Abstract
The search is underway for a catalytic RNA molecule capable of self-replication. Finding such a ribozyme would lend crucial support to the RNA World hypothesis, which holds that very early life-forms relied on RNA for both replicating and storing genetic information. We previously reported an RNA polymerase isolated from a pool of variants of an existing RNA ligase ribozyme. Here we report eight additional ligase-derived polymerase ribozymes isolated from this pool. Because each of them is a new potential starting point for further in vitro evolution and engineering, together they substantially enrich the set of candidates from which an RNA replicase ribozyme might eventually emerge.
Keywords: catalytic RNA, ribozymes, in vitro selection, polymerases, replicases, RNA World hypothesis
INTRODUCTION
In discussing the origins of life, current editions of many biology textbooks now sketch a description of the RNA World, a very early and hypothetical period of evolution during which it is imagined that ribo-organisms ruled the earth (Purves et al. 2004; Campbell and Reece 2005; Freeman 2005; Audesirk et al. 2005). These ancestral creatures had only one kind of encoded polymer: Both their enzymes and their genome were made of RNA (Pace and Marsh 1985; Gilbert 1986; Orgel 1986). The appeal of the RNA World hypothesis is rooted in its simplicity: Although the evolutionary innovation of such a sophisticated molecule as RNA would have been no small feat (Shapiro 1988; Larralde et al. 1995; see also Ricardo et al. 2004; Springsteen and Joyce 2004) and RNA might have been preceded by a simpler genetic polymer during the “pre-RNA World” (Nelson et al. 2000; Ichida et al. 2005), and although it has even been suggested that the earliest life-forms lacked genes of any kind (Shapiro 2000), the idea of a transitional RNA-only phase at some period during early evolution seems comfortably plausible in comparison to the alternative scenario in which oligonucleotides and coded protein synthesis emerged together in miraculous concert. Since the discovery that RNA, outside its well-established role as an information carrier, can also act as an enzyme to catalyze chemical reactions (Altman 1990; Cech 1990) and in fact catalyzes protein synthesis in all modern organisms (Steitz and Moore 2003), RNA has been the leading candidate for a polymer that “did it all” in our very early ancestors (Joyce 2002; Dworkin et al. 2003; Hughes et al. 2004; Orgel 2004).
Despite the popularity of the RNA World hypothesis, some of its crucial tenets remain assumptions. Chief among them is the notion that RNA can be replicated without proteins. In the early stages of the RNA World, nonenzymatic polymerization must have played a major role (Luther et al. 1998; Huang and Ferris 2003; Monnard et al. 2003; Franchi and Gallori 2005), but in most versions of the RNA World hypothesis, RNA at some point became genetically self-sufficient, taking over responsibility for its own synthesis. In order to substantiate the notion that RNA can be a good enough enzyme to catalyze RNA replication, considerable effort has been devoted to the search for an RNA polymerase ribozyme (Bartel 1999; McGinness and Joyce 2003).
The search has so far been dominated by efforts to extend the functionality of existing ribozymes. Some progress has been made using natural self-splicing introns as a starting point (Been and Cech 1988; Doudna and Szostak 1989; Bartel et al. 1991; Chowrira et al. 1993; Doudna et al. 1993; McGinness and Joyce 2002), while other approaches have focused on an artificial ribozyme called the Class I RNA ligase (Bartel and Szostak 1993; Ekland et al. 1995). Derivatives of this ligase are able to extend an RNA primer by several nucleotides, but they require specific base-pairing to the RNA template strand (Ekland and Bartel 1996; McGinness et al. 2002). This dependence on specific sequence elements prevents them from being general polymerases, capable of copying any template. In an attempt to convert the ligase into a general polymerase, we previously constructed a pool of more than 1015 ligase variants, to each of which was appended a 76-nt segment of random sequence (Johnston et al. 2001). This was called the Ligase+N76 pool (Fig. 1). It was hoped that some of these chimeric RNA molecules would be able to bind and extend a primer-template RNA duplex without base-pairing to it. The pool was enriched for molecules with the desired activity by repeated selection and amplification, and after 10 rounds of this in vitro selection, a single ribozyme was found showing robust template-dependent polymerization activity. This ribozyme, referred to here as Pol 1, was then subjected to eight further rounds of optimizing selection, improving its activity to the point where it could polymerize a whole turn of an RNA helix. The improved polymerase, referred to here as Evolved Pol 1, has been characterized with respect to its secondary structure and fidelity (Johnston et al. 2001), kinetics and processivity (Lawrence and Bartel 2003), and substrate recognition (Müller and Bartel 2003).
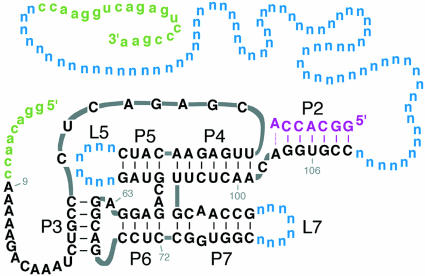
FIGURE 1.
Ligase+N76 starting pool, consisting of an RNA ligase ribozyme (black), plus P2-completing heptamer (purple) and two randomized loops (blue), concatenated to a 76-nt random region (blue), and flanked by primer-binding sites (green).
Here we report a new Ligase+N76 polymerase selection (called branch B), in which the selection protocol was modified using lessons learned from the original experiment (called branch A). The new selection led to the isolation of seven additional ligase-derived polymerase ribozymes, designated Pol 2 through Pol 8, each with another unique “auxiliary domain” derived from the randomized N76 segment. Additionally, a weak polymerase from the original selection was re-examined and named Pol 9. The whole collection of polymerase ribozymes was compared head-to-head in polymerization assays using a variety of primer-templates (PTs). Pols 1–4 had approximately equivalent levels of activity, while the activity of Pols 5–9 was less robust. In general, Evolved Pol 1 showed the strongest polymerization activity, although with some PTs it was nearly matched by Pols 1–4.
RESULTS
New selection
Having found Pol 1 as the only robust polymerase from the branch A selection, yet suspecting that the Ligase+N76 starting pool might still hold undiscovered polymerases, we designed a new selection strategy to look for ribozymes that the first experiment might have missed. We returned to an early point in the original selection and branched off along a new path to look for more polymerases. This new selection path, called branch B (Fig. 2), incorporated several modifications that were expected to alter the course of the in vitro evolution.
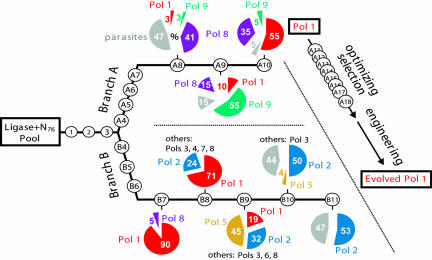
FIGURE 2.
Two branches of the polymerase selection, and breakdowns of pool populations. Pool evolution is represented as a line extending rightward, with each selection round indicated as a circled number. Pie graphs report the predominant families at certain rounds (as percent of total population). Families detected at low levels are reported as “others.”
In branch A, ribozymes had been incubated with 4-thioUTP, and ribozymes that tagged themselves with 4-thioU were isolated on the basis of their decreased mobility in a mercury gel. Pol 1 was later confirmed to utilize 4-thioUTP and unmodified UTP with comparable efficiency in polymerization assays (data not shown). However, in later rounds of branch A, an additional constraint had been imposed: Ribozymes were incubated with biotin-ATP, with the idea that ribozymes that tagged themselves with both 4-thioU and biotin-A would be isolated by successive purification using mercury gels and then streptavidin-coated magnetic beads (Table 1). Although branch A was eventually successful, Pol 1 was later found to reject biotin-ATP as a substrate, despite its efficient use of unmodified ATP. Moreover, the parental RNA ligase ribozyme was also shown to use ATP, but not biotin-ATP (data not shown), as a substrate in the single-nucleotide addition reaction it catalyzes using an internal template (Ekland and Bartel 1996). These findings suggested in hindsight that the use of biotin-ATP in some rounds of branch A had been ineffective in imposing additional selective pressure and that the survival of ribozymes at this step in those rounds was due merely to background binding to the streptavidin-coated beads. Moreover, some valuable polymerase ribozymes could have been lost or disfavored when the population was forced through this potential bottleneck. Therefore, in designing branch B to look for additional polymerases in the ligase-based pool, we eliminated the use of biotin-ATP (Table 1).
TABLE 1.
Polymerization templates and selection parameters used in polymerase evolution
| NTPs (mM) | |||||
| Round Template | 4SU | BA | A,C,G | Time (hr) | Selection criteria |
| Shared initial rounds | |||||
| 1 GGUCAGAUU | 2 | 0 | 0 | 36 | 4SU |
| 2 GGUCAGAACC | 2 | 0 | 0 | 20 | 4SU |
| 3 GGUCAGAA | 2 | 0 | 0 | 20 | 4SU |
| Branch A | |||||
| A4 CUUAGUUCAUU | 2 | 0 | 0 | 19 | 4SU |
| A5 CUUAGUUCAUU | 2 | 0 | 0 | 1 | 4SU |
| A6 GGUCAGAUU | 1 | 1 | 0 | 14 | BA, 4SU |
| A7 CUUAGUUCAUU | 1 | 1 | 0 | 17 | BA, 4SU |
| A8 GGUCAGAUU | 1 | 1 | 0 | 17 | BA, 4SU |
| A9 GGUCAGAUU | 1 | 1 | 0 | 4 | BA, 4SU |
| A10 CUUAGUUCAUU | 1 | 0 | 0 | 20 | 4SU |
| Branch B | |||||
| B4 ACAUACGGAUAUU | 2 | 0 | 0 | 24 | 4SU |
| B5 UCGACGGAACC | 2 | 0 | 0 | 18 | 2 × 4SU |
| B6 ACCUGAGaaCC | 0.5 | 0 | 0 | 18 | 2 × 4SU |
| B7 CAAGUCCaaGG | 0.1 | 0 | 2 ea. | 14 | 2 × 4SU |
| B8 ACCUGAGaaCC | 0.1 | 0 | 2 ea. | 15 | 2 × 4SU |
| B9 CAAGUCCaaGG | 0.1 | 0 | 2 ea. | 1 | 2 × 4SU |
| B10 CAAGUCCaaUGAUCGUA | 0.1 | 0 | 2 ea. | 5 | 2 × 4SU |
| B11 UCGACGGaaCCUGCGUC | 0.1 | 0 | 2 ea. | 0.4 | 2 × 4SU |
Polymerization primer was complementary to the underlined portion of each template (“a” indicates the adenine isomer 2-aminopurine). BA indicates N6-biotin-A, and 4SU indicates 4-thioU.
In branch A, ribozymes that added one nucleotide had been lumped together with ribozymes that added two or more. At each step of the selection, all ribozymes that managed to add the first 4-thioU were recovered and amplified, regardless of how much additional polymerization they had succeeded in catalyzing. Thus, Pol 1 was isolated on the basis of its ability to add merely a single nucleotide. During the Pol 1 optimization selection (rounds A11–A18) (Fig. 2), we learned that RNA molecules containing two 4-thioUs halted in a mercury gel, rather than merely slowing down, and that we could therefore increase the stringency of the mercury-gel technique to select for addition of two 4-thioUs by collecting only those molecules caught at the mercury interface. In branch A, this two–4-thioU technique had been used in the Pol 1 optimization phase (rounds A11–A18), but not in the original polymerase discovery phase (rounds 1–A10). In branch B, the two–4-thioU technique was implemented much earlier (Table 1), in hopes that the more stringent selection criterion (demanding addition of two 4-thioUs instead of just one) would shift the course of the in vitro evolution and reveal new polymerases.
Branch B was further enhanced by earlier application of several other selection techniques that had been employed only in the Pol 1 optimization phase (rounds 11–18) of branch A. These modifications included the use of competitor NTPs (unlabeled ATP, CTP, and GTP), to select for high-fidelity polymerization; the use of 2-aminopurine in the polymerization template, to improve Watson-Crick pairing geometry with 4-thioU (by avoiding steric clash of the 4-thioU sulfur atom and the adenine 6-amino group); the use of longer templates, to favor ribozymes that could accommodate them; the reduction of 4-thioUTP concentration, to favor ribozymes with stronger NTP binding; and the use of mutagenic PCR, to increase pool diversity and optimize active ribozymes. Finally, whereas branch A had essentially alternated one pair of primer-templates, sometimes repeating a PT in subsequent rounds (Table 1), branch B employed a greater diversity of PT sequences in order to enhance its selective power for polymerases that catalyze general (as opposed to sequence-specific) template-directed RNA polymerization.
Branch B began with pool 3 of the original selection (Fig. 2), a pool that had already undergone three rounds of selection for the ability to add a single 4-thioU but did not yet have detectable polymerization activity. This pool was subjected to eight new rounds of selection, incorporating the changes described above (Table 1). Pool activity was detected after the first new round of selection (round B4), with ~2% of the pool adding a single 4-thioU in 24 h. In every subsequent round, the selection criterion was addition of two 4-thioUs, and this activity was first detected after round B6. Following round B8, pool activity was robust, and was shown to require a correctly paired primer and template. After round B10, the pool was shown to catalyze polymerization in the untethered format (with no covalent linkage between pool and primer) using all three different PTs tested. After one more round, selection was stopped and pool populations were analyzed.
Dramatic population shifts
In the present study, 139 clones from pools B7–11 were isolated and sequenced, then grouped into 33 families, with the members of each family having nearly identical sequences, by virtue of descent from a single ancestral sequence in the starting pool. Immediately obvious was the early dominance of the Pol 1 family in the branch B selection: 90% of the isolates from pool B7 were identical or nearly identical to Pol 1 (Fig. 2). Had this result been apparent immediately after round B7, it might have led to the early abandonment of the branch B selection; despite our efforts to improve the selection protocol and uncover new polymerases, evolution seemed stuck in a rut. Fortunately, however, branch B had already been carried forward several more rounds, thereby revealing the first new polymerase family, represented by Pol 2. This new family gradually but completely displaced the Pol 1 family during rounds B8–B10, apparently indicating its superior fitness under branch B selection conditions. Several additional families were also detected during the population shift: The Pol 5 family dominated pool B9 but disappeared in the next round, while Pols 3, 4, 6, 7, and 8 were detected at small numbers in pools B7–B9. At round B10, about half of the clones had multiple lesions in their ligase domains. Clones such as these always failed in polymerization assays and were provisionally classified as “parasites,” because their garbled catalytic domain and consequent inactivity meant that they had been able to prosper during the selection by alternative, uncharacterized mechanisms. Most of the parasites in branch B belonged to a few large families.
New polymerases
A consensus clone was chosen to represent each new polymerase family (Table 2; Fig. 3). The new clones were evaluated alongside Pol 1 in polymerization assays using a variety of PTs (Fig. 4). On the basis of these comparisons, the new clones were named Pols 2–8 in order of their approximate overall activity. All eight polymerases extended PT A, which codes for addition of a single C. Pols 1–5 went further by adding an additional, untemplated nucleotide, as do most proteinaceous polymerases (Bausch et al. 1983; Clark et al. 1987). With a longer template (PT D), extension by at least 3 nt was detected using all eight polymerases. With other PTs, the performance of Pols 1–4 was consistently strong, and their activity levels were generally comparable. Pols 5 and 6 were usually much weaker, and Pols 7 and 8 extended only a few of the PTs tested.
TABLE 2.
Ligase loop sequences and other changes in each polymerase
| Polymerase | Isolate no. | Loop 5 | Loop 7 | Ligase changes |
| Pol 1 | A10.2 | UGUGAAUU | GCGAUUGC | |
| Pol 2 | B11.78 | ACUCAUAA | CAUCAUAA | A9C, A63C |
| Pol 3 | B9.41 | UAGUAUCG | AAUCUCUC | A63U |
| Pol 4 | B8.36 | ACAUUGGU | CUAAGUUG | |
| Pol 5 | B9.02 | AGUCCCAA | UCCGCUAA | |
| Pol 6 | B9.50 | AGUCCCAA | UCCGCUAA | A100G |
| Pol 7 | B8.64 | GCGUAUGU | ACGUGCCU | C72U |
| Pol 8 | B8.38 | CAUAUUCGG | GGGGUGCC | |
| Pol 9 | A9.1 | GCAGUAGC | UGAUACUA | |
| Ev Pol 1 | A18.12.23 | UUCG | GCGAUAGC | Δ3–8, U106C |
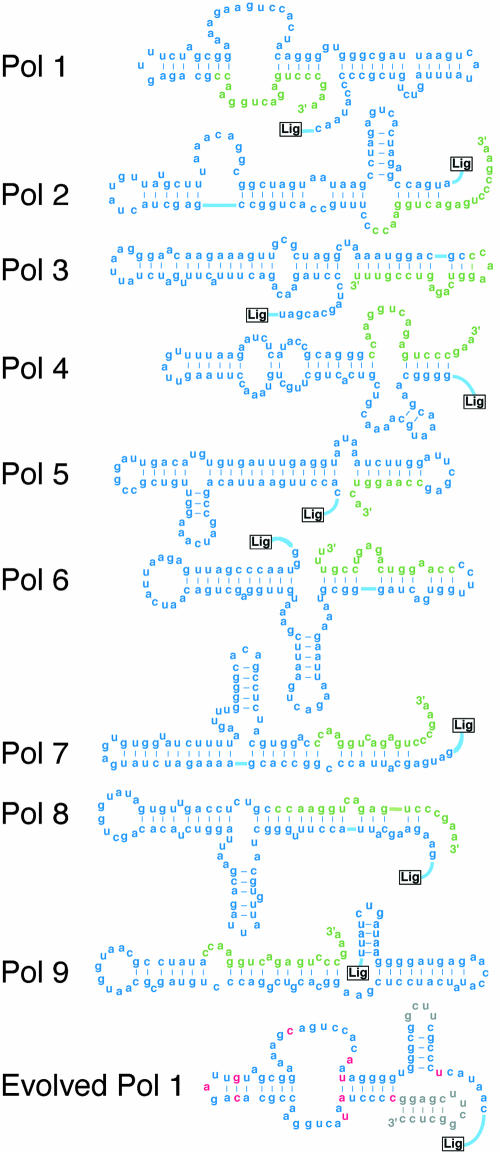
FIGURE 3.
Polymerase ribozymes. Each polymerase contains a ligase domain (shown as boxed “Lig”; see Table 2 for details) concatenated to an auxiliary domain derived from the pool N76 region (blue) and 3′ primer-binding site (green). Secondary structures of Pols 2–9 are speculative. Evolved Pol 1 is shown with changes from Pol 1 indicated in pink and in gray.
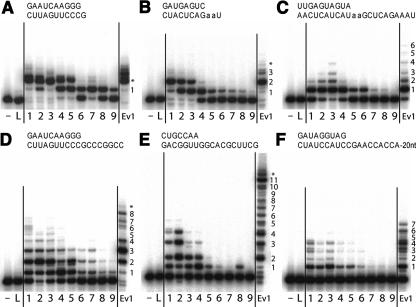
FIGURE 4.
Ribozyme-catalyzed primer extension. Each panel shows polymerization using a different primer-template (PT), shown at top (“a” indicates 2-aminopurine). In each panel, the first two lanes are negative controls, showing primer incubated with no ribozyme (−) or with ligase core only (L). Lanes 1–9 and Ev1 show the activity of Pols 1–9 and Evolved Pol 1. Extension products are numbered at right, with stars indicating full-length molecules extended by an additional, untemplated nucleotide.
Polymerization requires both domains
Without an auxiliary domain, the ligase core was completely inactive in all polymerization assays (Fig. 4, “L” lanes). Pol 2 activity was dependent on the P2-completing oligo (data not shown), as observed previously with Pol 1 (Johnston et al. 2001), indicating that the ligase P2 stem remained essential for polymerization. Clones with disrupted ligase domains (parasites) were always inactive in polymerization assays (data not shown). However, many families with intact ligase domains were also found to be inactive, indicating that not just any auxiliary domain will do.
Attempts were made to trim some of the new polymerases. Earlier work that had produced the ligase ribozyme itself (Ekland et al. 1995) showed that ribozymes isolated from in vitro selection experiments could often tolerate terminal truncations. We suspected that the ribozymes emerging from the branch B selection would not require their very 3′ ends, because the last four nt of the pool’s 3′ terminus had been systematically varied at each round of the selection in a deliberate effort to discourage the pool from relying on its 3′ terminus (for instance, by subversively using it as the polymerization primer in place of the primer ligated to the 5′ end of the pool at each round). However, in the case of Pol 2, deleting just 8 nt from its 3′ end was enough to nearly abolish its activity. Attempts to trim Pol 5 were slightly more successful: This polymerase tolerated the deletion of 10 nt from its 3′ terminus. (The truncated version of Pol 5, depicted in Fig. 3, was used in the Fig. 4 assays.) Further truncation was not tolerated, however, with Pol 5 completely inactivated by deletion of 20 or more nt from its 3′ end.
A closer look at Pol 1
During the original branch A selection, 74 variants were cloned and sequenced from pools A8–A10, then grouped into 23 families (Johnston et al. 2001). One family, represented by isolate 10.2, showed robust template-dependent polymerization and was renamed the round 10 ribozyme, referred to here as Pol 1. It was the starting point for further optimization (rounds A11–A18) and site-directed sequence engineering, yielding eventually the round 18 ribozyme or Evolved Pol 1. With one of the primer-templates examined, this polymerase can accurately synthesize a full turn of an RNA helix, extending this primer by 14 nt (Johnston et al. 2001). It also showed strong activity with a 3 nt-shorter version of this template (PT E) (Fig. 4). However, with other PTs, the activity of Evolved Pol 1 was more modest. PTs B and C are analogous to the PTs used during the selection in that they code first for the addition of two Us. With PT B, Evolved Pol 1 managed to add the first two Us to all of the PT molecules but added the final encoded A to only a tiny fraction of them (Fig. 4). With PT C, which codes for the addition of 12 nt, Evolved Pol 1 was able to add the first U to most of the PTs, but it usually stalled there; it added the second U to only a small fraction of the molecules; addition of 3–6 nt was barely detectable, and no polymerization was observed beyond 6 nt. With PT F, which codes for 30 nt, Evolved Pol 1 added at most 7 nt; similar results were seen with two other PTs of the same length (data not shown).
Race against hydrolysis
Evolved Pol 1 typically extends its PT by 4–8 nt, and never by more than 14 nt, even when the template codes for many more. Why doesn’t it do better? The reason is its very weak primer-template binding. The polymerization reaction uses micromolar concentrations of RNA, but the polymerase binds its PT substrate with only millimolar affinity, and simply increasing the PT concentration fails to improve the situation, because higher RNA concentrations actually inhibit the ribozyme (Lawrence and Bartel 2003). This extremely poor affinity makes PT binding the rate-limiting step. As the ribozyme slowly polymerizes NTPs, it suffers the constant hydrolytic onslaught of the high-magnesium, high-pH reaction buffer, and this causes the reaction to decelerate. We measured how long Evolved Pol 1 survives under polymerization assay conditions and found that its half-life was in close agreement with the halving time of reaction rate; after a day-long exposure to 200 mM magnesium and a pH of 8.5, only about half of the polymerase remained full-length.
Weak additional ribozymes from branch A
Pol 1 was the only robust polymerase discovered during branch A of the selection. However, a second polymerase family was also reported, represented by isolate 9.1 and comprising 55% of the pool after round A9 (Fig. 2). It had an unmutated ligase domain and was shown to catalyze a weak but template-dependent extension of a tethered primer (Johnston et al. 2001). Here we renamed this ribozyme Pol 9 (Fig. 3) and tested it with several untethered PTs, confirming that its polymerization activity is very weak, in fact undetectable with most PTs (Fig. 4). A third family that rose to prominence during branch A comprised 41% of pool A8 and 35% of pool A10 (Fig. 2), but despite its evolutionary success and intact ligase domain, it showed no activity in the initial tethered PT assay, and so was not reported. After the branch B selection, this family was recognized as including Pol 8 (Fig. 3), a very weak polymerase present at low frequency in several branch B pools (Fig. 2).
DISCUSSION
The Class I RNA ligase is one of the fastest ribozymes known (Bergman et al. 2000), and it catalyzes a reaction similar in many crucial respects to a single step of RNA polymerization: The reactants are aligned by Watson-Crick pairing to an RNA template, the 3′-hydroxyl of one reactant attacks the 5′-triphosphate of the other reactant, pyro-phosphate is released, and a new internucleotide linkage is created. When assisted by base-pairing, the ligase can extend an RNA primer by as many as 6 nt (Ekland and Bartel 1996). However, with PTs that don’t base-pair to it, the ligase can’t add even a single nucleotide (Fig. 4, “L” lanes). The Ligase+N76 pool (Fig. 1) was constructed in hopes of finding an auxiliary domain that, when appended to the ligase, would somehow help it bind the PT sequence nonspecifically, thus converting the ligase into a general polymerase (Johnston et al. 2001). Now nine such auxiliary domains have been identified. Thus it appears that there are multiple ways of binding the primer-template RNA duplex, and a corresponding diversity of ways to convert this RNA ligase into an RNA polymerase.
A bouquet of long stems
The secondary structures of the auxiliary domains of Pols 2–9 are not yet known. Speculative structures based on the predictions of the m-fold algorithm (Zuker 2003) are shown in Figure 3. In many cases, long paired regions (for example, a putative 13-base pair stem in Pol 3) lend credibility to the structures, because these long stems would be extremely stable in the presence of 200 mM magnesium, and they significantly constrain the rest of the fold. Moreover, m-fold correctly predicted the structure of the Evolved Pol 1 auxiliary domain; the predicted structure includes all the base pairs shown in the Figure 3 structure, which was modeled independently by comparative analysis of 25 diverse variants (Johnston et al. 2001). It is tempting to interpret these highly base-paired, largely linear structures as a general feature of successful polymerase auxiliary domains. The auxiliary domains of the most active polymerases, Pols 1–5, were each compared to their permuted cohort, a set of sequences in which the N76-derived sequence was randomly reordered, neutralizing the contributions of evolution but preserving base composition (Schultes et al. 1999). In each case, the evolved structure was more stable than at least 70% of its permuted cohort (95% in the cases of Pols 1 and 5). Thus, the predicted stability trend reflects conformational order accumulated during selection, and not merely the innate folding behavior of RNA. It would be interesting to explore whether this property directly aids polymerization, or whether it was favored during the selection for more general reasons, such as compactness or resistance to hydrolysis.
Polymerase ecology
The defining feature of in vitro evolution is the use of iterated selection and amplification to move from a starting pool in which sequences are distributed randomly, to an evolved pool in which they occur in proportion to their aptitude for the desired activity. In our ribozyme selections, however, we have often been frustrated by a perversely weak correlation between activity and multiplicity. For instance, the Pol 8 and Pol 9 families dominated the branch A pools numerically but showed only weak activity, whether assayed in the tethered format (analogous to the selection context) or untethered (requiring the activity ultimately desired). Conversely, Pols 3 and 4, two of the most active polymerases to emerge from the Ligase+N76 pool, never multiplied beyond a few percent of total population. This weak correlation exposes a fundamental inefficiency in the selection protocol. A related issue is the emergence of “parasites,” clones with ruined catalytic domains and no polymerization activity whatsoever, which emerge consistently in later rounds of our polymerase selections. We speculate that these inactive clones survive by efficiently inserting their primers into the active sites of working polymerases, thus exploitatively ensuring their own propagation while contributing nothing to the overall level of pool activity. This hypothesis explains their absence during early selection rounds, when there are not yet enough working polymerases around to exploit. Similar phenomena have been observed in other in vitro selection experiments (Hanczyc and Dorit 1998).
On the path to RNA self-replication?
Each of these nine rudimentary polymerases is a potentially promising evolutionary intermediate between ligase and replicase. In the case of Pol 1, the promise has already borne fruit: After eight rounds of optimizing selection and a little site-directed tinkering, it gave rise to Evolved Pol 1, the strongest polymerase ribozyme yet reported. Evolved Pol 1 can add 14 nt to one particular PT, but more typically it adds 4–8 nt (Fig. 4). Previous work demonstrated its sensitivity to PT sequence: A change as slight as adding or subtracting a single nucleotide from the starting primer altered the observed extension rate by as much as an order of magnitude (Lawrence and Bartel 2003). Such sequence-specific variation is not surprising, having been observed as well with proteinaceous polymerases (Echols and Goodman 1991; Kunkel 1992). Nonetheless, without exception, Evolved Pol 1 has extended every PT tested: It is truly a general RNA polymerase.
Pols 2–9 have not yet had the benefit of optimization, and it is interesting to imagine what Evolved Pols 2–9 might be like. One could imagine constructing a pool of variants of each ribozyme, then mixing them all together into one super pool and letting them all compete against each other during selection. By this approach, the problem of “declaring wild type” (Joyce 2004) would be softened slightly: Instead of forcing the population through the bottleneck of a single sequence, an increased genetic diversity of forms would be retained, and with it the possibility of productive recombination between unrelated structures. In any case, it will be interesting to see if an RNA replicase ribozyme can eventually be isolated, whether it descends from one of these ligase-derived polymerases, or from any other branch of the ribozyme family tree.
MATERIALS AND METHODS
In vitro selection
The starting pool for the branch B selection (Fig. 2) was an aliquot of pool 3 DNA archived during the original selection experiment (Johnston et al. 2001). RNA was transcribed using T7 RNA polymerase, purified by PAGE (polyacrylamide gel electrophoresis), and ligated to an RNA primer using T4 DNA ligase and a DNA splint, then PAGE-purified again and annealed to an RNA template. Primer and template sequences were changed in each round (Table 1). The pool was also annealed to the RNA heptamer GGCACCA, which completed the P2 stem of the ligase domain. The polymerization reaction was initiated by addition of selection buffer (60 mM MgCl2, 200 mM KCl, 50 mM EPPS [pH 8.0]) plus 4-thioUTP and competitor NTPs, then allowed to proceed at 22°C for 0.4–24 h (Table 1). The reaction was stopped by addition of 80 mM EDTA, and excess 4-thioUTP was removed by Centricon filtration. Molecules containing 4-thioU were isolated by mercury PAGE. From round B4 onward, RNA was excised from the interface between the −Hg and +Hg regions, enforcing selection for addition of two 4-thioUs. RNA was eluted from the gel slice, then used to seed the next round of amplification-selection.
Cloning and sequencing
Molecules were cloned from pools B7–B11 using TOPO-TA cloning (Invitrogen), then 72 were sequenced and grouped into nine families of nearly identical sequences. The population statistics shown in Figure 2 were computed from this first set of clones (for rounds 7–11, n = 18, 12, 15, 20, and 19, respectively). Additional minor families were then uncovered using a two-step colony hybridization screen. In the first step, thousands of cloned colonies from pools B8–B10 were transferred to nitrocellulose membranes and screened with 70-nt DNA probes corresponding to the auxiliary domains of the known major families. Clones that hybridized to any of these probes were ruled out as novel candidates. In the second step, the clones were rescreened using a probe that corresponded to the ligase domain. Clones that hybridized only in this second step were candidates of interest, because they had an intact ligase domain and an unknown auxiliary domain. Sixty-seven such clones were sequenced, bringing the total number of sequence families to 33.
Polymerization assays
Polymerase ribozymes (5 μM final concentration) were transcribed from cloned DNA templates, PAGE-purified, and annealed to a 1.25-fold molar excess of P2 oligo. Primer (0.1 μM, 5′-radiolabeled) and template (0.5 μM) were annealed separately, then mixed with assay buffer (200 mM MgCl2, 50 mM Tris-HCl [pH 8.5]) and NTPs (4 mM each). Polymerization reactions were initiated by mixing annealed ribozyme with annealed PT/buffer/NTPs. After a 24-h incubation at 22°C, reactions were mixed with 4 vol of gel loading buffer (8 M urea, 25 mM EDTA), template was separated from primer by addition of 5 molar equivalents of a competitor RNA identical to fully extended primer, and extension products were resolved by denaturing PAGE.
Measurement of polymerase survival time during polymerization
Three identical polymerization assays were carried out as above, varying only the position of the 5′ radiolabel: In one reaction the primer was end-labeled (as normally), whereas in the other two either the template or the ribozyme (Evolved Pol 1) was end-labeled. Aliquots were withdrawn and quenched at a series of time points, and RNAs of different sizes were separated by denaturing PAGE. At each time point, amounts of full-length ribozyme and degradation fragments were quantitated.
Acknowledgments
This work was supported by a grant from the NIH. We thank Wendy Johnston for invaluable technical advice and the sample of pool DNA. We thank Sarah Bagby and Ulrich Müller for helpful comments on the manuscript.
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.2110905.
REFERENCES
- Altman, S. 1990. Enzymatic cleavage of RNA by RNA. Biosci. Rep. 10: 317–337. [DOI] [PubMed] [Google Scholar]
- Audesirk, T., Audesirk, G., and Byers, B.E. 2005. Biology: Life on Earth, 7th ed. Pearson Prentice Hall, Upper Saddle River, NJ.
- Bartel, D.P. 1999. Re-creating an RNA Replicase. In The RNA World (eds. R.F. Gesteland et al.), 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Bartel, D.P. and Szostak, J.W. 1993. Isolation of new ribozymes from a large pool of random sequences. Science 261: 1411–1418. [DOI] [PubMed] [Google Scholar]
- Bartel, D.P., Douda, J.A., Usman, N., and Szostak, J.W. 1991. Template-directed primer extension catalyzed by the Tetrahymena ribozyme. Mol. Cell. Biol. 11: 3390–3394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bausch, J.N., Kramer, F.R., Miele, E.A., Dobkin, C., and Mills, D.R. 1983. Terminal adenylation in the synthesis of RNA by Q-β replicase. J. Biol. Chem. 258: 1978–1984. [PubMed] [Google Scholar]
- Been, M.D. and Cech, T.R. 1988. RNA as an RNA polymerase: Net elongation of an RNA primer catalyzed by the Tetrahymena ribozyme. Science 239: 1412–1416. [DOI] [PubMed] [Google Scholar]
- Bergman, N., Johnston, W., and Bartel, D.P. 2000. Kinetic framework for ligation by an efficient RNA ligase ribozyme. Biochemistry 39: 3115–3123. [DOI] [PubMed] [Google Scholar]
- Campbell, N.A. and Reece, J.B. 2002. Biology, 7th ed. Pearson/Benjamin Cummings, San Francisco, CA.
- Cech, T.R. 1990. Self-splicing and enzymatic activity of an intervening sequence RNA from Tetrahymena. Biosci. Rep. 10: 239–261. [DOI] [PubMed] [Google Scholar]
- Chowrira, B.M., Berzal-Herranz, A., and Burke, J.M. 1993. Novel RNA polymerization reaction catalyzed by a group I ribozyme. EMBO J. 12: 3599–3605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark, J.M., Joyce, C.M., and Beardsley, G.P. 1987. Novel blunt-end addition reactions catalyzed by DNA polymerase I of Escherichia coli. J. Mol. Biol. 198: 123–127. [DOI] [PubMed] [Google Scholar]
- Doudna, J.A. and Szostak, J.W. 1989. RNA-catalysed synthesis of complementary-strand RNA. Nature 339: 519–522. [DOI] [PubMed] [Google Scholar]
- Doudna, J.A., Usman, N., and Szostak, J.W. 1993. Ribozyme-catalyzed primer extension by trinucleotides: A model for the RNA-catalyzed replication of RNA. Biochemistry 32: 2111–2115. [DOI] [PubMed] [Google Scholar]
- Dworkin, J.P., Lazcano, A., and Miller, S.L. 2003. The roads to and from the RNA world. J. Theor. Biol. 222: 127–134. [DOI] [PubMed] [Google Scholar]
- Echols, H. and Goodman, M.F. 1991. Fidelity mechanisms in DNA replication. Annu. Rev. Biochem. 60: 477–511. [DOI] [PubMed] [Google Scholar]
- Ekland, E.H. and Bartel, D.P. 1996. RNA-catalysed RNA polymerization using nucleoside triphosphates. Nature 382: 373–376. [DOI] [PubMed] [Google Scholar]
- Ekland, E.H., Szostak, J.W., and Bartel, D.P. 1995. Structurally complex and highly active RNA ligases derived from random RNA sequences. Science 269: 364–370. [DOI] [PubMed] [Google Scholar]
- Franchi, M. and Gallori, E. 2005. A surface-mediated origin of the RNA world: Biogenic activities of clay-adsorbed RNA molecules. Gene 346: 205–214. [DOI] [PubMed] [Google Scholar]
- Freeman, S. 2005. Biological science, 2d ed. Prentice Hall, Upper Saddle River, NJ.
- Gilbert, W. 1986. The RNA world. Nature 319: 618. [Google Scholar]
- Hanczyc, M.M. and Dorit, R.L. 1998. Experimental evolution of complexity: In vitro emergence of intermolecular ribozyme interactions. RNA 4: 268–275. [PMC free article] [PubMed] [Google Scholar]
- Huang, W. and Ferris, J.P. 2003. Synthesis of 34–50 mers of RNA oligomers from unblocked monomers. A simple approach to the RNA world. Chem. Commun. (Camb.) 12: 1458–1459. [DOI] [PubMed] [Google Scholar]
- Hughes, R.A., Robertson, M.P., Ellington, A.D., and Levy, M. 2004. The importance of prebiotic chemistry in the RNA world. Curr. Opin. Chem. Biol. 8: 629–633. [DOI] [PubMed] [Google Scholar]
- Ichida, J.K., Zou, K., Horhota, A., Yu, B., McLaughlin, L.W., and Szostak, J.W. 2005. An in vitro selection system for TNA. J. Am. Chem. Soc. 127: 2802–2803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston, W.K., Unrau, P.J., Lawrence, M.S., Glasner, M.E., and Bartel, D.P. 2001. RNA-catalyzed RNA polymerization: Accurate and general RNA-templated primer extension. Science 292: 19–25. [DOI] [PubMed] [Google Scholar]
- Joyce, G.F. 2002. The antiquity of RNA-based evolution. Nature 418: 214–221. [DOI] [PubMed] [Google Scholar]
- ———. 2004. Directed evolution of nucleic acid enzymes. Annu. Rev. Biochem. 73: 791–836. [DOI] [PubMed] [Google Scholar]
- Kunkel, T.A. 1992. Biological asymmetries and the fidelity of eukaryotic DNA replication. Bioessays 14: 303–308. [DOI] [PubMed] [Google Scholar]
- Larralde, R. Robertson, M.P., and Miller, S.L. 1995. Rates of decomposition of ribose and other sugars: Implications for chemical evolution. Proc. Natl. Acad. Sci. 92: 8158–8160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence, M.S. and Bartel, D.P. 2003. Processivity of ribozyme-catalyzed RNA polymerization. Biochemistry 42: 8748–8755. [DOI] [PubMed] [Google Scholar]
- Luther, A., Brandsch, R., and von Kiedrowski, G. 1998. Surface-promoted replication and exponential amplification of DNA analogues. Nature 396: 245–248. [DOI] [PubMed] [Google Scholar]
- McGinness, K.E. and Joyce, G.F. 2002. RNA-catalyzed RNA ligation on an external RNA template. Chem. Biol. 9: 297–307. [DOI] [PubMed] [Google Scholar]
- ———. 2003. In search of an RNA replicase ribozyme. Chem. Biol. 10: 5–14. [DOI] [PubMed] [Google Scholar]
- McGinness, K.E., Wright, M.C., and Joyce, G.F. 2002. Continuous in vitro evolution of a ribozyme that catalyzes three successive nucleotidyl addition reactions. Chem. Biol. 9: 585–596. [DOI] [PubMed] [Google Scholar]
- Monnard, P.-A., Kanavarioti, A., and Deamer, D.W. 2003. Eutectic phase polymerization of activated ribonucleotide mixtures yields quasi-equimolar incorporation of purine and pyrimidine nucleobases. J. Am. Chem. Soc. 125: 13734–13740. [DOI] [PubMed] [Google Scholar]
- Müller, U.F. and Bartel, D.P. 2003. Substrate 2′-hydroxyl groups required for ribozyme-catalyzed polymerization. Chem. Biol. 10: 799–806. [DOI] [PubMed] [Google Scholar]
- Nelson, K.E., Levy, M., and Miller, S.L. 2000. Peptide nucleic acids rather than RNA may have been the first genetic molecule. Proc. Natl. Acad. Sci. 97: 3868–3871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orgel, L.E. 1986. RNA catalysis and the origins of life. J. Theor. Biol. 123: 127–149. [DOI] [PubMed] [Google Scholar]
- ———. 2004. Prebiotic chemistry and the origin of the RNA world. Crit. Rev. Biochem. Mol. Biol. 39: 99–123. [DOI] [PubMed] [Google Scholar]
- Pace, N.R. and Marsh, T.L. 1985. RNA catalysis and the origin of life. Origins Life 16: 97–116. [DOI] [PubMed] [Google Scholar]
- Purves, W.K., Sadava, D., Orians, G.H., and Heller, H.C. 2004. Life: The science of biology, 7th ed. Sinauer Associates, Sunderland, MA.
- Ricardo, A., Carrigan, M.A., Olcott, A.N., and Benner, S.A. 2004. Borate minerals stabilize ribose. Science 303: 196. [DOI] [PubMed] [Google Scholar]
- Schultes, E.A., Hraber, P.T., and LaBean, T.H. 1999. Estimating the contributions of selection and self-organization in RNA secondary structure. J. Mol. Evol. 49: 76–83. [DOI] [PubMed] [Google Scholar]
- Shapiro, R. 1988. Prebiotic ribose synthesis: A critical analysis. Orig. Life Evol. Biosph. 18: 71–85. [DOI] [PubMed] [Google Scholar]
- ———. 2000. A replicator was not involved in the origin of life. IUBMB Life 49: 173–176. [DOI] [PubMed] [Google Scholar]
- Springsteen, G. and Joyce, G.F. 2004. Selective derivitization and sequestration of ribose from a prebiotic mix. J. Am. Chem. Soc. 126: 9578–9583. [DOI] [PubMed] [Google Scholar]
- Steitz, T.A. and Moore, P.B. 2003. RNA, the first macromolecular catalyst: The ribosome is a ribozyme. Trends Biochem. Sci. 28: 411–418. [DOI] [PubMed] [Google Scholar]
- Zuker, M. 2003. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 31: 3406–3415. [DOI] [PMC free article] [PubMed] [Google Scholar]