Abstract
Chrysanthemum chlorotic mottle viroid (CChMVd) RNA (398–401 nucleotides) can form hammerhead ribozymes that play a functional role in its replication through a rolling-circle mechanism. In contrast to most other viroids, which adopt rod-like or quasi-rod-like secondary structures of minimal free energy, the computer-predicted conformations of CChMVd and Peach latent mosaic viroid (PLMVd) RNAs are branched. Moreover, the covariations found in a number of natural CChMVd variants support that the same or a closely related conformation exists in vivo. Here we report that the CChMVd natural variability also supports that the branched conformation is additionally stabilized by a kissing-loop interaction resembling another one proposed in PLMVd from in vitro assays. Moreover, site-directed mutagenesis combined with bioassays and progeny analysis showed that: (1) single CChMVd mutants affecting the kissing loops had low or no infectivity at all, whereas infectivity was recovered in double mutants restoring the interaction; (2) mutations affecting the structure of the regions adjacent to the kissing loops reverted to wild type or led to rearranged stems, also supporting their interaction; and (3) the interchange between 4 nucleotides of each of the two kissing loops generated a viable CChMVd variant with eight mutations. PAGE analysis under denaturing and nondenaturing conditions revealed that the kissing-loop interaction determines proper in vitro folding of CChMVd RNA. Preservation of a similar kissing-loop interaction in two hammerhead viroids with an overall low sequence similarity suggests that it facilitates in vivo the adoption and stabilization of a compact folding critical for viroid viability.
Keywords: Chrysanthemum chlorotic mottle viroid, Avsunviroidae, catalytic RNAs
INTRODUCTION
Viroids are unique systems for the study of RNA structural-functional relationships. Their minimal circular genome (246–401 nucleotides) does not code for any protein, yet these RNAs contain sufficient information to select certain host plants, to usurp part of their metabolism for the synthesis of the new viroid progeny and, as a consequence, to incite in most cases specific diseases (Diener 1999; Flores et al. 2004; Tabler and Tsagris 2004). Viroids are divided into two families, Pospiviroidae and Avsunviroidae, whose members replicate and accumulate in the nucleus and the chloroplast, respectively. The classification scheme is supported by other criteria which in members of the family Avsunviroidae include self-cleavage, mediated by hammer-head ribozymes, of the oligomeric RNA intermediates of both polarities resulting from replication through a rolling-circle mechanism (Flores et al. 2000, 2004).
From the structural point of view there is firm evidence that Potato spindle tuber viroid (PSTVd) (Diener 1972), the type species of the family Pospiviroidae (Flores et al. 2004), adopts in vitro a typical rod-like (or quasi-rod-like) secondary structure formed by alternating short double-stranded regions and single-stranded loops (Sogo et al. 1973; Sänger et al. 1976; Gross et al. 1978; Riesner et al. 1979; Dingley et al. 2003). Moreover, the artificial deletions introduced in PSTVd (Wassenegger et al. 1984) and the natural repetitions observed in other species of its family (Haseloff et al. 1982; Semancik et al. 1994; Fadda et al. 2003a) lead to variants preserving the rod-like structure, indicating that this structure is also significant in vivo. Based on sequence comparisons, the rod-like structure has been divided into five structural domains (Keese and Symons 1985; Sano et al. 1992), some of which contain characteristic conserved regions such as the central conserved region (CCR) (McInnes and Symons 1991), the terminal conserved region (TCR), and the terminal conserved hairpin (TCH) (Koltunow and Rezaian 1988; Flores et al. 1997).
This model, however, does not apply to members of the family Avsunviroidae (Flores et al. 2000). Avocado sunblotch viroid (ASBVd) (Symons 1981; Hutchins et al. 1986), the type species of this family, and Eggplant latent viroid (ELVd) (Fadda et al. 2003b) adopt quasi-rod-like secondary structures but do not contain the aforementioned structural domains or conserved regions. On the other hand, the computer-predicted branched conformation of Chrysanthemum chlorotic mottle viroid (CChMVd) (Navarro and Flores 1997) is supported by physical and biological data indicating that this, or a closely related conformation, also exists in vitro and in vivo. The insolubility of CChMVd in 2 M LiCl is consistent with a folding different from the rod-like or quasi rod-like structure proposed for most viroids, which are soluble in these saline conditions (Navarro and Flores 1997), and a collection of covariations and point mutations found in CChMVd natural variants also supports the proposed branched conformation (Navarro and Flores 1997; De la Peña et al. 1999; De la Peña and Flores 2002; this work). Peach latent mosaic viroid (PLMVd) (Hernández and Flores 1992), which is also insoluble in 2 M LiCl (Navarro and Flores 1997), is the only other viroid with a similar secondary structure in which the observed sequence variability can be accommodated (Ambrós et al. 1998, 1999; Pelchat et al. 2000; Malfitano et al. 2003). Additional credence for such a PLMVd secondary structure was obtained by in vitro nuclease mapping and oligonucleotide binding shift assays (Bussière et al. 2000), which also indicated the probable existence of a pseudoknot-like interaction between two hairpin loops of the proposed branched conformation. Moreover, based on a comparative analysis of PLMVd and CChMVd structures, the possibility of a similar interaction in CChMVd was advanced (Bussière et al. 2000). However, in vitro chemical probing does not necessarily entail base pairing in vivo, because the protected residues might be involved in complex tertiary interactions or in RNA-protein contacts.
Based on the structural analysis of numerous natural and experimental CChMVd variants, on bioassay in chrysanthemum to assess their biological properties, and on the genetic stability of the resulting progenies, we report here the existence of the predicted kissing-loop interaction, which, moreover, is critical for the in vitro folding and the in vivo viability of the viroid.
RESULTS
Covariations and point mutations in natural CChMVd variants suggest the existence in vivo of a kissing-loop interaction
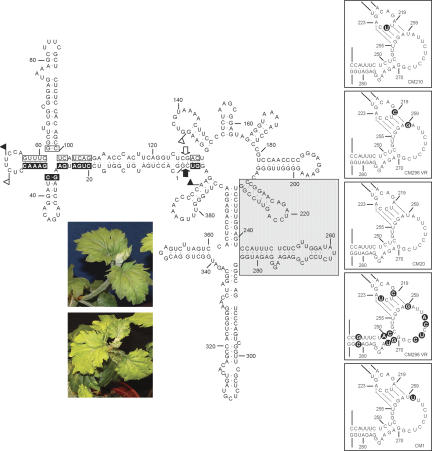
Figure 1 presents the computer-predicted conformation of the CChMVd reference variant CM20 (Navarro and Flores 1997; De la Peña et al. 1999) with the proposed kissing-loop interaction (Bussière et al. 2000). To obtain evidence supporting this interaction, we analyzed the genetic heterogeneity of natural CChMVd variants (Fig. 1, right insets). Four different mutations are located in the CChMVd nucleotides directly involved in the proposed kissing-loop interaction. Two consist of C→U transitions (positions 221 and 222), which substitute Watson-Crick by wobble pairs with the two Gs at positions 257 and 256, respectively. Interestingly, the transversions A→C (position 219) and U→G (position 259), which have been concurrently observed in a number of CChMVd natural variants, are covariations that convert the base pair between A219 and U259 into the pair between C219 and G259. A fifth change, A→U (position 260), could be interpreted as the replacement of a non-Watson-Crick base pair between G218 and A260 with a wobble pair between G218 and U260, suggesting that the kissing-loop interaction might be larger and even include the adjacent base pair between A217 and U261.
FIGURE 1.
Primary and predicted secondary structure of the CChMVd RNA (variant CM20). Plus and minus self-cleavage domains are delimited by flags, residues conserved in most natural hammerhead structures are within boxes, and the self-cleavage sites are indicated by arrows. Black and white backgrounds in flags, boxes, and arrows refer to plus and minus polarities, respectively. The gray square demarcates the domain that alternatively can form a loop–loop interaction. (Right insets) Proposed kissing-loop interaction between nucleotides 219–223 and 255–259 of CM20 and other natural CChMVd variants in which covariations and point mutations in the kissing loops or in their adjacent stems, shown on black circles, support the interaction. Watson-Crick and wobble base pairs are denoted by dots. Only the CChMVd hairpins engaged in this interaction are presented. (Left inset) Typical leaf symptoms induced by CChMVd variant CM20 (bottom) compared with a healthy control (top).
Furthermore, additional covariations were found affecting the nucleotides forming the stem adjacent to the kissing loop delimited by positions 253–269: In some variants, the base pair between U251 and A271 was substituted by an alternative C-G pair, and the base pair between C250 and G272 was substituted by an A-U pair, and in some others the base pair between A244 and U281 was substituted by a G-C pair (Fig. 1, right insets). These data, together with the covariations reported previously in positions 215 and 225, and 216 and 224 of the stem adjacent to the other kissing loop (Fig. 1; Navarro and Flores 1997; De la Peña et al. 1999), support the in vivo existence of the proposed kissing-loop interaction.
Single mutants affecting the kissing loops are less infectious than double mutants with the interaction restored
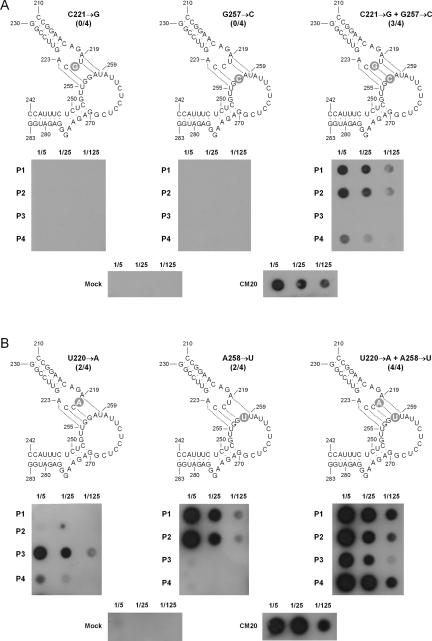
To provide further evidence in this direction, site-directed mutagenesis was used to construct from the CM20 variant the two single mutants C221→G and G257→C, which should impede the presumed base pair formed by C221 and G257, and the double mutant, which should restore the base pair between these two positions (Fig. 2A). When the monomeric CChMVd RNAs with the two single mutations, obtained from self-cleavage during transcription of linearized plasmids with the corresponding dimeric head-to-tail inserts, were bioassayed in chrysanthemum, no symptoms appeared up to 90 d postinoculation (dpi) (when the experiment was concluded), and dot-blot hybridization analysis failed to reveal any viroid accumulation (Fig. 2A). In contrast, in a control experiment with the wild-type CM20 RNA, symptoms (Fig. 1, left inset) and strong hybridization signals (Fig. 2A) were observed 7–10 dpi in all the inoculated plants. The double mutant RNA was also very infectious (3/4 plants), although symptom expression was delayed up to 21 dpi; at that time, the intensity of the hybridization signals paralleled those of the plants inoculated with the wild-type variant (Fig. 2A). Moreover, RT-PCR amplification, cloning, and sequencing of the progeny RNA from one of the symptomatic plants inoculated with the double mutant revealed that the two mutations were preserved in the three clones analyzed, indicating that the infectivity and accumulation of this double mutant did not result from reversion to wild type.
FIGURE 2.
Effects on CChMVd infectivity (indicated by the fraction of symptomatic plants within parentheses) of experimental single and double mutations disrupting and restoring, respectively, the central base pair (A) and an adjacent base pair (B) of the proposed kissing-loop interaction. Dot-blot hybridizations of individual chrysanthemum plants (P1–P4) inoculated with the corresponding CChMVd RNAs are displayed below each mutant. Control preparations from mock- and CChMVd (CM20)-inoculated plants are shown in the lower part of the two panels. Aliquots of plant RNA preparations (5 μL) of three dilutions (1/5, 1/25, and 1/125) were applied to membranes that were hybridized with a full-length CChMVd-specific riboprobe. Tissue was collected 14 dpi, with the fraction of symptomatic plants referring also to this time except in the single mutants U220→A and A258→U, and in the double mutant C221→G + G257→C that refer to 21 dpi. Other details are as in the Figure 1 legend. Mutations introduced experimentally are shown on gray circles.
In a second experiment series, the presumed neighboring base pair formed by U220 and A258 was tested by constructing the two single mutants U220→A and A258→U and the double mutant, which should disrupt and restore, respectively, the base pairing between these positions (Fig. 2B). Bioassay of the corresponding CChMVd RNAs showed no symptoms 14 dpi, but dot-blot hybridization signals were observed in 2/4 (mutant U220→A) and 2/4 (mutant A258→U) plants, which did express symptoms 21 dpi. Progeny analysis of one dot-blot-positive plant of each single mutant revealed reversion to wild type in the four and five sequenced clones, respectively. On the other hand, inoculation with the double mutant induced symptoms (4/4 plants) at the same time and viroid accumulation with the same titer as the wild-type CChMVd RNA (Fig. 2B), and progeny analysis of one of the symptomatic plants showed that the two mutations were preserved in the eight clones sequenced. These results reinforce the conclusions of the previous experiment and also suggest that disrupting the central base pair (C221-G257) of the proposed kissing-loop interaction annuls viroid replication, whereas disrupting the adjacent base pair (U220-A258) allows some basal replication that eventually leads to reversion to wild type. This differential effect may be related to the relative position and strength of the two base pairs.
Mutations in the stems flanking the kissing loops also support their interaction
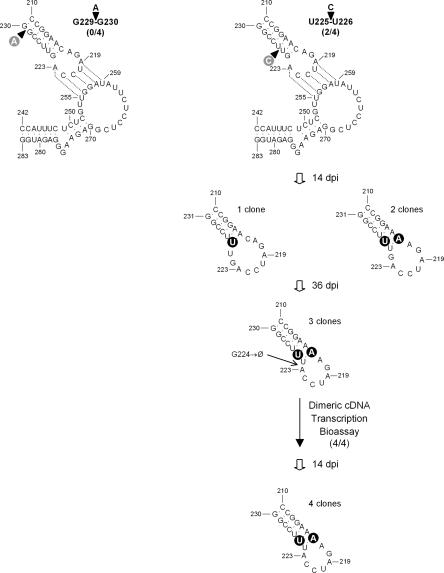
Considering the previous results, we anticipated that alterations in the length and structure of the stems adjacent to the kissing loops should affect their interaction. To check this hypothesis, the stem of the hairpin delimited by positions 210 and 230 was disrupted by inserting an A between G229 and G230 (Fig. 3). This bulged nucleotide at the base of the stem should induce a kink most likely disfavoring the kissing-loop interaction. The resulting CChMVd RNA failed to incite symptoms in chrysanthemum plants (and dot-blot hybridization confirmed that they were not infected), thus supporting the functional significance of this stem. The same hairpin stem was then disrupted in another position proximal to the hairpin loop. Insertion of a C between U225 and U226 generated a CChMVd RNA that induced symptoms in 2/4 inoculated plants, and analysis of the progeny of one of these plants 14 dpi showed that the original mutation was lost: The inserted C was substituted by a U in one clone (giving rise to a hairpin with a stem of six base pairs and a loop of 10 nucleotides), with the same change being accompanied in two other clones by a C216→A substitution (generating a stem of seven base pairs and a loop of 8 nucleotides) (Fig. 3). These two changes were also present in the three analyzed clones of tissue collected 36 dpi from the same plant together with the deletion of G224 that restored a hairpin with a size identical to the wild type: a stem of seven base pairs and a loop of 7 nucleotides (Fig. 3). Moreover, when the corresponding CChMVd RNA was in turn bioassayed, 4/4 inoculated plants displayed symptoms and the three mutations were preserved in the four clones analyzed from one of the symptomatic plants. Collectively these results, particularly the observed rearrangement, are consistent with a strong selective pressure favoring a hairpin with the ability to maintain the kissing-loop interaction.
FIGURE 3.
Effects of experimental mutations affecting the structure of the stem of the hairpin delimited by positions 210–230 on CChMVd infectivity (indicated by the fraction of symptomatic plants within parentheses). The structure of this hairpin in the viroid progeny from one of the mutants and from one of the clones of this mutant is displayed below. “Ø” designates deletion. No spontaneous mutations were observed in the region delimited by positions 242–283 (data not shown). Mutations introduced experimentally and those emerging spontaneously are shown on gray and black circles, respectively. Other details as in the legend to Figure 1.
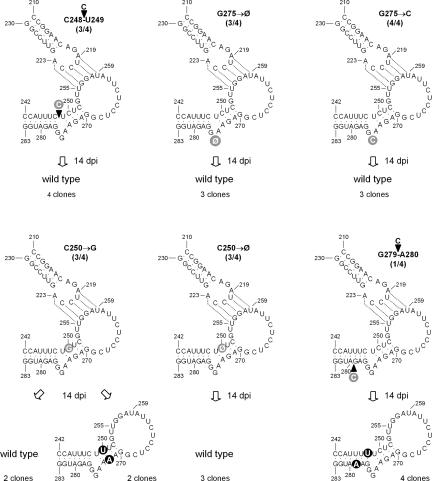
Alterations of the stem-bulge-stem structure adjacent to the other kissing loop also support this view. Insertion of a C between C248 and U249 should generate a 1-nucleotide bulge opposite the 3-nucleotide bulge (Fig. 4), most likely modifying the angle between the two flanking stems and, ultimately, the physical proximity between the two kissing loops. The corresponding CChMVd RNA was infectious and induced symptoms in 3/4 inoculated plants 7–10 dpi, but progeny analysis of one of the symptomatic plants 14 dpi showed that the mutation had reverted to wild type in the four sequenced clones, thus indicating that the presumed angle distortion was disfavored. Furthermore, the size and even the sequence of the 3-nucleotide bulge appear also important for infectivity, because deletion of G275, or substitution by C, led to CChMVd RNAs that were infectious and induced symptoms 7–10 dpi in 3/4 and in 4/4 inoculated plants, respectively (Fig. 4). Progeny analysis, however, of one symptomatic plant of each bioassay 14 dpi showed reversion to wild type in the three sequenced clones.
FIGURE 4.
Effects of experimental mutations affecting the stem-bulge-stem structure adjacent to positions 242–283 on CChMVd infectivity (indicated by the fraction of symptomatic plants within parentheses). The structure of this region in the viroid progeny from one symptomatic plant is shown below each mutant. No spontaneous mutations were observed in the hairpin delimited by positions 210–230 (data not shown). Other details as in the legends to Figures 1 and 3.
The RNA of another single mutant, C250→G, disrupting a predicted base pair in a position proximal to the 3-nucleotide bulge was also infectious and induced symptoms in 3/4 inoculated plants 7–10 dpi. However, progeny analysis of one of the symptomatic plants 14 dpi revealed that the mutation had reverted to wild type in two sequenced clones, and in two other the G250 had been substituted by a U concomitantly with another substitution, G272→A, which restored the base pair impeded by the initial C250→G mutation (Fig. 4). Furthermore, the RNA of a third single mutant with the same position deleted (C250→Ø) was also infectious and induced symptoms in 3/4 inoculated plants 7–10 dpi, but progeny examination of one of the symptomatic plants 14 dpi showed reversion to wild type in the three sequenced clones. These results, together with the sequence heterogeneity of natural variants in this same region (Fig. 1), give strong support to the existence of the four-base-pair stem adjacent to the loop delimited by positions 253–269.
The six-base-pair stem formed by CCAUUUC242–248 and GAGAUGG277–283, located at the other side of the 3-nucleotide bulge, was also probed by inserting a C between G279 and A280 that should interrupt the continuity of this stem. Although the corresponding RNA infected only 1/4 inoculated plants, symptom expression and viroid accumulation were not delayed. Sequencing of four clones of the progeny of this plant 14 dpi revealed deletion of the mutation introduced, concurrently with the substitutions G279→A (which transformed a G-U into an A-U base pair) and C248→U (which transformed a C-G into a U-G base pair) (Fig. 4). These data, together with the covariation in one natural variant in this same stem (Fig. 1), provide firm evidence for its existence in vivo.
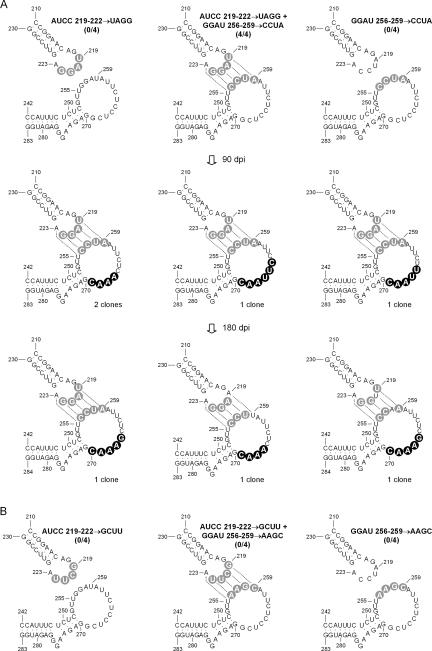
Interchanging 4 nucleotides between the two kissing loops generates a viable CChMVd variant with eight mutations
To make a stronger case for the in vivo significance of the proposed kissing loops, 4 contiguous nucleotides of each loop involved in this interaction were interchanged, generating a CChMVd variant with eight mutations (AUCC219–222→UAGG and GGAU256–259→CCUA) (Fig. 5). The controls for this experiment were the two quadruple mutants (AUCC219–222→UAGG) and (GGAU256–259→CCUA) in which the kissing-loop interaction could not be formed. The CChMVd RNAs corresponding to these controls were not infectious, as revealed by the lack of symptoms and dot-blot hybridization signals, in contrast to the CChMVd RNA harboring the eight mutations, which started inducing symptoms in the four inoculated plants ~50 dpi. Dot-blot hybridization of one of these plants 90 dpi, when symptoms had clearly developed, showed a viroid titer similar to the wild type, and progeny analysis of one of the plants demonstrated that the eight introduced mutations were preserved in the four clones examined. These clones contained additional base changes affecting several consecutive positions of one of the kissing loops (Fig. 5), which would reduce its size by extending the adjacent stem. When the progeny of the same plant was examined 180 dpi, one out of three clones retained the eight introduced mutations, but the two other clones only retained six because of the double reversion U219→A and A259→U in one, and the double reversion A220→U and U258→A in the other, which restored the wild-type interaction between these positions (Fig. 5). This double reversion was predominant in the progeny of this same plant 360 dpi (data not shown).
FIGURE 5.
Effects on CChMVd infectivity (indicated by the fraction of symptomatic plants within parentheses) of introducing separately two experimental quadruple mutations disrupting the proposed kissing-loop interaction, and concurrently the eight mutations restoring this interaction. Two experiment series (A, B) were performed. The structure of this region in viroid progeny from one symptomatic plant inoculated with the infectious CChMVd RNA containing eight mutations is shown below. Other details as in the legends to Figures 1 and 3.
To explore whether other combinations preserving the kissing-loop interaction were viable, a second CChMVd variant was constructed (AUCC219–222→GCUU and GGAU256–259→AAGC) in which the eight mutations were transitions. The controls for this experiment were the two quadruple mutants (AUCC219–222→GCUU) and (GGAU256–259→AAGC) in which the loops could not interact. None of the CChMVd RNAs corresponding to the three constructions was infectious: The inoculated plants did not express symptoms for 90 dpi, and dot-blot hybridization failed to reveal viroid replication and accumulation. Altogether, these results and those from the previous experiment series demonstrate the biological relevance of the proposed kissing loops: Even a variant with eight mutations is viable as long as the kissing-loop interaction is preserved. However, this appears to be a condition necessary but not sufficient, because another variant, with an alternative kissing-loop interaction of similar free energy (two G-C and two A-U pairs), was not infectious. This limited flexibility may result from additional restrictions such as having the two stronger and contiguous G-C pairs of the kissing loops located at specific positions. The delayed infectivity of the viable variant with eight mutations, and the gradual reversion to wild type of part of the positions involved in the kissing loops, also indicate that this interaction is optimized by some specific combination of nucleotides.
The kissing-loop interaction affects proper in vitro folding of CChMVd RNA
As anticipated, we did not observe any effect of the kissing loops on the extent of in vitro self-cleavage during transcription, because the hammerhead structures and the kissing loops are located in different domains of the CChMVd RNA (Fig. 1). In an attempt to obtain a mechanistic insight regarding the influence of the kissing loops, we applied two in vitro approaches. First, we determined the effect of this interaction on the solubility of CChMVd RNA in high-salt conditions. To this end, the products from in vitro transcriptions of linearized plasmids with head-to-tail dimeric CChMVd-cDNA inserts corresponding to the wild type, the quadruple mutant (GGAU256–259→CCUA) in which the kissing-loop interaction was disrupted, and the variant with eight mutations (AUCC219–222→UAGG and GGAU256–259→CCUA) in which the kissing-loop interaction was restored, were mixed with LiCl to a 2 M final concentration. PAGE analysis in denaturing gels showed that the monomeric CChMVd RNAs were found in the fraction insoluble in 2 M LiCl (data not shown), indicating that the kissing-loop interaction does not play a significant role in this physical property.
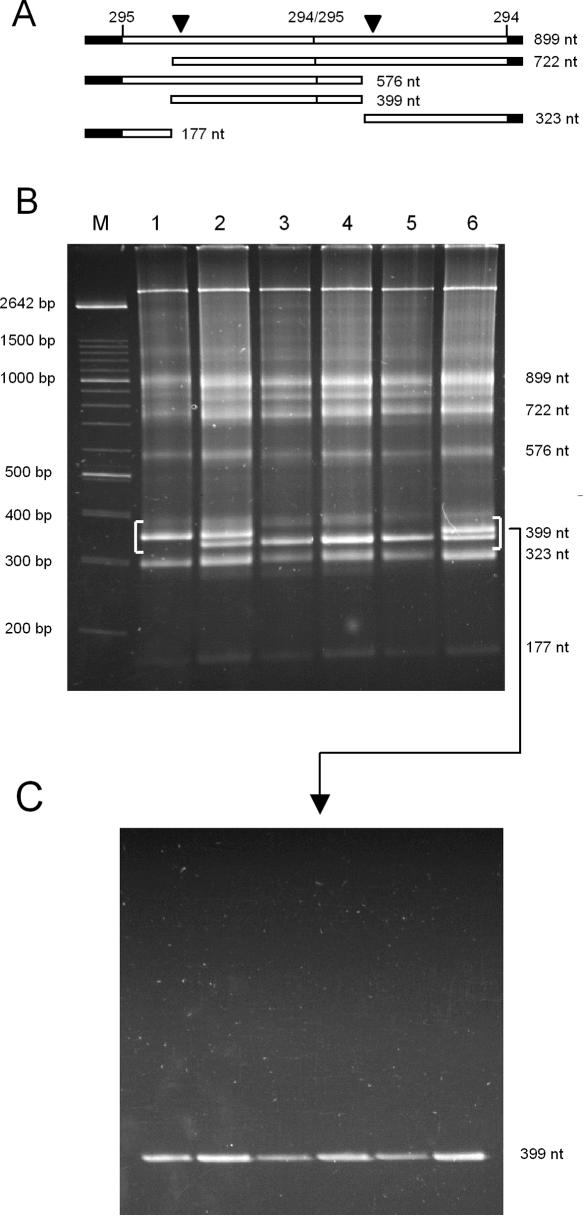
Second, the primary dimeric CChMVd (+) transcripts and their self-cleavage products from the same in vitro transcriptions (Fig. 6A) were directly analyzed by nondenaturing PAGE. This approach did provide a clear distinction between the monomeric RNA with four mutations, which showed a slower electrophoretic mobility, and the wild-type RNA and the variant RNA with eight mutations, which displayed a faster electrophoretic mobility (Fig. 6B, cf. lane 1 and lanes 3,5). Similar results were obtained when the same monomeric CChMVd RNAs, purified by elution from denaturing gels, were resuspended in the electrophoresis buffer before their separation through a nondenaturing gel (data not shown). Co-electrophoresis of mixed samples confirmed this point and also revealed no difference in mobility between the wild-type RNA and the variant RNA with eight mutations wherein the kissing-loop interaction was restored (Fig. 6B). Moreover, as anticipated, the mobility in denaturing conditions of the three CChMVd RNAs was the same, demonstrating that the differences observed in nondenaturing conditions were due to conformation and not to size (Fig. 6C). Interestingly, analysis in nondenaturing gels of the primary dimeric CChMVd (−) transcripts and the self-cleavage products of the same three variants failed to reveal any difference in mobility (data not shown), illustrating that the kissing-loop interaction is restricted to the (+) polarity strand. Overall, these results showed that the kissing loops promote in vitro, and most likely in vivo, a more compact folding of (+) strands that is critical for viroid viability.
FIGURE 6.
PAGE analysis of the in vitro transcription products of linearized plasmids containing head-to-tail dimeric CChMVd-cDNA inserts of the wild type (CM20), the noninfectious quadruple mutant (GGAU256–259→CCUA) impeding the kissing-loop interaction, and the infectious variant with eight mutations (AUCC219–222→UAGG and GGAU256–259→CCUA) in which the kissing-loop interaction is restored. (A) Schematic representation of a dimeric CChMVd (+) transcript and the self-cleavage products thereof. Vector and viroid sequences are denoted by black and white bars, respectively, and the self-cleavage sites are marked by arrowheads. Numbers on top of the bars refer to CChMVd positions (Navarro and Flores 1997), and numbers on the right indicate the size in nucleotides (nt). (B) Non-denaturing PAGE stained with ethidium bromide. Lanes 1,3,5 were loaded with aliquots of the quadruple mutant, the variant with eight mutations, and the wild-type RNAs, respectively. Lanes 2,4,6: Co-electrophoresis of mixed aliquots of the quadruple mutant and the variant with eight mutations, of the wild type and the variant with eight mutations, and of the wild type and the quadruple mutant, respectively. Lane M, DNA markers with their size in base pairs (bp) indicated. (C) Denaturing PAGE of the CChMVd RNAs migrating in the segment of the nondenaturing gel delimited by brackets, which was cut and applied directly on top of a urea gel. The identity of the RNAs stained with ethidium bromide was confirmed by Northern blot hybridization with a CChMVd-specific riboprobe (data not shown).
DISCUSSION
The rod-like secondary structure proposed initially for PSTVd and later assumed as a general model for viroids precludes the existence of long-distance interactions between loops (Sänger et al. 1976). This structural model does not apply to PLMVd (Hernández and Flores 1992) and CChMVd (Navarro and Flores 1997), which display branched conformations with the potential to form loop-loop interactions. In particular, we showed previously that the sequence heterogeneity of natural CChMVd variants strongly supports the biological significance of the computer-predicted branched conformation for this viroid, either because the changes map at loops or because when affecting a base pair, they are covariations or substitutions of Watson-Crick by wobble base pairs or vice versa (Navarro and Flores 1997; De la Peña et al. 1999; De la Peña and Flores 2002). Here we extended this analysis to assess the reliability of a kissing-loop interaction that, based on comparison with the situation found in vitro for PLMVd, was proposed in CChMVd between the loops delimited by positions 217–223 and 253–269 (Bussière et al. 2000). Examination of natural CChMVd variants for covariations and other point mutations in these loops and in their adjacent stems provided solid evidence for the in vivo significance of the kissing-loop interaction in CChMVd (Fig. 1) and indirectly for that proposed in PLMVd between loops GCGG178–181 and CCGC211–214 (numbering corresponds to the reference variant) (Hernández and Flores 1992; Ambrós et al. 1998), which is not disrupted by the only natural mutation C214→U reported in this region (Malfitano 2000).
Besides having a higher natural variability in the region encompassing the proposed kissing loops, CChMVd is more convenient than PLMVd for experiments of reverse genetics, because chrysanthemum is more easily propagated than peach and reacts to CChMVd by developing symptoms in a shorter time. To support the existence of the kissing-loop interaction with data other than those from natural covariations, we performed a compensatory mutational analysis. The underlying principle of both approaches is that if an RNA structure is functionally relevant, the sequences involved can vary but their ability to base-pair is preserved; thus, natural or experimental mutations that disrupt base pairing will annul function, which will be reinstated by compensatory sequence changes restoring base pairing. Construction of single mutants disrupting the central base pair of the presumed kissing-loop interaction and bioassay of their corresponding CChMVd RNAs showed that they were not infectious, in contrast to the double mutant with the interaction restored, which was infectious and retained the two mutations in the progeny. Similar results were obtained when the existence of a second base pair adjacent to the first was tested (Fig. 2), although infectivity of the single mutants was not abolished as a consequence of partial reversion to wild type. Moreover, mutations affecting the structure of the two regions flanking the kissing loops also support their interaction, leading in a particular case to a progeny with a rearranged stem that permits the interaction (Figs. 3, 4). Further and stronger evidence was obtained by our observation that the interchange between 4 nucleotides of each of the two kissing loops generated a viable CChMVd variant with eight mutations that were retained in the progeny, whereas the two quadruple mutants were not infectious (Fig. 5). Altogether, these results leave little doubt regarding the existence in vivo of the proposed kissing-loop interaction. It is also worth mentioning that, compared to the wild type, some of the experimental CChMVd mutants exhibited a delay in symptom expression, but symptom severity and viroid titer were very similar. Because the kissing-loop interaction does not affect the major pathogenicity determinant of CChMVd, which was mapped in a separate domain (De la Peña et al. 1999; De la Peña and Flores 2002), this suggests that the replication of these mutants is impaired but that they finally reach the normal in vivo titer that triggers the onset of symptoms.
The kissing-loop interaction entails the disruption of a short stem (formed by GGA256–258 and UCC264–266) between two loops and the generation of a large loop of 17 nucleotides (Fig. 1). Although the thermodynamic balance favors the kissing-loop interaction against the short stem (formation of five base pairs vs. the concurrent disappearance of three base pairs; Bussière et al. 2000; this work), and the electrophoretic mobility in nondenaturing gels of natural and artificial CChMVd variants is consistent with the kissing-loop interaction stabilizing a predominant compact folding (Fig. 6), the less stable folding may still be physiologically significant, at least transiently during replication, permitting a switch between two functionally non-equivalent conformations. The delay in symptom expression and viroid accumulation observed with the viable CChMVd variant containing eight mutations, which impede formation of the short stem, may reflect some crippling of this artificial variant to mimic the switch between the two conformations characteristic of the wild-type variant. Pertinent to this context is the observation that the progeny of this viable variant (Fig. 5) presented additional spontaneous mutations in several consecutive positions of one of the kissing loops that could extend the adjacent stem. Pseudoknot domains acting as molecular switches, with mutations resulting in a shift of the thermodynamic equilibrium between two RNA conformations, have been reported (Comolli et al. 2002; Theimer et al. 2003).
Evolutionary conservation of a structural RNA motif usually predicts conservation of biological function. In line with this idea, the conservation in PLMVd and CChMVd, two viroids with an overall low sequence similarity (Hernández and Flores 1992; Navarro and Flores 1997), of a similar kissing-loop interaction strongly suggests that this interaction helps to stabilize a compact folding critical for viroid viability. Such interactions may be needed in viroids with branched secondary structures to act as molecular clamps that delimit the conformation space, avoiding kinetic traps and facilitating the quick adoption of the biologically relevant RNA folding.
MATERIALS AND METHODS
Viroid natural variants
CChMVd variants CM1, CM20, and CM210 were obtained from a severe strain characterized previously in the chrysanthemum cultivar “Bonnie Jean” (Navarro and Flores 1997; De la Peña et al. 1999), and variants CM296-VR and CM298-VR were obtained from a second severe strain infecting the cultivar “Velvet Ridge.” Accession numbers of the EMBL Nucleotide Sequence Database are AJ878085–AJ878089 for variants CM20, CM1, CM210, CM296-VR, and CM298-VR, respectively.
Bioassay and detection of viroid RNA
Cuttings of chrysanthemum (Dendranthema grandiflora Tzvelez, cv. “Bonnie Jean”) were propagated in growth chambers (Navarro and Flores 1997). Plants were mechanically inoculated with the monomeric CChMVd RNAs (40 ng per plant) resulting from self-cleavage during in vitro transcription of linearized recombinant plasmids containing dimeric head-to-tail CChMVd-cDNA inserts, which were prepared by PCR amplification of their monomeric counterparts with Pwo DNA polymerase (Roche Diagnostics), and subsequently ligated and cloned in the EcoRV site of pBlueScript II KS+ (Stratagene). The primary transcripts and their self-cleavage products were separated by PAGE in 5% gels containing 1X TBE and 8 M urea, and the monomeric-length RNAs were eluted and quantified. CChMVd infection was assessed by symptom development and dot-blot hybridization of nucleic acids from young leaves (3 g) extracted with buffer-saturated phenol and enriched in viroid RNAs by chromatography on nonionic cellulose (CF11, Whatman) (De la Peña et al. 1999). RNAs were resuspended in a final volume of 80 μL, and aliquots (5 μL) of 1/5, 1/25, and 1/125 dilutions were applied to positively charged nylon membranes (Roche Diagnostics). The RNAs were fixed to the membranes by UV irradiation and then hybridized with a digoxigenin-labeled full-length riboprobe complementary to CChMVd variant CM20 obtained by transcription with T3 RNA polymerase of the corresponding linearized recombinant plasmid (De la Peña et al. 1999). After washing, the membranes were subjected to detection using an anti-digoxigenin antibody conjugated to alkaline phosphatase and the chemiluminescent substrate CSPD (Roche Diagnostics).
Analysis of mutant progenies by RT-PCR amplification, cloning, and sequencing
Unless otherwise indicated, young leaves were collected 14 d post inoculation (dpi), and nucleic acid preparations, enriched in CChMVd RNAs by chromatography on nonionic cellulose, were reverse-transcribed with Superscript II reverse transcriptase (Invitrogen) and primer RF-147 (5′-TCCAGTCGAGACCT GAAGTGGGTTTCC-3′), complementary to positions 133–107 of the CM20 variant. The resulting cDNA was PCR-amplified with Pwo DNA polymerase and primers RF-147 and RF-617 (5′-AGGTCGTAAAACTTCCCCTCTAAGCGG-3′), homologous to positions 134–160, and cloned (Navarro and Flores 1997). Inserts were sequenced automatically with an ABI Prism DNA apparatus (Perkin-Elmer).
Site-directed mutagenesis
A PCR-based protocol (Byrappa et al. 1995) was followed with minor modifications. Plasmid pCM20 (5 ng) was amplified with 250 ng each of pairs of adjacent and phosphorylated primers complementary and homologous to different segments of the CM20 variant, with the exception of some 5′-proximal positions in which appropriate changes were introduced to obtain the desired mutants (Table 1). The double mutants restoring the kissing-loop interaction were constructed by first introducing the disrupting mutation(s) and then by using the resulting plasmid to introduce the corresponding compensatory mutation(s). The PCR cycling profile, designed to amplify the complete plasmids with Pwo DNA polymerase, was as reported (De la Peña et al. 1999) with minor adjustments of the annealing temperature. After electrophoresis in 1% agarose gels, PCR-amplified products of plasmid length were eluted, purified with the QIAquick kit (QIAGEN), circularized with T4 DNA ligase, and used for transformation. Sequencing confirmed that the new plasmids contained only the expected mutations.
TABLE 1.
Primers for site-directed mutagenesis of CChMVd
| Mutanta | Primers | Nucleotide sequence (5′→3′)b | Positions |
| C221→G | RF 469 (sense) | GCAGTTCCGGTCCTTTGGAG | 221–240 |
| RF 470 (antisense) | ATCTGTTCCGGGTCCCAACC | 220–201 | |
| G257→C | RF 471 (sense) | CGTTGCATATTCTCCTCGGA | 252–271 |
| RF 472 (antisense) | AGAGAAATGGACTCCAAAGG | 251–232 | |
| U220→A | RF 608 (sense) | CCAGTTCCGGTCCTTTGGA | 221–239 |
| RF 609 (antisense) | TTCTGTTCCGGGTCCCAACC | 220–201 | |
| A258→U | RF 610 (sense) | CGTTGGTTATTCTCCTCGGA | 252–271 |
| RF 611 (antisense) | AGAGAAATGGACTCCAAAGG | 251–232 | |
| A | RF 635 (sense) | CCAGTTCCGAGTCCTTTGGA | 221–239 |
| G229↓G230 | RF 470 (antisense) | ATCTGTTCCGGGTCCCAACC | 220–201 |
| C | RF 473 (sense) | CCAGTCTCCGGTCCTTTGGA | 221–239 |
| U225↓U226 | RF 470 (antisense) | ATCTGTTCCGGGTCCCAACC | 220–201 |
| C | RF 516 (sense) | CGTTGGATATTCTCCTCGGA | 252–271 |
| C248↓U249 | RF 517 (antisense) | AGAGGAAATGGACTCCAAAGG | 251–232 |
| G275→Ø | RF 104 (sense) | GTCGGTTCGCTCTCGTAGTCACAGCC | 295–320 |
| RF 515 (antisense) | TGGGACTGGCCCCATCTCC-TTCTCC | 294–269 | |
| G275→C | RF 104 (sense) | GTCGGTTCGCTCTCGTAGTCACAGCC | 295–320 |
| RF 558 (antisense) | TGGGACTGGCCCCATCTCCGTTCTCC | 294–269 | |
| C250→G | RF 516 (sense) | CGTTGGATATTCTCCTCGGA | 252–271 |
| RF 559 (antisense) | ACAGAAATGGACTCCAAAGG | 251–232 | |
| C250→Ø | RF 516 (sense) | CGTTGGATATTCTCCTCGGA | 252–271 |
| RF 518 (antisense) | A-AGAAATGGACTCCAAAGG | 251–232 | |
| C | RF 104 (sense) | GTCGGTTCGCTCTCGTAGTCACAGCC | 295–320 |
| G279↓A280 | RF 519 (antisense) | TGGGACTGGCCCCATGCTCCCTTCTCC | 294–269 |
| AUCC219-222→UAGG | RF 612 (sense) | TAGGAGTTCCGGTCCTTTGGAG | 219–240 |
| RF 613 (antisense) | CTGTTCCGGGTCCCAACC | 218–201 | |
| GGAU256-259→CCUA | RF 614 (sense) | CGTTCCTAATTCTCCTCGGA | 252–271 |
| RF 611 (antisense) | AGAGAAATGGACTCCAAAGG | 251–232 | |
| AUCC219-222→GCUU | RF 615 (sense) | GCTTAGTTCCGGTCCTTTGGAG | 219–240 |
| RF 613 (antisense) | CTGTTCCGGGTCCCAACC | 218–201 | |
| GGAU256-259→AAGC | RF 616 (sense) | CGTTAAGCATTCTCCTCGGA | 252–271 |
| RF 611 (antisense) | AGAGAAATGGACTCCAAAGG | 251–232 |
aHorizontal arrows denote nucleotide substitutions; Ø, deletions; and vertical arrows, insertions with respect to the CM20 reference variant.
bNucleotide substitutions and insertions with respect to the CM20 reference variant are underlined, and deletions are indicated by a hyphen.
Solubility in 2 M LiCl and conformation analysis by nondenaturing and denaturing PAGE
The products from in vitro transcriptions of linearized recombinant plasmids with head-to-tail dimeric CChMVd-cDNA inserts of the wild type, the quadruple mutant (GGAU256–259→CCUA), and the variant with eight mutations (AUCC219–222→UAGG and GGAU256–259→CCUA) were made to contain 2 M LiCl and left overnight at 4°C. The soluble and insoluble fractions were collected by centrifugation at 8000g for 20 min and their RNAs examined by PAGE in denaturing gels.
The products from the three in vitro transcriptions were also analyzed by nondenaturing PAGE in 5% gels (with TAE buffer and run in the cold room) that were stained with ethidium bromide (Navarro and Flores 1997). The section of the gel containing the monomeric CChMVd RNAs was cut and directly applied on top of a denaturing 5% gel of the same dimensions (with 0.25X TBE buffer and 8 M urea) which, following electrophoresis, was stained with ethidium bromide.
Acknowledgments
We thank C. Hernández and J.A. Daròs for critical reading and A. Ahuir for excellent technical assistance. Work in R.F.’s laboratory was partially supported by grant BMC2002-03694 from the Ministerio de Ciencia y Tecnología (MCyT) and by the Generalidad Valenciana (GRUPOS03/064). During this work, S.G. was supported by the Ministerio de Educación, Cultura y Deporte, and by the Fundación “José y Ana Royo”, and M.D.P. by the MCyT.
Article published ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.2230605.
REFERENCES
- Ambrós, S., Hernández, C., Desvignes, J.C., and Flores, R. 1998. Genomic structure of three phenotypically different isolates of peach latent mosaic viroid: Implications of the existence of constraints limiting the heterogeneity of viroid quasi-species. J. Virol. 72: 7397–7406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambrós, S., Hernández, C., and Flores, R. 1999. Rapid generation of genetic heterogeneity in progenies from individual cDNA clones of peach latent mosaic viroid in its natural host. J. Gen. Virol. 80: 2239–2252. [DOI] [PubMed] [Google Scholar]
- Bussière, F., Ouellet, J., Côté, F., Lévesque, D., and Perreault, J.P. 2000. Mapping in solution shows the peach latent mosaic viroid to possess a new pseudoknot in a complex, branched secondary structure. J. Virol. 74: 2647–2654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrappa, S., Gavin, D.K., and Gupta, K.C. 1995. A highly efficient procedure for site-specific mutagenesis of full-length plasmids using Vent DNA polymerase. PCR Methods Applic. 5: 404–407. [DOI] [PubMed] [Google Scholar]
- Comolli, L.R., Smirnov, I., Xu, L., Blackburn, E.H., and James, T.L. 2002. A molecular switch underlies a human telomerase disease. Proc. Natl. Acad. Sci. 99: 16998–17003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De la Peña, M. and Flores, R. 2002. Chrysanthemum chlorotic mottle viroid RNA: Dissection of the pathogenicity determinant and comparative fitness of symptomatic and non-symptomatic variants. J. Mol. Biol. 321: 411–421. [DOI] [PubMed] [Google Scholar]
- De la Peña, M., Navarro, B., and Flores, R. 1999. Mapping the molecular determinant of pathogenicity in a hammerhead viroid: A tetraloop within the in vivo branched RNA conformation. Proc. Natl. Acad. Sci. 96: 9960–9965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diener, T.O. 1972. Potato spindle tuber viroid VIII. Correlation of infectivity with a UV-absorbing component and thermal denaturation properties of the RNA. Virology 50: 606–609. [DOI] [PubMed] [Google Scholar]
- ———. 1999. Viroids and the nature of viroid diseases. Arch. Virol. (Suppl.) 15: 203–220. [DOI] [PubMed] [Google Scholar]
- Dingley, A.J., Steger, G., Esters, B., Riesner, D., and Grzesiek, S. 2003. Structural characterization of the 69 nucleotide potato spindle tuber viroid left-terminal domain by NMR and thermodynamic analysis. J. Mol. Biol. 334: 751–767. [DOI] [PubMed] [Google Scholar]
- Fadda, Z., Darò s, J.A., Flores, R., and Duran-Vila, N. 2003a. Identification in eggplant of a variant of citrus exocortis viroid (CEVd) with a 96 nucleotide duplication in the right terminal region of the rod-like secondary structure. Virus Res. 97: 145–149. [DOI] [PubMed] [Google Scholar]
- Fadda, Z., Darò s, J.A., Fagoaga, C., Flores, R., and Duran-Vila, N. 2003b. Eggplant latent viroid (ELVd): Candidate type species for a new genus within family Avsunviroidae (hammerhead viroids). J. Virol. 77: 6528–6532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flores, R., Di Serio, F., and Hernández, C. 1997. Viroids: The non-coding genomes. Semin. Virol. 8: 65–73. [Google Scholar]
- Flores, R., Daròs, J.A., and Hernández, C. 2000. The Avsunviroidae family: Viroids with hammerhead ribozymes. Adv. Virus Res. 55: 271–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flores, R., Randles, J.W., Owens, R.A., Bar-Joseph, M., and Diener, T.O. 2004. Viroidae. In Virus taxonomy, 8th Rep. Intern. Com. Taxon. Viruses (eds. C.M. Fauquet et al.), pp. 1145–1159. Elsevier/Academic Press, London, UK.
- Gross, H.J., Domdey, H., Lossow, C., Jank, P., Raba, M., Alberty, H., and Sänger, H.L. 1978. Nucleotide sequence and secondary structure of potato spindle tuber viroid. Nature 273: 203–208. [DOI] [PubMed] [Google Scholar]
- Haseloff, J., Mohamed N.A., and Symons, R.H. 1982. Viroid RNAs of cadang-cadang disease of coconuts. Nature 229: 316–321. [Google Scholar]
- Hernández, C. and Flores, R. 1992. Plus and minus RNAs of peach latent mosaic viroid self-cleave in vitro via hammerhead structures. Proc. Natl. Acad. Sci. 89: 3711–3715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchins, C., Rathjen, P.D., Forster, A.C., and Symons, R.H. 1986. Self-cleavage of plus and minus RNA transcripts of avocado sun-blotch viroid. Nucleic Acids Res. 14: 3627–3640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keese, P. and Symons, R.H. 1985. Domains in viroids: Evidence of intermolecular RNA rearrangements and their contribution to viroid evolution. Proc. Natl. Acad. Sci. 82: 4582–4586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koltunow, A.M. and Rezaian, M.A. 1988. Grapevine yellow speckle viroid: Structural features of a new viroid group. Nucleic Acids Res. 16: 849–864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malfitano, M. 2000. “Distribuzione di varianti di sequenza del viroide del mosaico latente del pesco in isolati di campo italiani.” Ph.D thesis, Università Degli Studi di Napoli “Federico II”.
- Malfitano, M., Di Serio, F., Covelli, L., Ragozzino, A., Hernández, C., and Flores, R. 2003. Peach latent mosaic viroid variants inducing peach calico contain a characteristic insertion that is responsible for this symptomatology. Virology 313: 492–501. [DOI] [PubMed] [Google Scholar]
- McInnes, J.L. and Symons, R.H. 1991. Comparative structure of viroids and their rapid detection using radioactive and nonradioactive nucleic acid probes. In Viroids and satellites: Molecular parasites at the frontier of life (ed. K. Maramorosch), pp. 21–58, CRC Press, Boca Raton, FL.
- Pelchat, M., Levesque, D., Ouellet, J., Laurendeau, S., Levesque, S., Lehoux, J., Thompson, D.A., Eastwell, K.C., Skrzeczkowski, L.J., and Perreault, J.P. 2000. Sequencing of peach latent mosaic viroid variants from nine North American peach cultivars shows that this RNA folds into a complex secondary structure. Virology 271: 37–45. [DOI] [PubMed] [Google Scholar]
- Riesner, D., Henco, K., Rokohl, U., Klootz, G., Kleinschmidt, A.K., Domdey, H., Jank, P., Gross, H.J., and Sänger, H.L. 1979. Structure and structure formation of viroids. J. Mol. Biol. 133: 85–115. [DOI] [PubMed] [Google Scholar]
- Sänger, H.L., Klotz, G., Riesner, D., Gross, H.J., and Kleinschmidt, A. 1976. Viroids are single-stranded covalently closed circular RNA molecules existing as highly base-paired rod-like structures. Proc. Natl. Acad. Sci. 73: 3852–3856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sano, T., Candresse, T., Hammond, R.W., Diener, T.O., and Owens, R.A. 1992. Identification of multiple structural domains regulating viroid pathogenicity. Proc. Natl. Acad. Sci. 89: 10104–10108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semancik, J.S., Szychowski, J.A., Rakowski, A.G., and Symons, R.H. 1994. A stable 463 nucleotide variant of citrus exocortis viroid produced by terminal repeats. J. Gen. Virol. 75: 727–732. [DOI] [PubMed] [Google Scholar]
- Sogo, J.M., Koller, T., and Diener, T.O. 1973. Potato spindle tuber viroid. X. Visualization and size determination by electron microscopy. Virology 55: 70–80. [DOI] [PubMed] [Google Scholar]
- Symons, R.H. 1981. Avocado sunblotch viroid: Primary sequence and proposed secondary structure. Nucleic Acids Res. 9: 6527–6537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabler, M. and Tsagris, M. 2004. Viroids: Petite RNA pathogens with distinguished talents. Trends Plant Sci. 9: 339–348. [DOI] [PubMed] [Google Scholar]
- Theimer, C.A., Finger, L.D., and Feigon, J. 2003. YNMG tetraloop formation by a dyskeratosis congenita mutation in human telomerase RNA. RNA 12: 1446–1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wassenegger, M., Heimes, S., and Sänger, H.L. 1984. An infectious viroid RNA replicon evolved from an in vitro-generated non-infectious viroid deletion mutant via a complementary deletion in vivo. EMBO J. 13: 6172–6177. [DOI] [PMC free article] [PubMed] [Google Scholar]