Abstract
Dicer processes long double-stranded RNA (dsRNA) and pre-microRNAs to generate the functional intermediates (short interfering RNAs and microRNAs) of the RNA interference pathway. Here we identify features of RNA structure that affect Dicer specificity and efficiency. The data presented show that various attributes of the 3′ end structure, including overhang length and sequence composition, play a primary role in determining the position of Dicer cleavage in both dsRNA and unimolecular, short hairpin RNA (shRNA). We also demonstrate that siRNA end structure affects overall silencing functionality. Awareness of these new features of Dicer cleavage specificity as it is related to siRNA functionality provides a more detailed understanding of the RNAi mechanism and can shape the development of hairpins with enhanced functionality.
Keywords: Dicer, cleavage specificity, overhang structure, RNAi
INTRODUCTION
RNA interference (RNAi) is an endogenous cellular pathway involved in sequence specific post-transcriptional gene regulation. The key to the specificity of this process lies in the mechanism by which the associated protein machinery mediates enzymatic processing and silencing activity. At a key stage of the RNAi pathway, the ribonuclease (RNase) III-like enzyme Dicer processes long double-stranded RNA (dsRNA) or pre-microRNA hairpin precursors into small interfering RNAs (siRNAs) or microRNA (miRNAs) (for reviews, see Hannon and Rossi 2004; Bartel 2004). These short RNAs are then loaded into the RNA induced silencing complex (RISC) through the action of R2D2 and Dicer (Liu et al. 2003). The 21–23-nt products of Dicer digestion contain characteristic 2-nt 3′ overhangs and act as the functional intermediates of RNAi directing mRNA cleavage and translational attenuation.
Dicer has been found in nearly all eukaryotes studied thus far. In humans, Dicer is a large, 220-kDa protein that contains single PAZ, dsRNA binding (dsRBD), and ATPase/ RNA helicase domains, as well as two RNase III-like domains, and a domain of unknown function (DUF283). Dicer’s two RNase III-like domains form an intramolecular dimer to create a single active site responsible for cleavage of dsRNA (Zhang et al. 2004). In addition, detailed structure–function studies of the dsRBD and PAZ domains led to the current hypothesis that these two substructures play a role in recognition and binding of substrates. Dicer’s dsRBD can bind dsRNA and interfere with cleavage of RNA duplexes by the Dicer holoenzyme (Provost et al. 2002). Similarly, studies of a closely related PAZ domain derived from the human Argonaute eIF2c1 protein (Ago1) suggest that PAZ may function in the recognition of 2-nt 3′ overhangs that are generated during cleavage reactions by both Dicer and Drosha (Lee et al. 2003; Ma et al. 2004).
While details of Dicer’s functional domains are being elucidated, less is known about the contributions that the dsRNA substrate makes to position and efficiency of Dicer cleavage. Dicer generates ~20 base-pair (bp) siRNAs by entering the duplex from the termini (Zamore et al. 2000; Zhang et al. 2002) and cleaves both short (30 nt) and long (130 nt) dsRNA molecules with equal efficiency (Elbashir et al. 2001; Myers et al. 2003). In contrast, smaller duplexes (21 nt siRNAs) do not appear to bind Dicer in vitro (Provost et al. 2002). While substrates with 3′ overhangs are more efficiently processed than blunt ended molecules, the state of 5′ phosphorylation of the duplex does not play a role in efficiency (Zhang et al. 2002, 2004).
Gene silencing can be achieved by introduction of chemically synthesized siRNA or intracellularly expressed shRNAs. The silencing efficiency of siRNAs are readily predicted (Reynolds et al. 2004), but the necessity for Dicer processing of shRNAs adds an additional level of complexity for obtaining highly functional silencing molecules. The exact composition of the cleaved shRNA is essential for functionality, yet the structural requirements for specific Dicer processing of hairpin substrates are unknown. In the following work the contributions of substrate terminal structure in determining Dicer cleavage patterns are examined. The results of these studies show that Dicer cleaves dsRNA and shRNA containing blunt and overhang termini at distinct positions and that both overhang structure and composition play a key role in defining Dicer cleavage specificity and efficiency. The distinct positions of cleavage are the result of Dicer employing a 3′ end counting rule and measuring a distance of ~22 nt from the 3′ terminus to cleave both strands of RNA. Understanding the structural requirements for Dicer specificity and efficiency provides the tools for designing functional shRNA in the future.
RESULTS
Dicer cleavage of blunt and overhang ended dsRNA
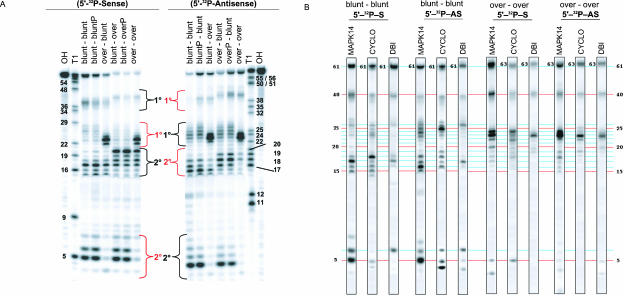
To evaluate the importance of dsRNA sequence and end structure in determining the position and pattern of Dicer cleavage, three 61mer dsRNAs (derived from human mitogen-activated protein kinase 14 [MAPK14] NM_001315, Cyclophilin B [CYCLO] NM_000942, and Diazepam binding inhibitor [DBI] NM_020548) were processed in vitro by Dicer. Dicer processing of blunt and overhang dsRNA resulted in distinct and unique patterns of cleavage. Figure 1 ▶ shows that Dicer cleavage of [32P] end-labeled 61mer dsRNA from MAPK14 having blunt or overhang termini generates unique sense strand cleavage fragments (four sites: 5–7, 15–18, 23–28, and ~39 nt for blunt ended molecules and 3 sites: 17–20, 22–24, and ~41 nt for overhang molecules). The relative appearance (and disappearance) of bands over the course of time distinguished primary and secondary Dicer cleavage products. For blunt–blunt MAPK14, the 39-nt and 23–28-nt bands appear early in the time course of the experiment and are depleted at later times points, suggesting that these are primary cleavage products generated by Dicer entering the 3′ (sense) and 5′ (sense) ends of the MAPK14 duplex, respectively. Secondary products of the reaction (the 15–18-nt and 5–6-nt bands) appear later in the reaction (t > 5 min) and result from digestion of the primary products (39 → 15–18; 23– 28 → 5–7) (Fig. 1 ▶). The ability of the 26mer fragment to generate smaller fragments was confirmed by a separate experiment using a small dsRNA corresponding to the primary cleavage product (see Supplementary Fig. 1 ▶ at www.dharmacon.com/tech/publications). Dicer generates a markedly different cleavage pattern for over–over MAPK14 with only three positions of cleavage. As was the case in experiments performed on blunt-ended molecules, the relative rate of appearance/disappearance of each band identifies the 41- and 22–24-nt fragments as primary cleavage sites, whereas the 17–20-nt band represents a secondary product (41 nt → 17–20 nt). Together, these findings are indicative of Dicer entering and processing the 61mer from both ends of the molecule and cleaving blunt and overhang end structures in distinctly different positions.
FIGURE 1.
61 mer MAPK14 dsRNA containing blunt and overhang ends are processed in distinctly different manners. (A) Dicer cleavage of [32P]-5′ sense labeled dsRNA with two blunt ends (blunt–blunt) and two 2-nt overhang ends (over–over) analyzed by denaturing PAGE. Incubation times in minutes. RNase T1 digest (T1) and hydroxyl digest (OH) are labeled at the tops of the lanes for both substrates. The primary and secondary cleavage product bands that result from Dicer entering from the 5′ sense (left) and 3′ sense (right) ends are indicated in red and black, respectively. (B) Diagrams of Dicer processing of blunt and/or overhang ended MAPK14 61mer dsRNA substrates. Red and black arrows show Dicer positions of cleavage from the 5′ sense (left) and 3′ sense (right) ends, respectively. Solid arrows were derived from Dicer digestion of [32P]-5′ sense or antisense labeled dsRNA. Length of arrows indicates intensity of cleavage. Processing positions with dashed arrows were derived from processing of blunt secondary products (Supplementary Fig. 1 at www.dharmacon.com/tech/publications).
To confirm the importance of end structure in Dicer specificity, all possible blunt and 2-nt 3′ overhang end structure combinations (e.g., blunt–blunt, blunt–overhang, overhang–blunt, overhang–overhang) were constructed for the MAPK14 61mer. A side-by-side comparison of Dicer digest patterns generated from these duplexes reiterates that terminal structures play a central role in determining Dicer specificity. Sense-labeled MAPK14 dsRNA having a blunt terminus on the 5′ sense (left) end of the molecule shows two cleavage products (5–7 and 23–28 nt; see Fig. 2A ▶ lanes labeled blunt–blunt and blunt–over) when Dicer enters the duplex from the 5′ sense (left) end of the molecule. In contrast, when dsRNA contains a 2-nt overhang on the left end of the duplex, only a single digest product (22–24 nt; see Fig. 2A ▶ lanes labeled over–blunt and over–over) is generated. The digest patterns are recapitulated when Dicer enters the opposite side (3′ sense end, right side) of the duplex (see Antisense labeled RNA, Fig. 2A ▶). From this we conclude that the cleavage patterns characteristic of each duplex result from dissimilar processing of blunt and overhang ends by Dicer. While the position of Dicer cleavage is primarily dependent on terminal structure, nucleotide content clearly influences specificity (Fig. 2B ▶). Comparison of Dicer cleavage patterns from MAPK14, CYCLO, and DBI show that while duplexes with similar end structure generate comparable populations of cleavage fragments, the preferred product varied among each dsRNA (Fig. 2B ▶; Supplementary Figs. 2–4 at www.dharmacon.com/tech/publications). For instance, the primary and secondary cleavage products resulting from Dicer processing of the 5′ sense (left) end of blunt MAPK14 dsRNA are 26 and 5 nt, respectively. In contrast, the equivalent products generated from the opposite (right) end of the molecule are 24 and 4 nt in length. This trend toward differential processing was observed in all of the substrates studied and supports similar observations made by Provost et al. (2002) and Zhang et al. (2004).
FIGURE 2.
End structure and sequence composition determine Dicer cleavage pattern. (A) Processing of MAPK14 61mer dsRNA containing blunt and/or 2-nt 3′ overhang ends by Dicer analyzed by denaturing PAGE. Lanes are labeled describing dsRNA termini oriented relative to sense strand from 5′ to 3′. Sense or antisense labeled strands are shown on the left and right, respectively. (P) 5′ end of terminus was 5′ (nonradio-actively) phosphorylated during synthesis. (B) The contribution of sequence to Dicer cleavage specificity. MAPK14, CYCLO, and DBI 61mer sequences containing (1) blunt ends (blunt–blunt) or (2) 2-nt 3′ overhang ends (over–over) were digested by Dicer. Both (1) sense and (2) antisense labeled duplexes are shown. Lanes from separate gels were sized to enable side-by-side comparison of bands for different digests. (Original gels are in Supplementary Figs. 2–4 at www.dharmacon.com/tech/publications.)
Dicer binding of blunt and overhang ended RNA
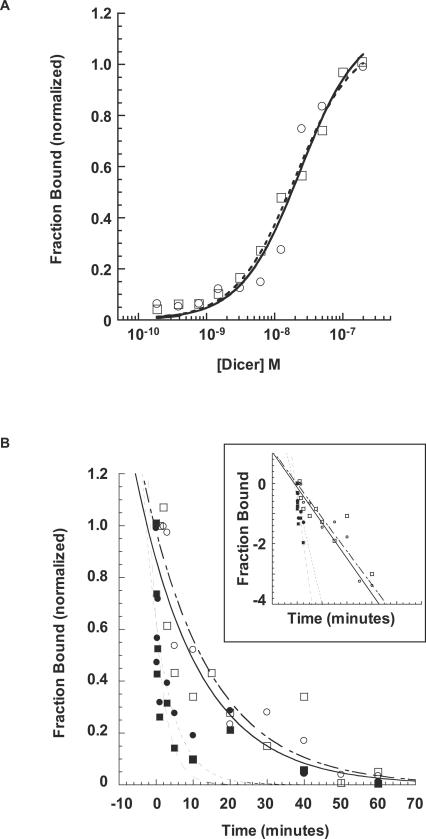
Since it has been shown that the 2-nt 3′ overhang structure is specifically recognized and bound by the PAZ domain (Lingel et al. 2003; Song et al. 2003; Ma et al. 2004) and duplexes containing blunt and overhang ends are distinctly processed by Dicer, we assessed whether binding plays a role in the observed differential processing by Dicer. Binding studies were performed on MAPK14 61mers containing blunt or overhang ends. As shown in Figure 3A ▶ both molecules have similar binding affinity for Dicer as measured by a nitrocellulose filter binding assay (dissociation constants [Kd] of 50 nM for both RNA species). The Kd obtained here is consistent with an apparent Kd of ~60 nM estimated from a gel shift assay for human Dicer to dsRNA (Provost et al. 2002). This equivalency of binding strength by the two species was observed over a range of pH and ionic strength (data not shown).
FIGURE 3.
Long dsRNA 61mers containing blunt or overhang ends have the same affinity to Dicer holoenzyme. (A) Determination of dissociation constant (Kd) of Dicer–RNA complex by nitrocellulose binding assay. Fraction [32P] RNA duplex bound versus Dicer concentration for 61-nt MAPK14 dsRNA duplex with 2-nt 3′ overhang (○) and blunt (□) ends. (B) Determination of Dicer–RNA complex t1/2 by pulse-chase experiment after a 1 min and 30 min preincubation. Fraction [32P]-labeled RNA duplex bound to 80 nM Dicer was measured as a function of time after addition of 100 nM unlabeled dsRNA (chase). One minute preincubation prior to addition of unlabeled chase: 61-nt MAPK14 dsRNA duplex with 2-nt 3′ overhang (▪); 61-nt MAPK14 blunt dsRNA duplex (•). Thirty minute preincubation: 61-nt MAPK14 dsRNA duplex with 2-nt 3′ overhang (□); 61-nt MAPK14 blunt dsRNA duplex (○). (Insert) Log-plot of pulse-chase experiment.
Interestingly, while overhang and blunt ended MAPK14 dsRNA complexes dissociated with the same rates, the pre-incubation time dramatically influenced the dissociation rate. In pulse chase experiments, short (1 min) and long (30 min) preincubation times led to distinctly different dsRNA–Dicer complex half-lives (t1/2 of ~3 sec and ~12 min respectively; Fig. 3B ▶). These findings cannot be explained by Dicer cleavage, since no RNA cleavage is observed in the absence of Mg2+ (data not shown and, e.g., Provost et al. 2002). Together, these findings suggest that Dicer–dsRNA complex formation likely proceeds through a two-step process, and that Dicer binding does not account for differences in Dicer cleavage specificity.
Influence of overhang structure and sequence on Dicer specificity and efficiency
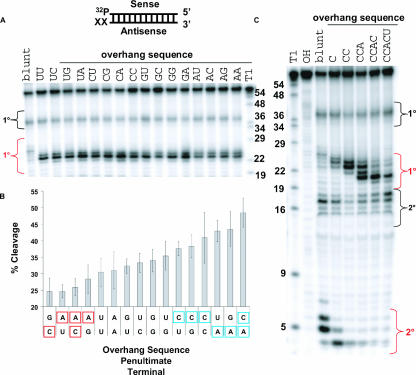
To identify which features of the overhang structure are important for Dicer recognition and specificity, 61mer MAPK14 dsRNA substrates having overhangs of varying lengths and composition were synthesized and processed by Dicer. Figure 4A ▶ illustrates Dicer cleavage of all possible 2-nt overhang sequences (UU, UC, UG, UA, CU, CG, CA, CC, GU, GC, GG, GA, AU, AC, AG, AA) and shows that while cleavage product sizes are not altered by overhang composition, nucleotide sequence does play a role in determining Dicer efficiency. Overhangs that contain a C and A in the penultimate and terminal positions (respectively) are processed most efficiently, while those containing an A in the penultimate position and a U in the terminal position are less optimal substrates. Overall, ranking the Dicer cleavage efficiency of substrates with different overhangs reveals a hierarchy with C > U = G > A being the preference at the penultimate position and A > G = U > C being the order of terminal nucleotides (Fig. 4B ▶).
FIGURE 4.
Length and sequence composition of overhangs affect Dicer processing of dsRNA. (A) Dicer cleavage of dsRNA with 2-nt 3′overhang of every possible sequence combination analyzed by denaturing PAGE. Diagram of MAPK14 61mer duplex indicates position of 5′-[32P]-labeling. (XX) position of 3′ overhang nucleotides. Lanes are labeled with sequence 5′-3′. (B) Quantification of primary Dicer cleavage products from the 2-nt 3′overhang end for different sequence combinations. The data are sorted from least to most efficient cleavage. Residues boxed in red result in less efficient cleavage while residues boxed in blue result in more efficient cleavage. The graph shows the average from three experiments and error bars indicate standard deviation. The vertical axis is scaled from 20% to 55% to emphasize differences in cleavage efficiency. (C) Dicer cleavage of dsRNA with 3′ overhang of length of 0–5 nt (natural sequence).
Studies of substrates with varying 3′ overhang lengths showed that the number of nucleotides in the overhang was a critical factor in Dicer specificity. Substrates with overhang lengths of 1, 2, and 3 nt showed concomitant shifts in the primary Dicer cleavage position. Interestingly, increasing the overhang length further reduced the diversity of cleavage products generating primarily a 21-nt product. The effects of overhang length on Dicer cleavage are illustrated in Figure 4C ▶, where RNA substrates with 3′ overhangs of 1–5 nt (derived from natural MAPK14 sequences, C, CC, CCA, CCAC, CCACU) were synthesized and processed by Dicer. As 3′ overhang length increases, the corresponding length of the primary cleavage product decreases. Thus, when overhang length is 0–3 nt it appears as though Dicer cleaves dsRNA by “counting” a distance of approximately 23 nt from the 3′ end of the overhang to cleave both strands. However, when the overhang length is ≥4 nt, the primary cleavage product is similar to that observed in duplexes containing 3-nt overhangs. Therefore, in cases where the overhang length extends beyond 3 nt, Dicer no longer uses the 3′ end of the overhang to determine cleavage position. These results suggest that Dicer confines its interaction with the three innermost nucleotides of the overhang region, perhaps enabling a critical interaction between Dicer and the junction between single-stranded and double-stranded RNA.
The length of the 3′ overhang also affects two additional attributes of duplex processing, cleavage efficiency and the generation of secondary cleavage products. With respect to efficiency, when the overhang length was 1–3 nt, roughly 40%–45% of the RNA cleavage products are generated from the overhang end after 1 h. In contrast, when the overhang lengths are increased to 4 nt and 5 nt, cleavage from the overhang end decreased to 36% and 22%, respectively, suggesting that an extended single-stranded overhang inhibits Dicer recognition and cleavage. In addition, the ability of Dicer to generate secondary cleavage products is dependent on overhang length. When shorter overhang sizes (0–1 nt) are incorporated into duplex design, both larger, primary (~24–36 nt) cleavage products and shorter, secondary (2–5 nt) products are generated (Fig. 4C ▶). However, for longer overhangs (2–5 nt) the primary cleavage products are smaller (<24 nt) and no secondary cleavage products result (see Supplementary Fig. 1 ▶ at www.dharmacon.com/tech/publications). Therefore, RNA end structure, particularly 3′ overhang length, plays a critical role in determining the position and efficiency of Dicer cleavage. This finding has profound implications for siRNA and shRNA design. shRNA expression by the Pol III promoter results in shRNA termini with variable overhang lengths (1–5 Us). As observed in Figure 4C ▶, this variation drastically affects the specificity of Dicer cleavage and consequently functionality of cleaved siRNA.
Dicer cleavage of blunt and overhang ended shRNA
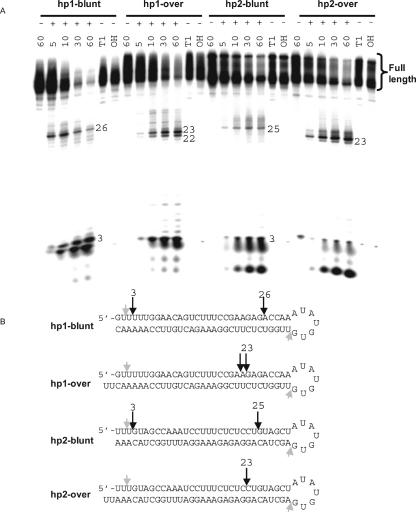
To determine whether Dicer cleavage of short hairpin RNA (shRNA) utilizes the same rules as those observed in processing of long dsRNA, shRNAs containing blunt and 2-nt 3′overhang ends were digested by Dicer. Blunt ended hairpins are cleaved by Dicer from the open dsRNA end and show primary cleavage products characteristic of blunt long dsRNA (~25–26 nt). Similarly, hairpins containing 2-nt 3′overhangs generate products 23–24 nt in length, similar to what is observed in Dicer digest of long dsRNA (Fig. 5 ▶). The results indicate that the laws that govern Dicer digestion of long dsRNA also apply to hairpin molecules.
FIGURE 5.
End structure has a strong impact on Dicer processing of shRNA. (A) Dicer processing of two cyclophilin directed shRNAs containing blunt or 2-nt 3′ overhang ends. shRNAs were incubated in the presence (+) and absence (−) of Dicer for varying periods of time (5–60 min). (B) Characteristic cleavage product sizes are indicated (26, 25, and 3 nt for blunt substrates, 23 and 22 nt for overhang substrates). Small gray arrows indicate products resulting from digestion of a small percentage of material having a nicked loop.
Effect of siRNA end structure on silencing efficiency
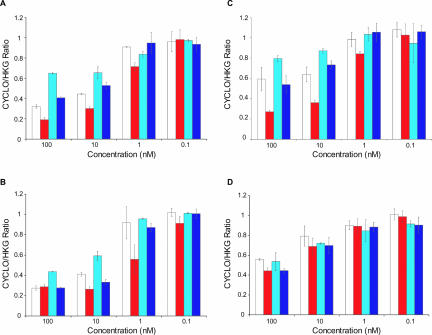
It has recently been suggested that Dicer is involved in siRNA RISC loading (Lee et al. 2004; Pham et al. 2004). As in vitro Dicer cleavage assays described here demonstrate that end structure influences Dicer efficiency, one prediction might be that siRNA end structure impacts overall silencing efficiency. To address this question, all possible end structure variants (overhang–overhang, overhang–blunt, blunt–overhang, blunt–blunt) for siRNA were synthesized and tested for the ability to silence the target gene (cyclophilin B). As shown in Figure 6A–C ▶, distinct end structure preferences were observed when mRNA knockdown of the duplexes was compared. In general, siRNAs with asymmetric overhangs on the antisense strand (i.e., a 3′ overhang on the antisense strand) performed better than siRNAs with asymmetric sense strand overhangs. When compared, the difference in functionality was more profound between structurally asymmetric siRNA (see blunt–over, over–blunt in Fig. 6A–C ▶) than symmetric molecules (see over–over, blunt–blunt in Fig. 6A–C ▶). This indicates that the effect on functionality may be the result of a shift in equilibrium between sense and antisense loading rather than overall RISC loading efficiency.
FIGURE 6.
siRNA activity is affected by asymmetric end structure. Silencing efficiency of siRNAs containing blunt and/or overhang ends at different concentrations (over–over [white], over–blunt [red], blunt–over [cyan], and blunt–blunt [blue] for siRNAs 1, 2, 3, and 4 in A,B,C,D, respectively). Silencing is expressed as a ratio of Cyclophilin B and house keeping gene (HKG) mRNA expression.
DISCUSSION
The studies presented here demonstrate that end structure, in particular 3′ overhang length and sequence, is the primary determinant of Dicer specificity and efficiency. As a result, blunt and overhang containing duplexes (and hair-pins) are cleaved in distinctly different manners. Although Dicer cleavage patterns observed by Zhang et al. (2004) for individual 50mer (blunt–blunt) and 30mer (over–over) substrates are consistent with observations here, we are the first to distinguish unique cleavage positions for dsRNA as a result of their containing overhang and blunt termini. While end structure was the main determinant for Dicer cleavage specificity, sequence composition appears to be a second substrate-associated attribute that contributes to Dicer cleavage position. Different substrates with similar terminal structures (e.g., blunt or overhang) exhibited slight 1–2-nt shifts in preferred cleavage positions. This trend (i.e., the contribution of sequence) was observed in all of the substrates in this study and most pronounced in duplexes with blunt ends, suggesting that in the absence of a 2-nt 3′ overhang end (presumably required for PAZ domain binding), the contributions made by sequence to Dicer specificity are more pronounced. While these differences appear small and possibly inconsequential, single nucleotide shifts have been shown to dramatically alter the thermodynamic stability of siRNA termini (Reynolds et al. 2004). As previous studies have clearly demonstrated that flexibility in the 5′AS end of the duplex is key for RISC entry and siRNA functionality, understanding the specificity of Dicer processing of dsRNA and shRNA at the single nucleotide level is critical for the development of highly functional shRNA (Holen et al. 2002; Reynolds et al. 2004). The observed influence of sequence on Dicer specificity could be the result of local sequence preferences in and around the site of cleavage as has been shown for an RNase III from Escherichia coli (Zhang and Nicholson 1997). Alternatively, the site of cleavage could be hardwired into the tertiary structure of the enzyme, thus opening the possibility that differences in cleavage patterns are the result of sequence composition that influence helix structure (i.e., base-pair stacking, helical bend) and flexibility. Finally, it should be noted that while primer extension assays performed by Siolas et al. (2004) have shown Dicer cleavage products found in vivo are very similar to those generated in vitro, it must be considered that additional proteins or factors in vivo may further alter Dicer processing.
Binding studies show that the strength of Dicer association with 61mer containing blunt and overhang ends is equivalent and therefore cannot account for differences in Dicer specificity. These findings contrast with those published by Ma et al. (2004), who demonstrated the association of the Ago1 PAZ domain with ~7mer dsRNA was drastically altered (>5000-fold stronger binding) by the presence of an overhang. It is possible that the difference in results reported by the two studies are the consequence of dissimilarities in experimental design (e.g., an isolated binding domain vs. a holoenzyme; a ~7-bp fragment vs. a 61mer; sequence variation between PAZ domains). In all likelihood, the Dicer dsRBD present in the full-length molecule contributes to the binding of long dsRNA and masks the smaller contributions made by the PAZ domain interaction with the RNA termini. These differences might also be responsible for the observed two-step dsRNA–Dicer association that has been identified here in studies of the holoenzyme, but was never described previously in work with an isolated PAZ or dsRBD.
Cleavage studies of substrates with varying overhang lengths revealed additional contributions of the substrate to Dicer specificity. We observed that as the length of the overhang increases from 1 to 3 nt, the position of the preferred Dicer cleavage site shifts. In contrast, as overhang length increased beyond 3 nt, the position of cleavage on the complementary strand remained constant (i.e., substrates with 3-nt, 4-nt, and 5-nt overhangs show roughly equivalent cleavage site preferences). These observations lend themselves to the hypothesis that when a molecule is blunt or when an overhang of 1–3 nt is present, Dicer recognizes the 3′ end of the molecule and cuts at a fixed distance from the end. When that overhang length is exceeded, the rules governing PAZ domain recognition are altered and other interactions (presumably mediated through the dsRBD) play a more dominant role in determining how Dicer orients itself. These findings are significant from the perspective of shRNA design and cloning. The polymerase III promoter/terminator is often used in vector-based shRNA expression design (Brummelkamp et al. 2002) and results in an shRNA containing a variable 3′ end of approximately 1–4 nt (Bogenhagen and Brown 1981; Miyagishi and Taira 2002). As shown in these studies, 3′ overhang lengths of 1–3 nt generate overlapping yet distinctly different preferred products; the use of current expression systems may lead to a range of desirable and undesirable cleavage products and impaired silencing. Our results may provide insights into the reason why silencing by shRNA has been more challenging to predict, compared to siRNAs.
The Dicer binding and cleavages studies presented here correlated well with what is currently known about the RNAi mechanism. The slow dissociation rate is consistent with results from studies in Drosophila that show Dicer remains associated with siRNA (and presumably siRNA resulting from Dicer cleavage) for subsequent assembly into RISC and target specific silencing (Lee et al. 2004; Pham et al. 2004). Moreover, we have shown that Dicer recognizes end structure and that terminal structure plays a role in overall siRNA functionality. Given Dicer’s putative role loading siRNA into RISC, we propose that the efficiency of this loading step is determined (in part) by duplex terminal structure, possibly in a highly coordinate and directional manner as proposed initially by Elbashir et al. (2001). If true, this would further advocate the need for the development of shRNA expression systems that generate shRNA of discrete sequence such as miRNA based expression that generates discrete hairpins from miRNA precursors (Zeng et al. 2002; Boden et al. 2004).
In this work it was also demonstrated that end structure of siRNAs play a role in determining siRNA functionality. Duplexes that contain an overhang on the 3′ antisense strand exhibit improved functionality, while an overhang on the 3′ end of the sense strand led to decreased silencing. This observation is consistent with results by Hohjoh (2004) that showed an improvement in functionality for over–blunt siRNAs versus over–over siRNAs. Asymmetric end structure did not enhance functionality of all molecules tested. For example, in cases where the antisense strand exhibited efficient loading into RISC, but poor silencing functionality, addition of an overhang on that strand failed to significantly improve duplex functionality (Fig. 6D ▶). These findings allude to a directionality in Dicer handoff of duplexes to RISC or possibly a yet unknown component of the RNAi pathway.
The studies presented here may also provide insight into the coordinated Dicer–Drosha processing of miRNA. While Dicer processing of pri-miRNA generates a wide range of products, the combination of Drosha and Dicer leads to the generation of a single, distinct, mature miRNA (Lee et al. 2003). One attribute of Drosha pri-miRNA cleavage is the generation of a pre-miRNA containing a 2-nt 3′ overhang. As we have shown here the overhang structure is important for Dicer recognition and specific cleavage. Thus, the coordinated action between Drosha and Dicer provide enhanced cleavage specificity and may contribute to enhanced functionality.
MATERIALS AND METHODS
RNA substrates
RNAs were chemically synthesized using 2′-ACE chemistry in house and PAGE purified. dsRNA sequences corresponding to the mitogen-activated protein kinase 14 (accession number NM_001315 position 1374–1435), human cyclophilin B (accession number NM_000942 positions 225–285), and Diazepam binding inhibitor (accession number NM_020548 positions 204–264) genes were used. shRNA were directed against human cyclophilin B. To test overhang length and overhang sequence, the antisense strand of MAPK14 was extended on the 3′ end. All RNA sequences are reported in Supplementary Materials at www.dharmacon.com/tech/publications. RNA was 5′ labeled using [32P]-γ-ATP (NEN) and polynucleotide kinase (Ambion), and purified by PAGE. For dsRNA, [32P]-labeled RNA was annealed to unlabeled RNA in 20 mM Tris, 0.1 mM EDTA (pH 8.0) by heating the samples at 95°C for 5 min and cooling gradually to 22°C over 10 min. [32P]-labeled shRNA was folded in 20 mM Tris, 0.1 mM EDTA (pH 8.0) by heating the samples at 95°C for 5 min, immediately cooling to 0°C on ice. A ratio of 1:10 labeled to unlabeled RNA was used to ensure that all labeled strands formed duplex RNA.
Human Dicer characterization
Commercially available recombinant human Dicer from Gene Therapy Systems, Stratagene, or Invitrogen contained 5%, ~15%, and 60% of the full-length protein as judged by SDS-electrophoresis, respectively. Dicer from Invitrogen containing at least 50% of full-length product was used in all binding experiments. No difference in cleavage specificity between different preparations of Dicer was found.
Dicer dsRNA cleavage assay
Dicer cleavage assay was performed in 20 mM Tris-HCl (pH 7.5), 250 mM NaCl, 2.5 mM MgCl2. Ten-microliter reactions were assembled using 0.05 units/μL human recombinant Dicer (Gene Therapy Systems, Stratagene, or Invitrogen) and duplex RNA (containing 0.05–0.1 μM labeled strand RNA) and incubated at 37°C for 0–3 h. Reactions were stopped by adding 10 μL 80% Formamide/10 mM EDTA with 10-fold excess RNA complementary to unlabeled strand for dsRNA or 10 μL 80% Formamide/10 mM EDTA for shRNA. Reactions were then heated for 5 min at 95°C before loading on 15% polyacrylamide/7 M urea gels. Data were collected using the Storm PhosphorImager 860 and quantified/analyzed using Imagequant 5.2 (Molecular Dynamics).
Nitrocellulose filter binding assays
For binding assays, Dicer purchased from Invitrogen was used (>50% full-length Dicer, as judged by SDS-electrophoresis). Protein concentration was determined using a Bradford Protein Assay (Bio-Rad). Active site titration was performed by varying labeled RNA concentration from 2 to 105 nM using the fixed protein concentration of 20 nM. According to active site titration, the protein was 50% active. To determine fraction RNA bound as a function of protein concentrations, filter binding assays were performed (on ice) in buffer containing 20 mM Tris (pH 7.5) with or without 2.5 mM magnesium chloride. Approximately 12 fmol of [32P]-labeled 61mer MAPK14 RNA was mixed with Dicer at a concentration of 2 or 400 nM in a reaction volume of 40 μL. After a 1-h incubation, the reactions were filtered through a 0.45 μm Millipore MF nitrocellulose membrane using the Schleicher and Schuell dot-blot apparatus (Wong and Lohman 1993). Subsequently, the amount of radioactivity retained on the filter was quantitated using a Molecular Dynamics PhosphorImager.
To determine the koff, approximately 300 pM [32P]-labeled 61mer MAPK14 was mixed with 40 nM Dicer in the buffer described above without magnesium. The RNA/protein complex was allowed to form on ice for (a) 1 min or (b) 30 min before an unlabeled RNA chase (100 nM) was added. Following the addition of the chase, aliquots were taken at various time points and filtered on nitrocellulose membrane as described previously.
Transfection of siRNAs
For transfection HEK293 cells were plated in 96-well plates at a density of 25,000 cells per well. siRNAs ranging in concentration between 0.032 and 100 nM were transfected using Lipofectamine 2000 (Invitrogen) at 0.4 μL per well. The cells were then grown at 37°C and harvested 48 h after transfection. mRNA levels were quantified by branched-DNA technology (Collins et al. 1997) using the QuantiGene High Volume Kit (Genospectra).
Supplemental Materials are available at www.dharmacon.com/tech/publications.
Acknowledgments
This project was partially funded by NSF Grant #DMI-0320480. We thank Dharmacon Production Staff, particularly Stephanie Hartsel, for oligonucleotide synthesis. In addition, we thank Kathryn Robinson for technical assistance and Emily Anderson, Yuriy Federov, Devin Leake, Elena Maksimova, and Barbara Robertson for helpful discussions.
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.7272305.
REFERENCES
- Bartel, D. 2004. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 116: 281–297. [DOI] [PubMed] [Google Scholar]
- Boden, D., Pusch, O., Silbermann, R., Lee, F., Tucker, L., and Ramratnam, B. 2004. Enhanced gene silencing of HIV-1 specific siRNA using microRNA designed hairpins. Nucleic Acids Res. 32: 1154–1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogenhagen, D. and Brown, D. 1981. Nucleotide sequences in Xenopus 5S DNA required for transcription termination. Cell 24: 261–270. [DOI] [PubMed] [Google Scholar]
- Brummelkamp, T.R., Bernards, R., and Agami, R. 2002. A system for stable expression of short interfering RNAs in mammalian cells. Science 296: 550–553. [DOI] [PubMed] [Google Scholar]
- Collins, M.L., Irvine, B., Tyner, D., Fine, E., Zayati, C., Chang, C., Horn, T., Ahle, D., Detmer, J., Shen, L.P., et al. 1997. A branched DNA signal amplification assay for quantification of nucleic acid targets below 100 molecules/ml. Nucleic Acids Res. 25: 2979–2984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbashir, S.M., Lendeckel, W., and Tuschl, T. 2001. RNA interference is mediated by 21- and 22-nucleotide RNAs. Genes & Dev. 15: 188–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannon, G.J. and Rossi, J.J. 2004. Unlocking the potential of the human genome with RNA interference. Nature 431: 371–378. [DOI] [PubMed] [Google Scholar]
- Hohjoh, H. 2004. Enhancement of RNAi activity by improved siRNA duplexes. FEBS Lett. 557: 193–198. [DOI] [PubMed] [Google Scholar]
- Holen, T., Amarzguioui, M., Wiiger, M.T., Babaie, E., and Prydz, H. 2002. Positional effects of short interfering RNAs targeting the human coagulation trigger Tissue Factor. Nucleic Acids Res. 30: 1757–1766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, Y., Ahn, C., Han, J., Choi, H., Kim, J., Yim, J., Lee, J., Provost, P., Radmark, O., Kim, S., et al. 2003. The nuclear RNase III Drosha initiates microRNA processing. Nature 425: 415–419. [DOI] [PubMed] [Google Scholar]
- Lee, Y., Nakahara, K., Pham, J., Kim, K., He, Z., Sontheimer, E., and Carthew, R. 2004. Distinct roles for Drosophila Dicer-1 and Dicer-2 in the siRNA/miRNA silencing pathways. Cell 117: 69–81. [DOI] [PubMed] [Google Scholar]
- Lingel, A., Simon, B., Izaurralde, E., and Sattler, M. 2003. Structure and nucleic-acid binding of the Drosophila Argonaute 2 PAZ domain. Nature 426: 465–469. [DOI] [PubMed] [Google Scholar]
- Liu, Q., Rand, T.A., Kalidas, S., Du, F., Kim, H.-E., Smith, D.P., and Wang, X. 2003. R2D2, a bridge between the initiation and effector steps of the Drosophila RNAi pathway. Science 301: 1921–1925. [DOI] [PubMed] [Google Scholar]
- Ma, J.B., Ye, K., and Patel, D.J. 2004. Structural basis for overhang-specific small interfering RNA recognition by the PAZ domain. Nature 429: 318–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyagishi, M. and Taira, K. 2002. U6 promoter-driven siRNAs with four uridine 3′ overhangs efficiently suppress targeted gene expression in mammalian cells. Nat. Biotechnol. 20: 497–500. [DOI] [PubMed] [Google Scholar]
- Myers, J.W., Jones, J.T., Meyer, T., and Ferrell, J.E. 2003. Recombinant Dicer efficiently converts large dsRNAs into siRNAs suitable for gene silencing. Nat. Biotechnol. 21: 324–328. [DOI] [PubMed] [Google Scholar]
- Pham, J., Pellino, J., Lee, Y., Carthew, R., and Sontheimer, E. 2004. A Dicer-2-dependent 80S complex cleaves targeted mRNAs during RNAi in Drosophila. Cell 117: 83–94. [DOI] [PubMed] [Google Scholar]
- Provost, P., Dishart, D., Doucet, J., Frendewey, D., Samuelsson, B., and Radmark, O. 2002. Ribonuclease activity and RNA binding of recombinant human Dicer. EMBO J. 21: 5864–5874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds, A., Leake, D., Scaringe, S., Marshall, W.S., Boese, Q., and Khvorova, A. 2004. Rational siRNA design for RNA interference. Nat. Biotechnol. 22: 326–330. [DOI] [PubMed] [Google Scholar]
- Siolas, D., Lerner, C., Burchard, J., Ge, W., Linsley, P.S., Paddison, P.J., Hannon, G.J., and Cleary, MA. 2004. Synthetic shRNAs as potent RNAi triggers. Nat. Biotechnol. 23: 227–231. [DOI] [PubMed] [Google Scholar]
- Song, J., Liu, J., Tolia, N., Schneiderman, J., Smith, S., Martienssen, R., Hannon, G., and Joshua-Tor, L. 2003. The crystal structure of the Argonaute2 PAZ domain reveals an RNA binding motif in RNAi effector complexes. Nat. Struct. Biol. 10: 1026–1032. [DOI] [PubMed] [Google Scholar]
- Wong, I. and Lohman, T.M. 1993. A double-filter method for nitro-cellulose-filter binding: Application to protein–nucleic acid interactions. Proc. Natl. Acad. Sci. 90: 5428–5432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamore, P.D., Tuschl, T., Sharp, P.A., and Bartel, D.P. 2000. RNAi: Double-stranded RNA directs the ATP-dependent cleavage of mRNA at 21 to 23 nucleotide intervals. Cell 101: 25–33. [DOI] [PubMed] [Google Scholar]
- Zeng, Y., Wagner, E.J., and Cullen, B.R. 2002. Both natural and designed micro RNAs can inhibit the expression of cognate mRNAs when expressed in human cells. Mol. Cell 9: 1327–1333. [DOI] [PubMed] [Google Scholar]
- Zhang, K. and Nicholson, AW. 1997. Regulation of ribonuclease III processing by double-helical sequence antideterminants. Proc. Natl. Acad. Sci. 94: 13437–13441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, H., Kolb, F.A., Brondani, V., Billy, E., and Filipowicz, W. 2002. Human Dicer preferentially cleaves dsRNAs at their termini without a requirement for ATP. EMBO J. 21: 5875–5885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, H., Kolb, F., Jaskiewicz, L., Westhof, E., and Filipowicz, W. 2004. Single processing center models for human Dicer and bacterial RNase III. Cell 118: 57–68. [DOI] [PubMed] [Google Scholar]