribosome is stalled at the 3′ end of mRNA lacking a stop codon. Aminoacylated tmRNA binds to the ribosomal A site assisted by EF-Tu, SmpB, and S1. Helix 5 as well as pk2 and pk3 stay outside of the ribosome and are wrapped around the beak on the head of the 30S ribosomal subunit. Elongation: Amino-acylated-tmRNA accepts the polypeptide from the peptidyl-tRNA and is translocated to the ribosomal P site. Aminoacylated tRNA starts decoding the MLD. Helix 5 unfolds, and stop codon(s) enter the mRNA channel. Termination: The TLD exits the ribosome. Pk2 unfolds as the stop codon enters the ribosomal A site. Releasing factors recognize the stop codon at the A site, and the trans-translational complex dissociates. Elements of the trans-translational apparatus are illustrated in the box.
Unfolding of helix 5 and the abutting pseudoknots is likely to be facilitated by ribosomal protein S1 in Gram-negative and by RNA helicases in Gram-positive bacteria (Thomas et al. 1979; Subramanian 1983; Kossen and Uhlenbeck 1999). Ribosomal protein S1 was shown to bind single-stranded and pseudoknotted RNA with high affinity (Bear et al. 1976; Ringquist et al. 1995). Recent studies show that the N-terminal domains of protein S1 bind primarily to the MLD and pk2 (Okada et al. 2004; Wower et al. 2004), suggesting, in agreement with cryo-electron microscopy (Valle et al. 2003), that protein S1 unfolds tmRNA at an early stage of trans-translation.
MATERIALS AND METHODS
The sources of enzymes, [32P]-labeled nucleotides, and antibodies are given in (Wower et al. 2000). E. coli strain XL1-B was the host for purification and maintenance of plasmids. Expression strain IW363 was constructed by P1 transduction of gene ssrA deletion from W3110ΔssrA::kanr into BL21(DE3)/pLysS (Komine et al. 1994). Plasmid pETrpmA-At-3 was used for the co-expression of tmRNA and proteins SmpB and T7-L27-At in vivo. Expression of the ssrA and smpB genes was controlled by their native promoters, while the truncated rpmA gene was under control of the T7 promoter as described (Wower et al. 2004). Derivatives of the tmRNA lacking stop codons in helix 5 and pk2 through pk4 were obtained by PCR-directed mutagenesis as described (Wower et al. 2004). Processing of tmRNA precursors was analyzed by Northern blots (Wower et al. 2004) probed with [32P]-labeled oligonucleotide (5′-CGAGGCCAACCGCCC-3′) complementary to E. coli tmRNA residues 54–67. Hybridization signals were quantified using a Typhoon Phosphorimager and ImageQuant Version 5.2 software (Amersham). The molecular mass of intact fusion proteins was measured by MALDI-TOF in the linear mode ion of an Axima-CRF (Kratos Analytical) using 2 mg/mL ferulic acid (Fluka) and 5 mg/mL gentisic acid (Sigma) as a matrix.
Acknowledgments
We thank Jiaming Yin for persistent technical assistance in carrying out the MALDI-TOF experiments. We are grateful to Frank F. Bartol for reading this manuscript and for his invaluable assistance. This research was supported by National Institutes of Health Grant GM58267 to J.W.
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.7269305.
REFERENCES
- Barends, S., Wower, J., and Kraal, B. 2000. Kinetic parameters for tmRNA binding to alanyl-tRNA synthetase and elongation factor Tu from Escherichia coli. Biochemistry 39: 2652–2658. [DOI] [PubMed] [Google Scholar]
- Barends, S., Karzai, A.W., Sauer, R.T., Wower, J., and Kraal, B. 2001. Simultaneous and functional binding of SmpB and EF-Tu-GTP to the alanyl acceptor arm of tmRNA. J. Mol. Biol. 314: 9–21. [DOI] [PubMed] [Google Scholar]
- Bear, D.G., Ng, R., Van Derveer, D., Johnson, N.P., Thomas, G., Schleich, T., and Noller, H.F. 1976. Alteration of polynucleotide secondary structure by ribosomal protein S1. Proc. Natl. Acad. Sci. 73: 1824–1828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chauhan, A.K. and Apirion, D. 1989. The gene for a small stable RNA (10Sa RNA) of Escherichia coli. Mol. Microbiol. 3: 1481–1485. [DOI] [PubMed] [Google Scholar]
- Felden, B., Himeno, H., Muto, A., Atkins, J.F., and Gesteland, R.F. 1996. Structural organization of Escherichia coli tmRNA. Biochimie 78: 979–983. [DOI] [PubMed] [Google Scholar]
- Felden, B., Himeno, H., Muto, A., McCutcheon, J.P., Atkins, J.F., and Gesteland, R.F. 1997. Probing the structure of the Escherichia coli 10Sa RNA (tmRNA). RNA 3: 89–103. [PMC free article] [PubMed] [Google Scholar]
- Gallant, J., Bonthuis, P., and Lindsley, D. 2003. Evidence that the bypassing ribosome travels through the coding gap. Proc. Natl. Acad. Sci. 100: 13430–13435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gueneau de Novoa, P. and Williams, K.P. 2004. The tmRNA website: Reductive evolution of tmRNA in plastids and other endosymbionts. Nucleic Acids Res. 32: D104–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallier, M., Ivanova, N., Rametti, A., Pavlov, M., Ehrenberg, M., and Felden, B. 2004. Pre-binding of small protein B to a stalled ribosome triggers trans-translation. J. Biol. Chem. 279: 25978–25985. [DOI] [PubMed] [Google Scholar]
- Karzai, A.W., Susskind, M.M., and Sauer, R.T. 1999. SmpB, a unique RNA-binding protein essential for the peptide-tagging activity of SsrA (tmRNA). EMBO J. 18: 3793–3799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keiler, K.C., Waller, P.R., and Sauer, R.T. 1996. Role of a peptide tagging system in degradation of proteins synthesized from damaged messenger RNA. Science 271: 990–993. [DOI] [PubMed] [Google Scholar]
- Komine, Y., Kitabatake, M., Yokogawa, T., Nishikawa, K., and Inokuchi, H. 1994. A tRNA-like structure is present in 10Sa RNA, a small stable RNA from Escherichia coli. Proc. Natl. Acad. Sci. 91: 9223–9227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kossen, K., and Uhlenbeck, O.C. 1999. Cloning and biochemical characterization of Bacillus subtilis Yxin, a DEAD protein specifically activated by 23S rRNA: Delineation of a novel sub-family of bacterial DEAD proteins. Nucleic Acids Res. 27: 3811–3820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Z., Pandit, S., and Deutscher, M.P. 1998. 3′ exoribonucleolytic trimming is a common feature of the maturation of small, stable RNAs in Escherichia coli. Proc. Natl. Acad. Sci. 95: 2856–2861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin-Chao, S., Wei, C.L., and Lin, Y.T. 1999. RNase E is required for the maturation of ssrA RNA and normal ssrA RNA peptide-tagging activity. Proc. Natl. Acad. Sci. 96: 12406–12411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Major, L.L., Edgar, T.D., Yip, P.Y., Isaksson, L.A., and Tate, W.P. 2002. Tandem termination signals: Myth or reality? FEBS Lett. 514: 84–89. [DOI] [PubMed] [Google Scholar]
- Makarov, E.M. and Apirion, D. 1992. 10Sa RNA: Processing by and inhibition of RNase III. Biochem Int. 26: 1115–1124. [PubMed] [Google Scholar]
- McGinness, KE. and Sauer, R.T. 2004. Ribosomal protein S1 binds mRNA and tmRNA similarly but plays distinct roles in translation of these molecules. Proc. Natl. Acad. Sci. 101: 13454–13459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada, T., Wower, I.K., Wower, J., Zwieb, C.W., and Kimura, M. 2004. Contribution of the second OB fold of ribosomal protein S1 from Escherichia coli to the recognition of tmRNA. Biosci. Biotech. BioChem. 68: 2319–2325. [DOI] [PubMed] [Google Scholar]
- Ringquist, S., Jones, T., Snyder, E.E., Gibson, T., Boni, I., and Gold, L. 1995. High-affinity RNA ligands to Escherichia coli ribosomes and ribosomal protein S1: Comparison of natural and unnatural binding sites. Biochemistry 34: 3640–3648. [DOI] [PubMed] [Google Scholar]
- Rudinger-Thirion, J., Giege, R., and Felden, B. 1999. Aminoacylated tmRNA from Escherichia coli interacts with prokaryotic elongation factor Tu. RNA 5: 989–992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steitz, J.A. 1969. Polypeptide chain initiation: Nucleotide sequences of the three ribosomal binding sites in bacteriophage R17 RNA. Nature 224: 957–964. [DOI] [PubMed] [Google Scholar]
- Subramanian, A.R. 1983. Structure and functions of ribosomal protein S1. Prog. Nucleic Acid Res. Mol. Biol. 28: 101–142. [DOI] [PubMed] [Google Scholar]
- Thomas, J.O., Boublik, M., Szer, W., and Subramanian, A.R. 1979. Nucleic acid binding and unfolding properties of ribosomal protein S1 and the derivatives S1-F1 and m1-S1. Eur. J. Biochem. 102: 309–314. [DOI] [PubMed] [Google Scholar]
- Tu, G.F., Reid, G.E., Zhang, J.G., Moritz, R.L., and Simpson, R.J. 1995. C-terminal extension of truncated recombinant proteins in Escherichia coli with a 10Sa RNA decapeptide. J. Biol. Chem. 270: 9322–9326. [DOI] [PubMed] [Google Scholar]
- Valle, M., Gillet, R., Kaur, S., Henne, A., Ramakrishnan, V., and Frank, J. 2003. Visualizing tmRNA entry into a stalled ribosome. Science 300: 127–130. [DOI] [PubMed] [Google Scholar]
- Williams, K.P. and Bartel, D.P. 1996. Phylogenetic analysis of tmRNA secondary structure. RNA 2: 1306–1310. [PMC free article] [PubMed] [Google Scholar]
- Wower, I.K., Zwieb, C.W., Guven, S.A., and Wower, J. 2000. Binding and cross-linking of tmRNA to ribosomal protein S1, on and off the Escherichia coli ribosome. EMBO J. 19: 6612–6621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wower, I.K., Zwieb, C., and Wower, J. 2004. Contributions of pseudoknots and protein SmpB to the structure and function of tmRNA in trans-translation. J. Biol. Chem. 279: 54202–54209. [DOI] [PubMed] [Google Scholar]
- Yusupova, G.Z., Yusupov, M.M., Cate, J.H., and Noller, H.F. 2001. The path of messenger RNA through the ribosome. Cell 106: 233–241. [DOI] [PubMed] [Google Scholar]
- Zwieb, C., Muller, F., and Wower, J. 1999a. Comparative three-dimensional modeling of tmRNA. Nucleic Acids Symp. Ser. 41: 200–204. [PubMed] [Google Scholar]
- Zwieb, C., Wower, I., and Wower, J. 1999b. Comparative sequence analysis of tmRNA. Nucleic Acids Res. 27: 2063–2071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwieb, C., Gorodkin, J., Knudsen, B., Burks, J., and Wower, J. 2003. tmRDB (tmRNA database). Nucleic Acids Res. 31: 446–447. [DOI] [PMC free article] [PubMed] [Google Scholar]