Abstract
Numerous problems exist with the current thinking of RNA as the first genetic material. No plausible prebiotic processes have yet been demonstrated to produce the nucleosides or nucleotides or for efficient two-way nonenzymatic replication. Peptide nucleic acid (PNA) is a promising precursor to RNA, consisting of N-(2-aminoethyl)glycine (AEG) and the adenine, uracil, guanine, and cytosine-N-acetic acids. However, PNA has not yet been demonstrated to be prebiotic. We show here that AEG is produced directly in electric discharge reactions from CH4, N2, NH3, and H2O. Electric discharges also produce ethylenediamine, as do NH4CN polymerizations. AEG is produced from the robust Strecker synthesis with ethylenediamine. The NH4CN polymerization in the presence of glycine leads to the adenine and guanine-N9-acetic acids, and the cytosine and uracil-N1-acetic acids are produced in high yield from the reaction of cyanoacetaldehyde with hydantoic acid, rather than urea. Preliminary experiments suggest that AEG may polymerize rapidly at 100°C to give the polypeptide backbone of PNA. The ease of synthesis of the components of PNA and possibility of polymerization of AEG reinforce the possibility that PNA may have been the first genetic material.
Keywords: prebiotic synthesis, first genetic material, pre–RNA world, RNA world, chemical evolution
The discovery of the catalytic activity of RNA (1, 2) brought the concept of an RNA world (3–7) into wide acceptance. However, the instability of ribose and other sugars (8), the great difficulty of prebiotic synthesis of the glycosidic bonds of the necessary nucleotides (9, 10), and the inability to achieve two-way non-enzymatic template polymerizations (11, 12) have raised serious questions about whether RNA could have been the first genetic material (13), although there are dissenting opinions (6, 14). A pre-RNA world in which the backbone of the first genetic material would have been different from the ribose phosphate seems more likely, but the nature of this backbone is unknown. One proposal offers peptide nucleic acids (PNA) as a possible precursor to RNA (15) because PNA binds DNA and forms double and triple helical structures that are related to the Watson-Crick helix (16–18).
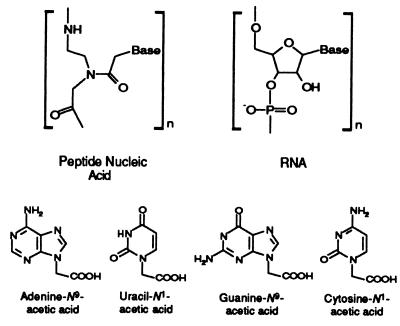
The backbone of PNA is a polymer of N-(2-aminoethyl)glycine (AEG), which is sometimes referred to as ethylenediamine monoacetic acid. It is interesting to note that Westheimer listed AEG as one of a number of possible backbones to replace ribose phosphate (19). This was four years before the invention of peptide nucleic acids. The bases are attached to the backbone by a base-substituted acetyl unit, as shown in Fig. 1. The simplicity of the components of PNA suggests that prebiotic syntheses might be feasible. We therefore examined a number of prebiotic syntheses, including electric discharges and NH4CN polymerizations for ethylenediamine (ED) and AEG, as well as the adenine and guanine-N9-acetic acids and the cytosine and uracil-N1-acetic acids. We show here that the components of PNA are synthesized under potentially prebiotic conditions. This finding makes a plausible case that PNA might have been the first genetic material.
Figure 1.
In peptide nucleic acids, the ribose phosphate backbone is replaced with a polyamide backbone of N-(2-aminoethyl)glycine (AEG or ED monoacetic acid), and the four bases are connected through an acetic acid linker.
Materials and Methods
Materials.
All chemicals and solvents were obtained from the Aldrich, Fisher, or Sigma. Sample purity was assayed by HPLC, and the products were identified by melting point, UV absorption spectra, and 1H NMR spectra. HPLC analysis was performed using a YMC (Kyoto) ODS-AQ column (4.5 × 250 mm) and two Beckman 110B pumps with UV detection by a Kratos Spectroflow 757 variable wavelength detector set to 260 nm or with fluorescence detection by a Gilman Spectra/glo filter fluorescence detector.
AEG was synthesized from ethylenediamine and chloroacetic acid (20). Adenine-N9-acetic acid was synthesized from adenine and methyl-bromoacetate (21). Cytosine and uracil-N1-acetic acids were synthesized from the pyrimidine bases and chloroacetic acid (22, 23). Cytosine-N3-acetic acid was synthesized from cytosine and chloroacetic acid anhydride (24) followed by purification to homogeneity by reversed phase HPLC. The melting point, UV absorption spectra, and 1H NMR spectra of each compound were consistent with literature values.
The synthesis of guanine-N9-acetic acid was carried out from 2-amino-6-chloropurine and bromoacetic acid in dry dimethylformamide under N2 (25). The colored product was suspended in 10.5 M NaOH and was hydrolyzed at room temperature for 96 h to give the modified guanine. The nucleobase derivative was purified by acidification of the reaction mixture to precipitate the product followed by reversed phase HPLC purification to homogeneity. The modified guanine was characterized by reversed phase HPLC, UV absorption spectra, and 1H NMR spectra.
The cyanoacetaldehyde was prepared from the reaction of isoxazole with sodium methoxide (26). The concentration was determined by the molar absorptivity of 15,200 M−1⋅cm−1 at 248 nm for the sodium salt.
Reactions.
The spark discharge reactions using CH4, NH3, N2, and H2O were carried out as previously described (27, 28). The reaction of 8.3 M NH4CN was prepared from solid KCN and NH4Cl and was heated in a round bottom flask under reflux for at 72°C for 20 h. The polymer was centrifuged and the supernatant hydrolyzed in 6 M HCl for 25 h. The reaction of 8.3 M NH4CN with 0.8 M H2CO was heated in a sealed glass ampoule at 81°C for 18 h followed by hydrolysis of the supernatant in 6 M HCl for 20 h. The reaction of 10 M HCN, 7.5 M NH4OH, and 2.5 M glycine was prepared from glycine, NH4OH, and gaseous HCN with the addition of 2.5 ml of 10 M NaOH to bring the pH to 9.8. This reaction was heated at 80°C for 18 h followed by hydrolysis of the supernatant in 6 M HCl for 25 h.
The Strecker synthesis with ED and reactions with the nucleobases were carried out in sealed glass ampoules at 25°C and were analyzed after acid hydrolysis with HCl at 100°C for 20 h. The nucleobases (1 mM) were reacted with 30 mM NaCN and 70 mM H2CO or 70 mM NaCN and 30 mM H2CO at pH 9.6 for 1 year. The reaction of the nucleobases (1 mM) with glyoxal (1 mM) was carried out at 25°C and pH 8 for 1 month.
The reaction of cyanoacetaldehyde (1 mM) with hydantoic acid (2 M) and urea (2 M) was carried out in sealed glass ampoules heated at 100°C for varying lengths of time up to 30 days. The pH was initially 7 (1 mM sodium phosphate) but drifted to 9 over the course of reaction.
Identification.
The yield and identity of ED, AEG, 2,3-diaminopropionic acid, and glycine from the spark discharge and NH4CN polymerization samples were determined by chromatography on a Dowex 50 (H+) column (29). Fractions collected from this column were then analyzed by reversed phase HPLC. Samples were derivatized before analysis with the fluorescent labels OPA-NAC (30) or dansyl chloride (31). The dansyl derivatives were eluted isocratically with 0.1 M sodium phosphate (pH 4.8) containing 70% methanol. The eluent for the dansyl-glycine and all OPA-NAC derivatives was 0.1 M sodium phosphate (pH 4.8) containing 45% methanol.
The yield and identity of the purine-N9-acetic acids and pyrimidine-N1-acetic acids were determined by chromatography on a Dowex 50 (H+) column using 4 M HCl as the eluent. Fractions collected from this column were analyzed by reversed phase HPLC (26). For the guanine-N9-acetic acid analysis, the eluent was 0.1% trifluoroacetic acid (vol/vol) in water with UV detection at 254 nm. Identification was based on elution time, coelution with a known sample, and UV absorption spectra.
Results and Discussion
N-(2-Aminoethyl)glycine and Ethylenediamine.
We used both the high temperature apparatus used in the first electric discharge experiments (27) and a room temperature apparatus (28) that gives a greater variety of amino acids. The yields are given in Table 1.
Table 1.
Percent yields of glycine, AEG, and ED from the electric discharge and ammonium cyanide polymerization
| Percent yield
|
|||
|---|---|---|---|
| Glycine | AEG | ED | |
| Spark discharge, 25°C | 0.23 | 1.5 × 10−5 | 7.5 × 10−5 |
| Spark discharge, 100°C | 0.58 | 6.2 × 10−6 | 5.5 × 10−5 |
| NH4CN polymerization | 2.1 | <1 × 10−5 | 4.4 × 10−5 |
| NH4CN + 10% H2CO | 9.5 | <1 × 10−5 | 0.05 |
The spark discharge yields are based on the CH4, the NH4CN polymerization is based on the carbon in CN−, and the NH4CN polymerization with H2CO is based on the concentration of H2CO.
In addition, we also examined an NH4CN polymerization, which produces both adenine (32) and amino acids, mostly glycine (33). The reaction gave 0.03% adenine, 2.1% glycine, and 4.4 × 10−5% ED but no AEG (<1 × 10−5%). Although these yields of ED and AEG are low, they might be sufficient because the high solubility of AEG monohydrochloride (11.5 mol/kg of H2O at 25°C) would allow it to be concentrated on drying beaches and lagoons.
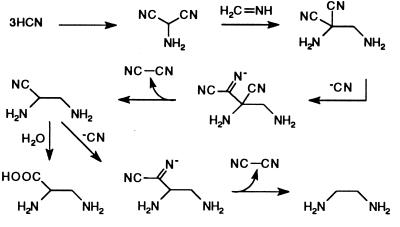
However, it seemed likely that more efficient syntheses of AEG would be possible in an NH4CN polymerization in the presence of H2CO, which is a catalyst for HCN oligomerization to adenine (34). Polymerization of 8.3 M NH4CN in the presence of 0.8 M H2CO gave satisfactory yields of ED (0.05%) together with 9.5% glycine, 0.1% 2,3-diaminopropionic acid, and 0.041% adenine. Such high concentrations of NH4CN and H2CO could be achieved on the primitive earth by freezing dilute solutions of NH3, HCN, and H2CO produced by electric discharges (35) or photochemical processes (36–38). Shown in Fig. 2 is a possible mechanism for the production of ED and diaminopropionic acid from ammonium cyanide and HCN trimer (39).
Figure 2.
A proposed mechanism for production of ED and 2,3-diaminopropionic acid from NH4CN and HCN trimer with consequent production of cyanogen from higher HCN oligomers.
We could detect no AEG in the NH4CN-H2CO reaction. However, ED is almost equivalent to AEG because AEG is produced via the Strecker synthesis: ![]()
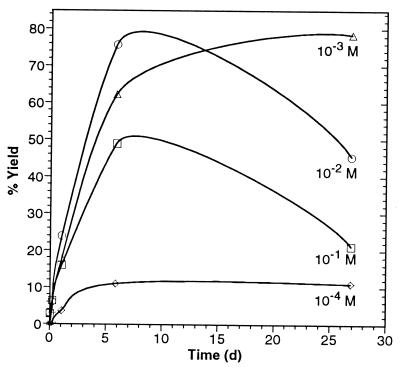
This reaction is quite robust and efficient at high dilution, as shown from the yields in Table 2 and Fig. 3. We have found that the AEG cyclizes rapidly at 100°C and neutral pH to form the lactam, monoketopiperazine, with an equilibrium constant of 5.1 and a half life of 2.5 h. (K.E.N. and S.L.M., unpublished results).
Table 2.
The percent yield of AEG from the given concentration of ED, HCN, and H2CO
| Concentration, M | Percent yield |
| 10−1 | 21 |
| 10−2 | 45 |
| 10−3 | 78 |
| 10−4 | 11 |
| 10−5 | 18 |
| 10−6 | 33 |
Yields are for samples reacted at 25°C and pH 7 for 27 days and hydrolyzed with 0.5 M HCl at 100°C.
Figure 3.
Yield of AEG versus time for the reaction of 10−1 to 10−4 M ED, HCN, and H2CO at 25°C and pH 7 after acid hydrolysis. The yields of AEG decline after 5 days because of the formation of ethylenediamine diacetic acid.
There may have been considerably more AEG on the primitive earth than indicated by the electric discharge or cyanide oligomerization reactions. The relatively low yield of AEG in the electric discharge experiment is apparently attributable to the low yields of ED. On the primitive earth, this problem could have been overcome if UV light is taken into account in the prebiotic process. Ogura et al. (40) showed that UV light acting on mixtures of CH4, NH3 and water gives high yields of ED (33%), with methylamine being the most abundant product (66%). Because there are efficient UV syntheses of H2CO (36, 37) and HCN (38), the prebiotic synthesis of AEG may have been far more favorable than indicated by the electric discharge experiments because of ED production through the Ogura process. Thus, the photochemical lability of CH4 and NH3, which is claimed to make a strongly reducing atmosphere untenable (41), may have been the dominant source of ED and AEG.
Adenine and Guanine-N9-Acetic Acids.
The next step in the prebiotic synthesis of the PNA components is the formation of the N9-acetic acid derivatives of adenine and guanine and the N1-acetic acid derivatives of uracil and cytosine. A promising reaction is that of formaldehyde and hydrogen cyanide with the free bases as shown below with adenine: ![]()
Using 1 mM portions of each base and 30–70 mM HCN and H2CO gave disappointing yields ranging from 0.04% to 0.15%. Attempts at an internal Cannizzaro rearrangement using 1 mM glyoxal and 1 mM of each base gave similar yields (0.01–0.18%). Because these reactions were run under favorable conditions, the results indicate that a new approach was needed for the modified bases.
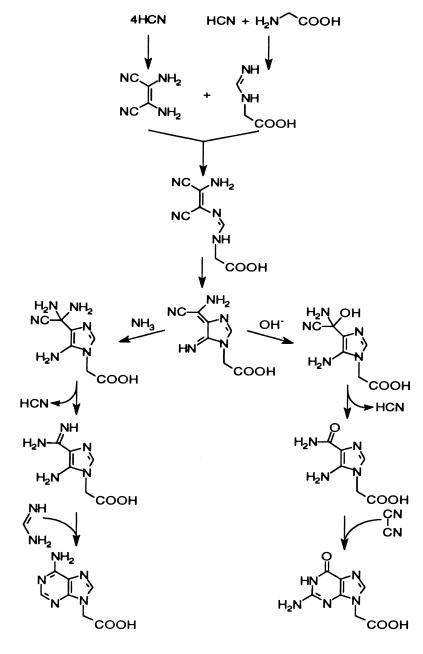
One possibility would be the synthesis of modified bases in the course of cyanide polymerizations in the presence of glycine. These solutions would arise from atmospheric syntheses of NH3 and HCN dissolved in ocean water that contained glycine from previous electric discharge syntheses or from extraterrestrial sources. The dilute solutions could be concentrated by freezing. Thus, heating mixtures of 2.5 M glycine, 7.5 M NH4OH, and 10 M NaCN gave 0.0062% adenine-N9-acetic acid, 0.0013% adenine-N6-acetic acid, and 0.0051% adenine. In the same mixture, guanine-N9-acetic acid was produced in 0.011% yield along with 5 × 10−4% guanine. These yields are much more encouraging than the results of the Strecker synthesis with the bases because the purine-N-acetic acids were isolated from the complex mixture of products in the NH4CN-glycine polymerization in yields comparable to adenine and guanine. The efficient synthesis of adenine and guanine have been demonstrated previously by using the frozen ocean model (42, 43). Shown in Fig. 4 is a proposed mechanism (39) for the production of adenine and guanine-N9-acetic acids from HCN tetramer and glycine. Such a mechanism would apply in the absence of UV light because of scattering or absorption by surface ice, thus precluding photochemical rearrangements during adenine synthesis (44).
Figure 4.
A proposed mechanism for the production of the purine-N9-acetic acids from HCN and glycine.
Cytosine and Uracil-N1-Acetic Acids.
The pyrimidine N1-acetic acids were obtained by a modification of the efficient prebiotic synthesis of the pyrimidine bases under dry beach conditions from reaction of cyanoacetaldehyde and high concentrations of urea (26). The reaction is the scheme shown here: ![]()
Hydantoin, the cyclic form of hydantoic acid, has been identified as a component of the Murchison meteorite (45) and as a product of the polymerization of HCN (46). Yields as high as 18% for cytosine-N1-acetic acid and 1.8% for uracil-N1-acetic acid based on cyanoacetaldehyde were observed in the reaction of 1 mM cyanoacetaldehyde with 2 M hydantoic acid at 100°C. About 14% cytosine-N3-acetic acid was also produced. We also examined this reaction in the presence of a competing nucleophile. Using 2 M urea in addition to 2 M hydantoic acid gave cytosine and cytosine-N1-acetic acid in an 8:1 ratio.
Polymerizability and Suitability as the First Genetic Material.
The above results show that the components of PNA are likely prebiotic compounds and, under favorable conditions, could be major constituents of the primitive milieu. Still to be worked out are the prebiotic syntheses of the monomers and mechanisms for their polymerization, but prebiotic polymerizations are imposing problems for any potential early genetic system (11, 12, 47). Our preliminary experiments indicate that AEG polymerizes readily at 100°C to give AEG oligomers and does so much more efficiently than mixtures of α-amino acids at higher temperatures (48). Although PNA also has stability problems of its own (49), they are highly sequence-dependent and may be alleviated by blocking or acetylating the N terminus. There is also the more difficult problem of PNA replication, which may be complicated by cyclization of the monomers. Nevertheless, this demonstration that the PNA components are prebiotic suggests the possibility that PNA or similar molecules may have been the first genetic material. However, other possibilities need to be considered because there may be other backbones and bases that were more abundant and more efficient for prebiotic replication.
Acknowledgments
We thank Jim Cleaves for sample preparation and technical discussions and Jeff Bada, John Eisch, Charles Salerno, and Jason Dworkin for helpful comments. We also wish to thank the NASA Specialized Center of Research and Training for fellowships (to K.E.N. and M.L.) and grant support (to S.L.M.).
Abbreviations
- PNA
peptide nucleic acid
- AEG
N-(2-aminoethyl)glycine
- ED
ethylenediamine
References
- 1.Kruger K, Grabowski P J, Zaug A J, Sands J, Gottschling D E, Cech T R. Cell. 1982;31:147–157. doi: 10.1016/0092-8674(82)90414-7. [DOI] [PubMed] [Google Scholar]
- 2.Guerrier-Takada C, Gardiner K, Marsh T, Pace N, Altman S. Cell. 1983;35:849–857. doi: 10.1016/0092-8674(83)90117-4. [DOI] [PubMed] [Google Scholar]
- 3.Woese C R. The Genetic Code: The Molecular Basis for Genetic Expression. New York: Harper & Row; 1967. pp. 179–195. [Google Scholar]
- 4.Crick F H C. J Mol Biol. 1968;38:367–379. doi: 10.1016/0022-2836(68)90392-6. [DOI] [PubMed] [Google Scholar]
- 5.Orgel L E. J Mol Biol. 1968;38:381–393. doi: 10.1016/0022-2836(68)90393-8. [DOI] [PubMed] [Google Scholar]
- 6.Gilbert W. Nature (London) 1986;319:618. [Google Scholar]
- 7.White H B., III J Mol Evol. 1976;7:101–104. doi: 10.1007/BF01732468. [DOI] [PubMed] [Google Scholar]
- 8.Larralde R, Robertson M P, Miller S L. Proc Natl Acad Sci USA. 1995;92:8158–8160. doi: 10.1073/pnas.92.18.8158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fuller W D, Sanchez R A, Orgel L E. J Mol Evol. 1972;1:249–257. doi: 10.1007/BF01660244. [DOI] [PubMed] [Google Scholar]
- 10.Fuller W D, Sanchez R A, Orgel L E. J Mol Biol. 1972;67:25–33. doi: 10.1016/0022-2836(72)90383-x. [DOI] [PubMed] [Google Scholar]
- 11.Orgel L E. Cold Spring Harbor Symp Quant Biol. 1987;52:9–16. doi: 10.1101/sqb.1987.052.01.004. [DOI] [PubMed] [Google Scholar]
- 12.Joyce G F. Cold Spring Harbor Symp Quant Biol. 1987;52:41–51. doi: 10.1101/sqb.1987.052.01.008. [DOI] [PubMed] [Google Scholar]
- 13.Miller S L. Nat Struct Biol. 1997;4:167–169. doi: 10.1038/nsb0397-167. [DOI] [PubMed] [Google Scholar]
- 14.Ertem G, Ferris J. Viva Origino. 1998;26:203–218. [Google Scholar]
- 15.Nielsen P E. Origins Life Evol Biosphere. 1993;23:323–327. doi: 10.1007/BF01582083. [DOI] [PubMed] [Google Scholar]
- 16.Nielsen P E, Egholm M, Berg R H, Buchardt O. Science. 1991;254:1497–1500. doi: 10.1126/science.1962210. [DOI] [PubMed] [Google Scholar]
- 17.Kim S K, Nielsen P E, Egholm M, Buchardt O, Berg R H, Norden B. J Am Chem Soc. 1993;115:6477–6481. [Google Scholar]
- 18.Hyrup B, Nielsen P E. Bioorg Med Chem. 1996;4:5–23. doi: 10.1016/0968-0896(95)00171-9. [DOI] [PubMed] [Google Scholar]
- 19.Westheimer F H. Science. 1987;235:1173–1178. doi: 10.1126/science.2434996. [DOI] [PubMed] [Google Scholar]
- 20.Byk G, Gilon C. J Org Chem. 1992;57:5687–5692. [Google Scholar]
- 21.Dueholm K L, Egholm M, Behrens C, Christensen L, Hansen H F, Vulpius T, Petersen K H, Berg R H, Nielsen P E, Buchardt O. J Org Chem. 1994;59:5767–5773. [Google Scholar]
- 22.Jones A S, Lewis P, Withers S F. Tetrahedron. 1973;29:2293–2296. [Google Scholar]
- 23.Jacobsen J R, Cochran A G, Stephans J C, King D S, Schultz P G. J Am Chem Soc. 1995;117:5453–5461. [Google Scholar]
- 24.Goody R S, Walker R T. J Org Chem. 1971;36:727–730. doi: 10.1021/jo00804a027. [DOI] [PubMed] [Google Scholar]
- 25.Meltzer P C, Liang A Y, Matsudaira P. J Org Chem. 1995;60:4305–4308. [Google Scholar]
- 26.Robertson M P, Miller S L. Nature (London) 1995;375:772–774. doi: 10.1038/375772a0. [DOI] [PubMed] [Google Scholar]
- 27.Miller S L. J Am Chem Soc. 1955;77:2351. [Google Scholar]
- 28.Ring D, Wolman Y, Friedmann N, Miller S L. Proc Natl Acad Sci USA. 1972;69:765–768. doi: 10.1073/pnas.69.3.765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wall J S. Anal Chem. 1953;25:950–953. [Google Scholar]
- 30.Zhao M X, Bada J L. J Chromatogr A. 1995;690:55–63. doi: 10.1016/0021-9673(94)00927-2. [DOI] [PubMed] [Google Scholar]
- 31.Tapuhi Y, Schmidt D E, Lindner W, Karger B L. Anal Biochem. 1981;115:123–129. doi: 10.1016/0003-2697(81)90534-0. [DOI] [PubMed] [Google Scholar]
- 32.Oró J. Biochem Biophys Res Commun. 1960;2:407–412. doi: 10.1016/j.bbrc.2011.07.075. [DOI] [PubMed] [Google Scholar]
- 33.Oró J, Kamat S S. Nature (London) 1961;190:442–443. doi: 10.1038/190442a0. [DOI] [PubMed] [Google Scholar]
- 34.Schwartz A W, Goverde M. J Mol Evol. 1982;18:351–353. doi: 10.1007/BF01733902. [DOI] [PubMed] [Google Scholar]
- 35.Schlesinger G, Miller S L. J Mol Evol. 1983;19:383–390. doi: 10.1007/BF02101643. [DOI] [PubMed] [Google Scholar]
- 36.Zahnle K J. J Geophys Res. 1986;91:2819–2834. [Google Scholar]
- 37.Pinto J P, Gladstone G R, Yung Y L. Science. 1980;210:183–185. doi: 10.1126/science.210.4466.183. [DOI] [PubMed] [Google Scholar]
- 38.Kasting J F, Pollack J B, Crisp D. J Atmos Chem. 1984;1:403–428. doi: 10.1007/BF00053803. [DOI] [PubMed] [Google Scholar]
- 39.Shuman R F, Shaerin W E, Tull R J. J Org Chem. 1979;44:4532–4536. [Google Scholar]
- 40.Ogura K, Migita C T, Yamada T. J Photochem Photobiol A. 1989;49:53–61. [Google Scholar]
- 41.Sagan C, Chyba C. Science. 1997;276:1217–1229. doi: 10.1126/science.276.5316.1217. [DOI] [PubMed] [Google Scholar]
- 42.Schwartz A W, Joosten H, Voet A B. BioSystems. 1982;15:191–193. doi: 10.1016/0303-2647(82)90003-x. [DOI] [PubMed] [Google Scholar]
- 43.Levy, M., Miller, S. L., Brinton, K. & Bada, J. L. (2000) Icarus, in press. [DOI] [PubMed]
- 44.Levy M, Miller S L, Oró J. J Mol Evol. 1999;49:165–168. doi: 10.1007/pl00006539. [DOI] [PubMed] [Google Scholar]
- 45.Cooper G W, Cronin J R. Geochim Cosmochim Acta. 1995;59:1003–1015. doi: 10.1016/0016-7037(95)00018-6. [DOI] [PubMed] [Google Scholar]
- 46.Ferris J P, Wos J D, Lobo A P. J Mol Evol. 1974;3:311–316. doi: 10.1007/BF01796046. [DOI] [PubMed] [Google Scholar]
- 47.Keefe A D, Miller S L. J Mol Evol. 1995;41:693–702. doi: 10.1007/BF00173147. [DOI] [PubMed] [Google Scholar]
- 48.Fox S W, Dose K. Molecular Evolution and the Origin of Life. San Francisco: Freeman; 1972. pp. 133–195. [Google Scholar]
- 49.Eriksson M, Christensen L, Schmidt J, Haaima G, Orgel L, Nielsen P E. New J Chem. 1998;22:1055–1059. doi: 10.1039/a803214i. [DOI] [PubMed] [Google Scholar]