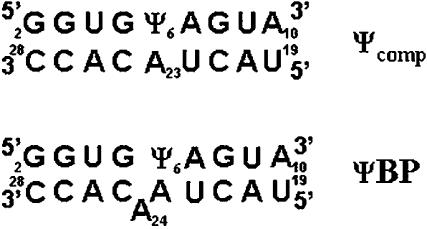Abstract
We have performed NMR experiments in supercooled water in order to decrease the temperature-dependent exchange of protons in RNA duplexes. NMR spectra of aqueous samples of RNA in bundles of narrow capillaries that were acquired at temperatures as low as −18°C reveal resonances of exchangeable protons not seen at higher temperatures. In particular, we detected the imino protons of terminal base pairs and the imino proton of a non-base-paired pseudouridine in a duplex representing the eukaryotic pre-mRNA branch site helix. Analysis of the temperature dependence of chemical shift changes (thermal coefficients) for imino protons corroborated hydrogen bonding patterns observed in the NMR-derived structural model of the branch site helix. The ability to observe non-base-paired imino protons of RNA is of significant value in structure determination of RNA motifs containing loop and bulge regions.
Keywords: pseudouridine, branch site, RNA, supercooled water, NMR, imino protons
INTRODUCTION
Determination of nucleic acid structures by NMR methodology relies upon numerous inter- and intramolecular distances and angular constraints. Interactions involving exchangeable imino and amino protons provide important information about hydrogen bonds and stabilizing interactions with water molecules, particularly for analysis of unusual structural motifs of folded RNA molecules. A sizable fraction of labile protons in RNA (-NH, -NH2, and -2′OH), however, exchange rapidly with solvent at T > 0°C. As a result, they are exchange broadened beyond detection, thus escaping use in structural studies. Sufficiently decreased temperature slows chemical exchange between these protons and water, allowing for their direct detection and recruitment for structural study. Borer and colleagues (Kerwood et al. 2001) detected resonance peaks of imino protons of terminal nucleotides at −6°C that were not observed at 0°C. In order to monitor non-base-paired imino protons, however, a method to achieve temperatures as low as −20°C without freezing is desirable.
Exploiting the empirical observation that the freezing point of water decreases proportionally with volume (Angell 1982), Poppe and van Halbeek (1994) studied sucrose in glass capillary tubes at −17°C, allowing measurement of –OH proton chemical shifts and 3JHH couplings. The capillary technique was extended to BPTI, ubiquitin, dATP, and dGTP in supercooled water at about −18°C (Skalicky et al. 2000, 2001).
In this study, we have investigated the structural role of exchangeable protons of pseudouridine (Ψ), a rotational isomer of uridine attached to its ribose through C5, in RNA duplexes. Ψ has two imino nitrogen atoms, Ψ N1H and Ψ N3H, both of which are protonated at physiological pH (Hall and McLaughlin 1991). Presence of Ψ in RNA helices has been shown to increase thermal stability without altering structure (Davis and Poulter 1991; Hall and McLaughlin 1991; Arnez and Steitz 1994; Kintanar et al. 1994; Durant and Davis 1999; Yarian et al. 1999), postulated to be the result of a water-mediated hydrogen bond involving the Ψ N1H (Arnez and Steitz 1994; Newby and Greenbaum 2002a) and/or improved base stacking (Yarian et al. 1999; Chui et al. 2002). In the case of the pre-mRNA branch site helix of the yeast spliceosome, the presence of a conserved Ψ induces a strikingly different structure than that observed with uridine (Newby and Greenbaum 2001, 2002b). Structural models of the branch site duplex suggested that Ψ did not appear to form a base pair with the opposing adenine (A23, adjacent to the bulged base, A24), and no resonance attributable to Ψ N3H was visible (Fig. 1). Identification of this resonance would contribute important information about the environment of this imino proton with respect to surrounding bases.
FIGURE 1.
Sequences of the two RNA duplexes Ψcomp (top) and ΨBP (bottom). ΨBP represents short sequences from the pairing of U2 snRNA (top strand) and the intron (bottom strand) from the yeast S. cerevisiae. The numbering scheme was that used in previous structure determination studies (Newby and Greenbaum, 2001, 2002a,b). The position of Ψ (Ψ6 in this sequence) corresponds to a phylogenetically conserved Ψ residue in U2 snRNA (Ψ35 in yeast). Ψcomp is a similar sequence without the bulged A.
In order to slow the exchange of imino protons, we have acquired NMR spectra of two RNA duplexes containing Ψ (Fig. 1) in aqueous solutions at supercooled temperatures by implementation of the capillary technique (Poppe and van Halbeek 1994). In addition to detecting imino protons corresponding to terminal base pairs of an RNA duplex, we observed the upfield-shifted N3H imino proton of a non-base-paired Ψ in the branch site duplex. Determining the role of Ψ in stabilizing RNA structures may explain its phylogenetic conservation in the branch site helix and elsewhere in structural RNA molecules.
RESULTS AND DISCUSSION
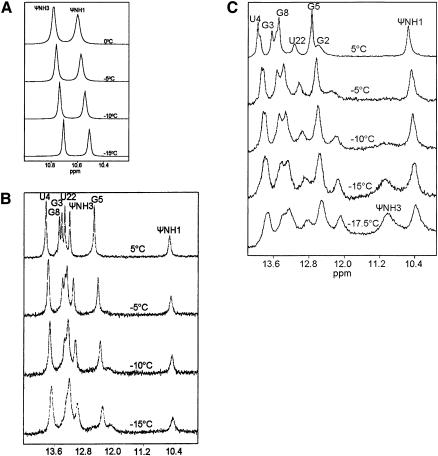
We first acquired NMR spectra of pseudouridine monophosphate (ΨMP) in order to characterize the resonances of its two imino protons, Ψ N1H and Ψ N3H, in a non-base-paired environment. Spectra of imino protons (including line widths) were identical for samples in capillaries and as a bulk sample at 0°C and 5°C (data not shown). The Ψ N1H and Ψ N3H proton chemical shifts were 10.6 and 10.8 ppm and line-widths at half-height (ν1/2) were −3.8 and ~ 5.1 Hz at 0°C, respectively (Fig. 2A). Upon cooling to −15°C, ν1/2 decreased monotonically (−2.0 and −2.7 Hz for N1H and N3H, respectively), consistent with line narrowing associated with slower solvent exchange. ν1/2 of N1H was slightly larger than that of N3H, presumably as a result of dipolar broadening from the proximal H6 proton of Ψ (−2.5 and 4.8 Å between H6 and N1H and N3H, respectively). We also observed a slight temperature dependence of chemical shift (thermal coefficient) for the imino protons. Positive thermal coefficients (i.e., more upfield chemical shift as a function of decreased temperature) correlate with rapid exchange, whereas negative thermal coefficients correlate with slower exchange, and are therefore consistent with hydrogen bonding (Nonin et al. 1995). We plotted the chemical shift with respect to temperature for each imino proton. The thermal coefficients for Ψ N1H and Ψ N3H were 6.1 and 5.3 ppb/°C, respectively, consistent with a non-base-paired environment for each proton (data not shown).
FIGURE 2.
Imino proton spectra of Ψ-monophosphate (ΨMP) (A), Ψcomp (B), and ΨBP (C) acquired at different temperatures between 5°C and −17.5°C. (A) ΨMP (10 mM in 10 mM Na phosphate at pH 6.4, 50 mM NaCl, 0.1 mM EDTA, in 90% H2O/10% D2O) was taken up into six capillary tubes, which were placed in a 5-mm NMR tube (fill factor of −0.25). (B) Ψcomp (2.7-mM duplex) was prepared as in A and was taken up into 10 capillary tubes, (fill factor of −0.4). (C) ΨBP (2.5 mM duplex) was prepared as in A and taken up into nine capillary tubes (fill factor of −0.3). Data were collected using a jump-return-echo pulse sequence (Sklenár and Bax 1987) on a Varian 720-MHz and 600-MHz spectrometer at the National High Magnetic Field Laboratory.
By comparison, NMR spectra of a complementary duplex (Ψcomp) (Fig. 1) indicate that all imino protons, including N3H of Ψ, participate in Watson-Crick base pairs (Fig. 2B). When Ψ is in the anti-conformation, its N3H forms a hydrogen bond with the opposing adenine N1, reflected in the downfield shift of the imino proton resonance to −13.1 ppm (Fig. 2B; Hall and McLaughlin 1992; Durant and Davis 1999). Further confirmation of this assignment came from comparison of chemical shifts of a similar duplex in which Ψ were replaced by uridine (U). We assigned the resonance at 10.6 ppm to Ψ N1H, which is on the major groove edge of the base when in an anti-conformation, based upon the chemical shift for this imino proton in ΨMP and from observation of an NOE between it and the H6 proton (−2.5 Å away) (Hall and McLaughlin 1992; Newby and Greenbaum 2001). In order to determine how this proton is protected from rapid exchange with solvent, our previous studies made use of a CLEANEX-PM pulse sequence (Hwang et al. 1997) to characterize the interaction between Ψ N1H and water molecules (Newby and Greenbaum 2002a). For protons undergoing chemical exchange, this experiment determines whether there is a component of cross-relaxation. The spectrum acquired using a CLEANEX-PM pulse sequence revealed a strong negative peak at the resonance location of Ψ N1H (but no other imino proton), indicating that the interaction with water is characterized by a significant component of cross-relaxation. This observation is consistent with participation of Ψ N1H in a water-mediated hydrogen bond with a phosphate oxygen atom of the same or a neighboring nucleotide. This hydrogen bonding status was predicted by Durant and Davis (1999), and visualized in a crystal structure of tRNA containing pseudouridine (Arnez and Steitz 1994).
At about −10°C, a broad peak appeared at −12.2 ppm with a shoulder at −12.5 ppm. All imino protons of Ψcomp had previously been assigned except for those belonging to terminal base pairs (Fig. 2B); therefore, these new peaks were attributed to the imino protons of terminal residues. In contrast with the narrowing of peaks of the monophosphate at lower temperatures, broadening of peaks of the complementary duplex was observed below 0°C (line widths increased gradually from −50 Hz to −130 Hz). The exact reasons for this behavior may be a complex combination of slower tumbling of the larger molecule as a result of increased solvent viscosity, salt and buffer effects, or may represent the beginning of cold denaturation.
We then performed similar experiments on a minimal pre-mRNA branch site duplex of Saccharomyces cerevisiae (ΨBP), which represents the pairing between a short consensus region of the U2 snRNA and the intron (Fig. 1). In contrast with the complementary duplex, presence of a Ψ residue in the U2 snRNA strand of ΨBP (top strand in Fig. 1) in its conserved position (Yu et al. 1998; Ma et al. 2003) results in a very different conformation than in its unmodified counterpart (Newby and Greenbaum 2001, 2002b). In the novel ΨBP motif, the unpaired adenosine is extruded from the helix and forms a base triple with the minor groove edge of A7 in the A7-U22 base pair. The 2′OH of this extrahelical adenosine is the nucleophile in the first cleavage step of splicing. The structural motif preferred in the presence of this Ψ (Ψ35 in the native yeast sequence) may explain the strong phylogenetic preservation of this modified base in this location. In ΨBP, the chemical shift of Ψ N1H is −10.5 ppm (vs. −10.6 ppm in ΨMP and Ψcomp). As was observed in Ψcomp, previous NMR investigation of the interaction of Ψ N1H of ΨBP indicated that this proton undergoes cross-relaxation with water molecule(s) in the major groove of the duplex (Newby and Greenbaum 2002a). Unlike the case with Ψcomp, proton spectra of ΨBP revealed no resonance attributable to Ψ N3H, and structural models did not indicate formation of a base pair between Ψ and the opposing adenine (A23, adjacent to the branch site base A24).
In order to identify the resonance location of the absent (and presumably exchange broadened) Ψ N3H in the branch site duplex at 0°C–5°C, we acquired spectra of ΨBP in supercooled water (Fig. 2C). As in spectra of the complementary duplex, a broad new peak emerged at −12.2 ppm below −5°C, corresponding to the G2 terminal base pair. The imino proton of the other terminal base pair may be degenerate with a resonance at −13.3 ppm. Unique to the branch site duplex, a broad peak emerged at −11.2 ppm at −15°C. Because assignments of all other imino protons of the branch site helix were made by systematic comparison with other duplexes (Newby and Greenbaum 2002b), we identified this upfield-shifted resonance as that of Ψ N3H. No NOEs were observed from this broad peak. The Ψ N3H chemical shift was close to that of the unpaired N3H of ΨMP, which is not base paired (Fig. 2A) and very different from that of the Watson-Crick paired Ψ of Ψcomp(Fig. 2B). The upfield location of Ψ N3H of ΨBP (Fig. 2C) further supports our original finding that Ψ6 does not form a canonical base pair with A23 (Newby and Greenbaum 2002b). As with Ψcomp, line widths began to broaden below 0°C, increasing from −50 Hz to −200 Hz.
We noted that chemical shifts of each imino proton varied slightly with a decrease in temperature. We therefore generated a thermal coefficient for each imino proton resonance by plotting the chemical shifts as a function of temperature (Fig. 3). For Ψcomp, negative thermal coefficients (more downfield shifts with respect to decreased temperature) were observed for all protons except the terminal imino proton (Fig. 3, top). The thermal coefficient for ΨN1H and ΨN3H was −6.2 ppb/°C and −0.7 ppb/°C, respectively, suggesting that both form hydrogen bonds (Newby and Greenbaum 2001, 2002a,b). The thermal coefficient for G2 was positive, consistent with rapid exchange expected for a solvent-exposed terminal base pair.
FIGURE 3.
Thermal coefficients for Ψcomp (top) and ΨBP (bottom). The chemical shift for each proton was plotted as a function of temperature at which the spectrum was acquired. The value (in ppb/°C) for each imino proton is shown next to its position in the sequence. Positive thermal coefficients correlate with rapid exchange and negative thermal coefficients correlate with hydrogen bonding (Nonin et al. 1995). See text for further details.
In ΨBP, protons with negative thermal coefficients belonged to G8, G5, G3, U4, and Ψ N1H, and those with positive thermal coefficients were G2, U22, and Ψ N3H (Fig. 3, bottom). The negative thermal coefficient for Ψ N1H (−3.7 ppb/°C) reinforced the conclusion that this imino proton is involved in a hydrogen bond, as suggested by the results of earlier NMR studies (Newby and Greenbaum 2002a). The thermal coefficient of U22 was 1.03 ppb/°C and that of Ψ N3H was 0.08 ppb/°C. Although U22 forms a Watson-Crick base pair with A7, its involvement in the base triple with the branch site A and its position adjacent to the unpaired Ψ exposes U22 N3H to solvent, which apparently increases its exchange rate. For Ψ N3H, this positive value is in accord with rapid exchange, suggesting the proton is not involved in a hydrogen bond. This observation, combined with its upfield-shifted position and lack of NOEs to surrounding residues, especially the adjacent A2H, is entirely consistent with our original conclusion about this Ψ’s non-base-paired status (Newby and Greenbaum 2002b) and assists in further refinement of the ΨBP structure.
In conclusion, NMR data at low temperatures allow us to identify imino protons that are undetected at higher temperatures due to rapid exchange. This technique may be helpful in structure determination of non-base-paired regions of novel RNA motifs and for extending the temperature range over which chemical shift data can be acquired. Applying the capillary technique appears to be promising for NMR in supercooled water using other interesting biomolecules.
MATERIALS AND METHODS
Design and synthesis of samples
The solutions of RNA we used were the pseudouridine mono-phosphate (ΨMP), a complementary RNA duplex (Ψcomp, 5′-GGUGΨAGUA-3′ vs. 5′-UACUACACC-3′) (shown in Fig. 1, top), and an RNA duplex with a bulged adenosine, representing a minimal form of the pre-mRNA branch site from S. cerevisiae (ΨBP, 5′-GGUGΨAGUA-3′ vs. 5′-UACUAACACC-3′; bulged A is underlined) (Fig. 1, bottom). Pseudouridine monophosphate was purchased from Berry and Associates (no. PYA11080). RNA oligomers were purchased from Dharmacon Research and deprotected according to company protocol (Wincott et al. 1995). Samples were precipitated with ethanol, partially desalted by three washes of the pelleted RNA with 80% ethanol, and lyophilized to dryness. The RNA pellets were then resuspended in diethyl pyrocarbonate–treated (DEPC) water, and strand concentrations were calculated from the absorbance at 260 nm. Equimolar amounts of the strands were combined to obtain duplexes of the Ψcomp and ΨBP and verified by mobility on a nondenaturing gel. The duplexes were lyophilized to dryness and resuspended in 250 μL of NMR buffer consisting of 10 mM sodium phosphate (pH 6.4), 50 mM sodium chloride, and 0.1 mM EDTA in 90% H2O/10% D2O (99.96%; Cambridge Isotope Laboratories). Final concentration of RNA strands in NMR samples was −2.7 mM for Ψcomp and −2.5 mM for ΨBP. NMR assignments of imino protons of ΨBP were reported previously (Newby and Greenbaum 2001, 2002b).
Sample preparation
Open-ended glass capillaries of 1.0 mm OD were purchased from Fisher (no. 34500-99). The capillaries were prepared by soaking in 100% methanol overnight and then dried at 170°C. Solutions of RNA were centrifuged at 14,000 rpm for 30 min in order to remove particles that could nucleate ice crystals. The samples were then taken up by capillary action with −27 μL in each capillary. The ends of the open capillaries were flamed sealed, and the capillaries were placed in a 5-mm NMR tube (Poppe and van Halbeek 1994; Skalicky et al. 2000, 2001). Bundles of 9–10 capillaries have a filling factor of 30%–40% to yield an effective concentration of −1 mM for the two RNA duplexes; ΨMP, with a starting concentration of 10 mM in six capillaries, had an effective concentration of −2.5 mM.
NMR spectroscopy
NMR spectra were acquired on a 720-MHz and 600-MHz Varian Unity Plus spectrometers (National High Magnetic Field Laboratory). Samples were cooled at a rate of −1.5°C/h to −2.5°C/h with an increase in rate as the temperature decreased. The jump-return echo pulse sequence (Sklenár and Bax 1987) was used to array the temperature decrease in order to observe the imino protons at different temperatures, and freezing of one or more capillaries was monitored by a proportional decrease in signal intensity, but no capillary tubes broke. We used 64 steady-state scans prior to each acquisition. In order to improve signal quality at lower temperatures, we doubled the number of acquisitions for each incremental temperature decrease of 2.5°C: 128 at −5°C, 512 scans for −10°C, and up to 4096 scans at −17.5°C. Processing of NMR data and simulation line-width at half height were accomplished using Felix 2.3 (Biosyn).
Acknowledgments
We thank the National High Magnetic Laboratory and the Nuclear Magnetic Resonance Laboratory in the Department of Chemistry and Biochemistry at FSU for the use of their NMR facilities. This material is based upon work supported by the National Science Foundation under Grant No. 0316494 and by the National Institutes of Health Grant GM54008.
Article and publication are at http://www.rnajournal.org/cgi/doi/10.1261/rna.2270205.
REFERENCES
- Angell, C.A. 1982. In water: A comprehensive treatise. Plenum Press, New York.
- Arnez, J.G. and Steitz, T.A. 1994. Crystal structure of unmodified tRNA(Gln) complexed with glutaminyl-tRNA synthetase and ATP suggests a possible role for pseudouridines in stabilization of RNA structure. Biochemistry 33: 7560–7567. [DOI] [PubMed] [Google Scholar]
- Chui, H.M., Desaulniers, J.P., Scaringe, S.A., and Chow, C.S.2002.Synthesis of helix 69 of Escherichia coli 23S rRNA containing its natural modified nucleosides, m(3) Ψ and Ψ. J. Org. Chem. 67: 8847–8854. [DOI] [PubMed] [Google Scholar]
- Davis, D.R. and Poulter, C.D. 1991. 1H-15N NMR studies of Escherichia coli tRNA(Phe) from hisT mutants: A structural role for pseudouridine. Biochemistry 30: 4223–4231. [DOI] [PubMed] [Google Scholar]
- Durant, P.C. and Davis, D.R. 1999. Stabilization of the anticodon stem-loop of tRNALys,3 by an A+-C base-pair and by pseudouridine. J. Mol. Biol. 285: 115–131. [DOI] [PubMed] [Google Scholar]
- Hall, K.B. and McLaughlin, L.W. 1991. Properties of a U1/mRNA 5′ splice site duplex containing pseudouridine as measured by thermodynamic and NMR methods. Biochemistry 30: 1795–1801. [DOI] [PubMed] [Google Scholar]
- ———. 1992. Properties of pseudouridine N1 imino protons located in the major groove of an A-form RNA duplex. Nucleic Acids Res. 20: 1883–1889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang, T.L., Mori, S., Shaka, A.J., and vanZijl, P.C.M. 1997. Application of phase-modulated CLEAN chemical EXchange spectroscopy (CLEANEX-PM) to detect water–protein proton exchange and intermolecular NOEs. J. Am. Chem. Soc. 119: 6203–6204. [Google Scholar]
- Kerwood, D.J., Cavaluzzi, M.J., and Borer, P.N. 2001. Structure of SL4 RNA from the HIV-1 packaging signal. Biochemistry 40: 14518–14529. [DOI] [PubMed] [Google Scholar]
- Kintanar, A., Yue, D., and Horowitz, J. 1994. Effect of nucleoside modifications on the structure and thermal stability of Escherichia coli valine tRNA. Biochimie 76: 1192–1204. [DOI] [PubMed] [Google Scholar]
- Ma, X., Zhao, X., and Yu, Y.T. 2003. Pseudouridylation (Ψ) of U2 snRNA in S. cerevisiae is catalyzed by an RNA-independent mechanism. EMBO J. 22: 1889–1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newby, M.I. and Greenbaum, N.L. 2001. A conserved pseudouridine modification in eukaryotic U2 snRNA induces a change in branch-site architecture. RNA 7: 833–845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ———. 2002a. Investigation of Overhauser effects between pseudouridine and water protons in RNA helices. Proc. Natl. Acad. Sci. 99: 12697–12702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ———. 2002b. Sculpting of the spliceosomal branch site recognition motif by a conserved pseudouridine. Nat. Struct. Biol. 9: 958–965. [DOI] [PubMed] [Google Scholar]
- Nonin, S., Leroy, J.L., and Gueron, M. 1995. Terminal base pairs of oligodeoxynucleotides: Imino proton exchange and fraying. Biochemistry 34: 10652–10659. [DOI] [PubMed] [Google Scholar]
- Poppe, L. and van Halbeek, H. 1994. NMR spectroscopy of hydroxyl protons in supercooled carbohydrates. Nat. Struct. Biol. 1: 215–216. [DOI] [PubMed] [Google Scholar]
- Skalicky, J.J., Sukumaran, D.K., Mills, J.L., and Szyperski, T. 2000. Toward structural biology in supercooled water. J. Am. Chem. Soc. 122: 3230–3231. [Google Scholar]
- Skalicky, J.J., Mills, J.L., Sharma, S., and Szyperski, T. 2001. Aromatic ring-flipping in supercooled water: Implications for NMR-based structural biology of proteins. J. Am. Chem. Soc. 123: 388–397. [DOI] [PubMed] [Google Scholar]
- Sklenár, V. and Bax, A. 1987. Spin-echo water suppression for the generation of pure-phase two-dimensional NMR-spectra. J. Magn. Reson. 74: 469–479. [Google Scholar]
- Wincott, F., DiRenzo, A., Shaffer, C., Grimm, S., Tracz, D., Workman, C., Sweedler, D., Gonzalez, C., Scaringe, S., and Usman, N. 1995. Synthesis, deprotection, analysis and purification of RNA and ribozymes. Nucleic Acids Res. 23: 2677–2684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yarian, C.S., Basti, M.M., Cain, R.J., Ansari, G., Guenther, R.H., Sochacka, E., Czerwinska, G., Malkiewicz, A., and Agris, P.F. 1999. Structural and functional roles of the N1- and N3-protons of Ψ at tRNA’s position 39. Nucleic Acids Res. 27: 3543–3549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, Y.T., Shu, M.D., and Steitz, J.A. 1998. Modifications of U2 snRNA are required for snRNP assembly and pre-mRNA splicing. EMBO J. 17: 5783–5795. [DOI] [PMC free article] [PubMed] [Google Scholar]