Abstract
The heterodimeric nuclear cap-binding complex (CBC) binds to the mono-methylated 5′ cap of eukaryotic RNA polymerase II transcripts such as mRNA and U snRNA. The binding is important for nuclear maturation of mRNAs and possibly in the first round of translation and nonsense-mediated decay. It is also essential for nuclear export of U snRNAs in metazoans. We report characterization by fluorescence spectroscopy of the recognition of 5′ capped RNA by human CBC. The association constants (Kas) for 17 mono- and dinucleotide cap analogs as well as for the oligomer m7GpppAm2′ pUm2′pAm2′ cover the range from 1.8 × 106 M−1 to 2.3 × 108 M−1. Higher affinity for CBC is observed for the dinucleotide compared with mononucleotide analogs, especially for those containing a purine nucleoside next to m7G. The mRNA tetramer associates with CBC as tightly as the dinucleotide analogs. Replacement of Tyr138 by alanine in the CBP20 subunit of CBC reduces the cap affinity except for the mononucleotide analogs, consistent with the crystallographic observation of the second base stacking on this residue. Our spectroscopic studies showed that contrary to the other known cap-binding proteins, the first two nucleotides of a capped-RNA are indispensable for its specific recognition by CBC. Differences in the cap binding of CBC compared with the eukaryotic translation initiation factor 4E (eIF4E) are analyzed and discussed regarding replacement of CBC by eIF4E.
Keywords: CBC, mRNA 5′, cap, molecular recognition, fluorescence, binding energy
INTRODUCTION
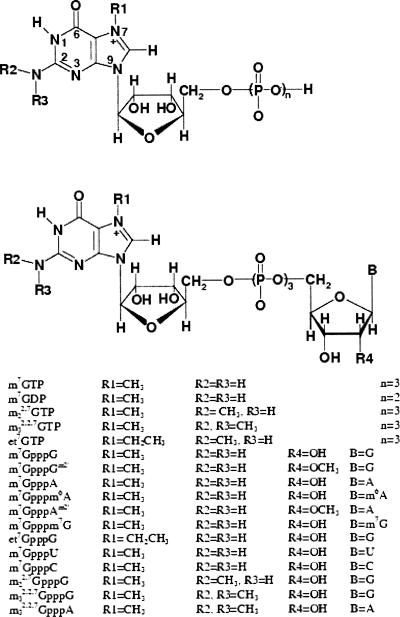
The 5′ terminal cap structure m7G(5′)ppp(5′)N (Scheme 1), N= G, A, U, or C, of nascent eukaryotic RNA polymerase II transcripts, e.g., pre-mRNA and U snRNA, is bound in the nucleus by the nuclear cap-binding complex (CBC). CBC is a tight heterodimer composed of two polypeptides, CBP80 and CBP20, highly conserved from yeast to man. This RNA– protein complex plays an active role in pre-mRNA splicing, 3′-end formation, and RNA nuclear export (for review, see Lewis and Izaurralde 1997). In particular, CBC is required for efficient splicing of the cap proximal intron by facilitating U1 snRNP association with the 5′ splice site (Izaurralde et al. 1994; Lewis et al. 1996) and enhances 3′-end formation by increasing the efficiency of the first step of that process, i.e., endonucleolytic poly(A) site cleavage (Gorlich et al. 1996). In multicellular organisms CBC is also required for nuclear export of U snRNA (Hamm and Mattaj 1990) as some steps of the U snRNP assembly occur in the cytoplasm. The CBC/capped U snRNA complex interacts with the PHAX protein (PHosphorylated Adapter for RNA eXport), which, when phosphorylated, binds to the receptor Crm1-RanGTP leading to nuclear export (Izaurralde et al. 1995; Ohno et al. 2000). Although CBC is exported with mRNA in yeast and vertebrate cells, it is not required for the transport process but has a stimulatory effect on it (Visa et al. 1996; Shen et al. 2000). Cytoplasmic CBC is rapidly reimported to the nucleus, leading to a steady-state localization that is predominantly nuclear. CBC recycling occurs by the importin-α pathway (Flaherty et al. 1997).
SCHEME 1.
Structures of the mono- and dinucleotide cap-analogs. In the tetranucleotide m7GpppAm2′pUm2′pAm2′the additional 2′-Omethylated nucleosides, U and A, are linked to m7GpppAm2′ through the 3′-to-5′ phosphodiester bonds.
Both subunits of CBC, CBP80 (780 residues in human) and CBP20 (156 residues in human), are essential for binding cap (Izaurralde et al. 1994). The binding specificity of CBC purified from HeLa nuclear extract was tested by using different cap analogs as competitive inhibitors (Izaurralde et al. 1994). Both m7GpppG and et7GpppG were able to compete for CBC with a capped RNA much more efficiently than were m22,7GpppG and m32,2,7GpppG, and markedly more efficiently than were mononucleotide cap analogs, m7GMP, m7GDP, and m7GTP. The results showed that human CBC and cytoplasmic cap-binding protein eIF4E exhibit different recognition specificity for the cap. eIF4E binds almost equally well to m7GpppG, m22,7GpppG, and m7GDP, while m7GTP has the highest observed affinity (Cai et al. 1999; Niedzwiecka et al. 2002a).
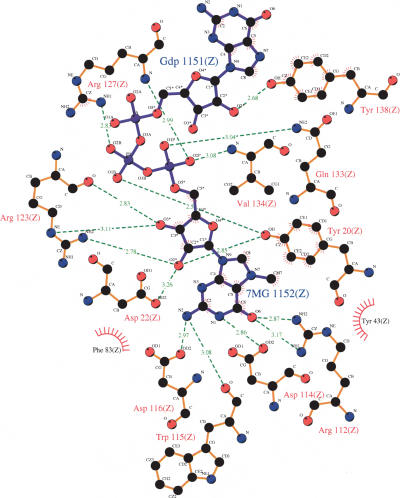
The three-dimensional structure of human CBC has been solved at about 2 Å resolution for apo-CBC (Mazza et al. 2001; Calero et al. 2002) and for the m7GpppG-bound protein (Calero et al. 2002; Mazza et al. 2002a). CBP80 consists of three entirely helical domains (called MIF4G domains), connected by two extended linkers. Each domain is arranged in five or six successive helical hairpins. The smaller subunit, CBP20, binds the mRNA 5′ cap only in the presence of CBP80 (Izaurralde et al. 1994). CBP20 has a core RNP domain that binds via its helical surface to CBP80. Binding of m7GpppG induces a remarkable cooperative folding of ~50 residues belonging to the N- and C-terminal parts around the cap, which lies on the β-sheet surface of the RNP domain. Cap binding is stabilized by numerous hydrogen bonds with the base and ribose moieties of m7G and with the phosphate oxygen atoms, by sandwich stacking of the m7G in between Tyr43 and Tyr20, and by stacking of the second G with Tyr138 (Scheme 2) (Calero et al. 2002; Mazza et al. 2002a). CBP20 shares a fundamental feature of the m7G recognition with other cap-binding proteins, eIF4E (Marcotrigiano et al. 1997; Matsuo et al. 1997; Niedzwiecka et al. 2002a; Tomoo et al. 2002) and vaccinia virus 2′-O-methyltransferase VP39 (Hu et al. 1999), this being the sandwiching of 7-methylguanine between two aromatic amino acid sidechains.
SCHEME 2.
Schematic representation of the stabilizing contacts in the CBC cap-binding center (Mazza et al. 2002a), Protein Data Bank accession number 1h2t, using LIGPLOT (Wallace et al. 1995). The interatomic distances are in Å.
To gain detailed insight into the formation and stability of the CBC-capped RNA complex, precise measurements in solution are necessary in addition to the three-dimensional structure derived from crystallography. Our previous studies on interactions of eukaryotic translation initiation factor 4E (eIF4E) with structurally selected cap analogs (Niedzwiecka et al. 2002a) and analysis of the crystal structures of the eIF4E-cap complexes (Marcotrigiano et al. 1997; Matsuo et al. 1997; Niedzwiecka et al. 2002a; Tomoo et al. 2002) revealed contributions of single structural modifications to the standard Gibbs free energy of the binding (ΔG°). This, together with an environmental (Niedzwiecka et al. 2002a), thermodynamic (Niedzwiecka et al. 2002b, 2004), and kinetic (Blachut-Okrasinska et al. 2000) characterization of complex formation and stability revealed a two-step mechanism of the intermolecular recognition between eIF4E and the mRNA 5′ terminus (Niedzwiecka et al. 2002a), as well as the influence of eIF4E phosphorylation on the eIF4E-cap association (Zuberek et al. 2003). In this article we present the first quantitative study in solution on binding of structurally modified cap analogs to the wild-type and Y138A mutated CBCs using the fluorescence titration approach (Niedzwiecka et al. 2002a).
RESULTS
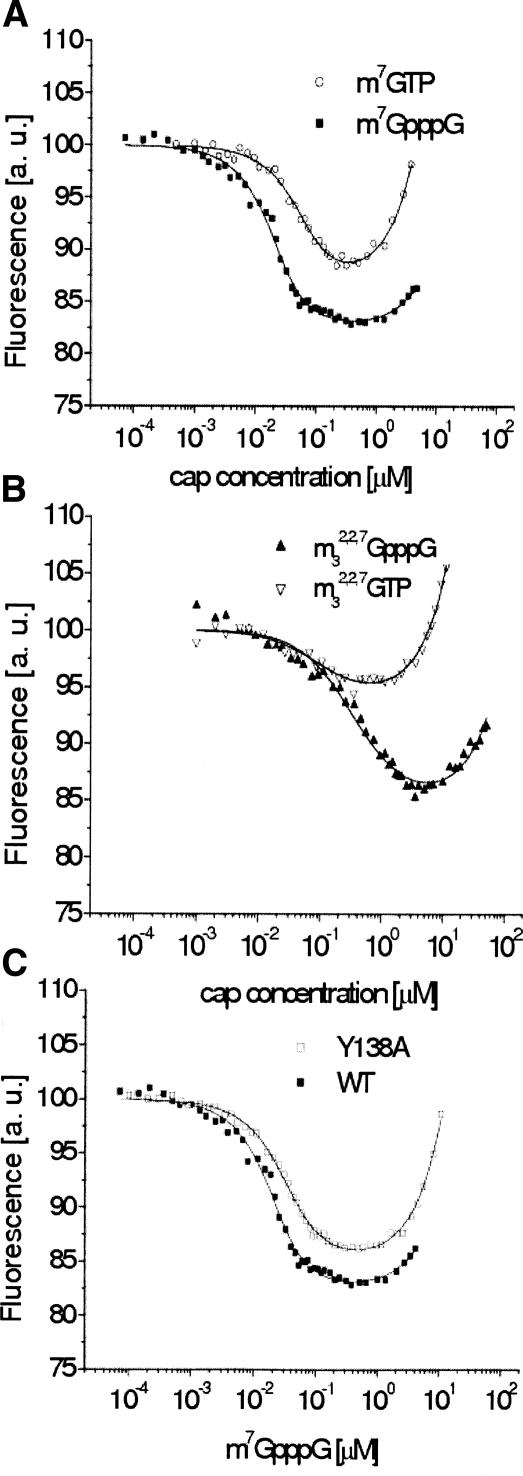
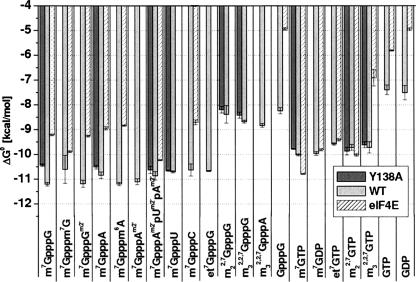
Interaction between the cap analogs and CBC results in quenching of the protein fluorescence, as shown in Figure 1. The association constants (Kas) and the corresponding Gibbs free energies of binding (ΔG°) are collected in Table 1, and the latter are graphically visualized in Figure 2. It is striking, that CBC significantly prefers to bind the dinucleotide, N7- methylated cap analogs compared with the mononucleotide analogs (Fig. 1A), except for the di- and trimethylated caps that contain one or two methyl substituents at the N2 amino group of 7-methylguanosine, respectively (Fig. 1B). The average binding energy of the dinucleotides in the bipartite CBC binding center is less by ~1 kcal/mol than that for m7GTP. This corresponds to increase of Kas by one order of magnitude. Replacement of methyl at N7 by a larger ethyl group decreases the binding affinity by twofold for both mono- and dinucleotide caps, probably due to steric hindrance. Methylation at the 2′-hydroxyl of the second nucleoside has no effect on the m7GpppG affinity and only leads to a slight increase of the Kas value in the case of m7GpppA.
FIGURE 1.
Comparison of the titration curves for selected cap analogs: (A) wild-type CBC, mono- and dinucleotide; (B) wild-type CBC, N2-methylated mono- and dinucleotide; and (C) Y138A CBC, m7GpppG. Titrations were performed in 50 mM Hepes/NaOH (pH 7.5), 200 mM NaCl, 0.2 mM EDTA, and 10 mM DTT. The point on the X-axis where the fluorescence curve attains plateau determines Kas. An increasing fluorescence signal at higher cap concentrations originates from the free-cap emission. Value of maximal quenching Q (Y-axis) reflects changes of the molecular environment of the fluorescent protein residues due to the binding of a cap analog, including conformational rearrangement of the protein.
TABLE 1.
Equilibrium association constants (Kas) and binding Gibbs free energies (ΔG°) for the cap–wild-type CBC and cap–Y138A CBC complexes at 20°C
| Wild type | Y138A | |||
| Cap analog | Kas·10−6 (M−1) | ΔG° (kcal/mol) | Kas·10−6 (M−1) | ΔG° (kcal/mol) |
| Dinucleotides | ||||
| m7GpppG | 231 ± 28 | −11.21 ± 0.07 | 61 .4 ± 5.5 | −10.43 ± 0.05 |
| m7GpppGm2′ | 223 ± 55 | −11.18± 0.14 | ||
| m7GpppA | 123± 28 | −10.84 ± 0.1 3 | 69.5 ± 9.5 | −10.51 ± 0.08 |
| m7Gpppm6A | 226 ± 26 | −11.19± 0.07 | ||
| m7GpppAm2′ | 193 ± 40 | −11.10± 0.12 | ||
| m7Gpppm7G | 82 ± 80 | −10.60 ± 0.56 | ||
| et7GpppG | 90.9 ± 4.2 | −10.70 ± 0.10 | ||
| m7GpppU | 97.1 ± 6.9 | −10.70 ± 0.04 | 89.0 ± 4.2 | −10.65 ± 0.03 |
| m7GpppC | 85 ± 36 | −10.62 ± 0.24 | ||
| m22,7GpppG | 1.8± 1.1 | −8.38 ± 0.36 | 1.3 ± 0.3 | −8.19 ± 0.1 3 |
| m32,2,7GpppG | 3.0 ± 0.3 | −8.68 ± 0.06 | 1.9 ± 0.4 | −8.41 ± 0.12 |
| m32,2,7GpppA | 3.7 ± 0.7 | −8.83 ± 0.08 | ||
| GpppG | 1.4 ± 0.3 | −8.23 ± 0.1 2 | ||
| Tetranucleotide | ||||
| m7GpppAm2′pUm2′pAm2′ | 128± 38 | −10.86 ± 0.1 7 | 85 ± 21 | −10.62 ± 0.1 4 |
| Mononucleotides | ||||
| m7GTP | 29.7 ± 2.3 | −10.01 ± 0.05 | 19.6 ± 0.9 | −9.77 ± 0.03 |
| m7GDP | 27.0 ± 3.0 | −9.96 ± 0.08 | ||
| m22,7GTP | 17.6 ± 3.5 | −9.71 ± 0.12 | 22.8 ± 6.0 | −9.85 ± 0.1 5 |
| m32,2,7GTP | 17.7 ± 6.8 | −9.71 ± 0.22 | 14.8 ± 3.0 | −9.61 ± 0.12 |
| et7GTP | 14.0± 1.1 | −9.57 ± 0.05 | ||
| GTP | 0.33 ± 0.11 | −7.39 ± 0.1 9 | ||
| GDP | 0.40 ± 0.20 | −7.50 ± 0.29 |
FIGURE 2.
Graphical presentation of the Gibbs free energies of the association of the cap analogs with wild-type CBC, the Y138A CBC mutant, and eIF4E. Titrations of eIF4E (Niedzwiecka et al. 2002a, Zuberek et al. 2003) were performed in 50 mM Hepes/NaOH (pH 7.2), 100 mM NaCl vs. 50 mM Hepes/NaOH (pH 7.5), 200 mM NaCl in the case of CBC.
Efficient stacking of the second base with Tyr138 correlates with the highest affinity of the dinucleotide cap analogs for the protein. The average binding free energy (ΔG°) of the dinucleotide caps containing C or U is by ~0.4 kcal/mol greater than for the dinucleotides containing A or G (approximately twofold decrease of Kas), unless the second guanosine is substituted with a methyl group at N7 (m7Gpppm7G). Moreover, the m7GpppG analog is bound stronger than m7GpppA, butmethylation of adenosine at the N6-amino group causes a marked increase of Kas.
The mononucleotide versus dinucleotide analogs that are additionally methylated at the exocyclic amino group (N2) differ distinctly in the binding to CBC (Table 1). Whereas for the dinucleotides the methyl substitution of either one proton or two protons at the amino group results in a dramatic ~100-fold reduction of Kas (decrease of the binding energy ΔG° by ~2.5 kcal/mol), the changes of Kas due to the methylation of the corresponding mononucleotides are less than twofold (decrease of ΔG° only by ~0.3 kcal/mol). Thus, N2-methylateddinucleotide analogs bind less tightly to CBC than to their mononucleotide counterparts. The binding parameters (Kas, ΔG°) for m22,7GpppG and m32,2,7GpppG are the same as for entirely unmethylated GpppG. To examine a possibility that m22,7GpppG and m32,2,7GpppG bind to CBP20 in a reversed orientation, with the unmethylated G in the place of m7G (like GpppG), we have run the CBC titration with m32,2,7GpppA. Adenosine possesses a different and unfavorable arrangement of hydrogen bond donors and acceptors for interaction with CBP20 compared with guanosine. CBC is found to bind both m32,2,7GpppG and m32,2,7GpppA with similar affinity (Table 1), which seems to preclude the possibility of the reverse type of binding. In contrast to the di-nucleotide analogs, methylation of the N2 amino group in the mononucleotide analogs only slightly affects their affinities to CBC. The total fluorescence-quenching pattern confirms this difference. A weak fluorescence quenching Q of 5% is observed for m32,2,7GTP, while it is up to 12% for m32,2,7GpppG (Fig. 1B). These observations show that CBC binds mono- and di-nucleotide cap analogs quite differently. The absence of the second nucleotide appears to weaken the discrimination by CBP20 of bases other than the cognate m7G. This is likely due to the incomplete folding of the CBP20 N-and C-terminal extensions in the absence of the second nucleotide, allowing more latitude in the orientation of the first base so as to avoid unfavorable contacts.
To shed some light on the nature of the interaction of the first transcribed nucleotide in the mRNA chain with Tyr138 of CBP20 (Scheme 1) we have run titration of the Y138A CBC mutant with some selected cap analogs (Table 1; Fig. 1C). The affinity of m7GpppG binding for Y138A CBC is fourfold lower than that for the wild-type protein, leading to 0.80 ± 0.36 kcal/mol decrease of ΔG°, whereas form7GTP the changes are negligible. The m7GpppU analog binds equally well to wild-type or mutated CBC, most probably due to lower strength of stacking between Tyr138 and U than between Tyr138 and G. There are also no significant differences in the Kas values for binding of either di- or trimethylated cap analogs to wild-type or Y138A CBC.
To examine a possible role of the nucleotides next to the second in the mRNA chain, we have titrated CBC with a tetranucleotide mRNA fragment, m7GpppAm2′pUm2′pAm2′. The binding free energy is slightly smaller than for the corresponding dinucleotide, m7GpppAm2′ (Table 1). This proves that no more than the first two nucleotides at the mRNA 5′ terminus are responsible for the tight specific interaction with CBC. However, further nucleotides may be involved in nonspecific contacts with the protein. The resulting small difference in the affinity of dinucleotide and that of the tetramer can be ascribed to an entropic cost of ordering the oligonucleotide chain upon the binding.
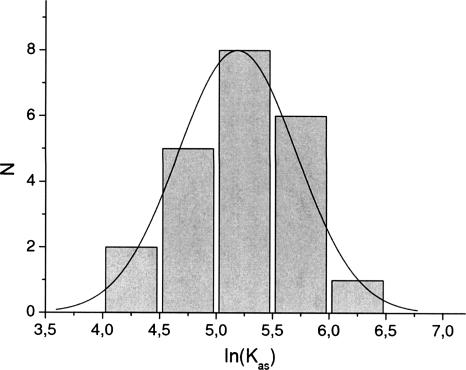
The observed changes of fluorescence intensity of CBC upon the binding of a cap analog are up to ~15% (Fig. 1). The quenching of the CBC fluorescence in the titration experiments most probably arises from changes of the emission of the tyrosines involved in the stacking interactions with cap and/or of the tryptophan emission due to conformational rearrangements of the protein upon formation of the complex. Determination of Kas from the fitting procedure is possible due to high sensitivity of the intrinsic protein fluorophors to their local environment. Our method of averaging lnKas (see Materials and Methods) provides reliable values of Kas and ΔG° even for few series of titration, due to the symmetric Gaussian type of distribution (Fig. 3).
FIGURE 3.
Statistics of the equilibrium association constant values (N) for m7GpppG with the imposed Gaussian distribution. The mean Kas = (194 ± 22) × 106 M−1 of the Gaussian distribution and the Kas value obtained by our averaging procedure, (231 ± 28) × 106 M−1 (Table 1) are very close to each other.
DISCUSSION
Application of precise spectroscopic methods to study the CBC–cap interaction enable us to complement and extend the crystallography-based structural view by a quantitative approach in terms of free energy of the association. Analysis of the crystallographic structures of the apo- and m7GpppG-bound CBC (Mazza et al. 2001, 2002a; Calero et al. 2002) revealed a large induced fit that involves ordering and stabilization of C- and N-terminal amino acids of CBP20 around the bound dinucleotide, accompanied by numerous additional inter- and intra-subunit interactions. A possible mechanism of the cap-binding process was proposed on the basis of the crystallographic data (Calero et al. 2002; Mazza et al. 2002a). In short, the conformationally stable part of CBP20, the RNP domain containing Tyr43, is an anchoring platform for the initial binding of m7G, and the folding of the CBP20 domains leads to crucial stacking of 7-methylguanine with Tyr20, followed by the final stabilization of the second nucleotide (Mazza et al. 2002a).
The fluorescence data support this model but additionally show that full discrimination of the first nucleotide is only achieved upon binding of the second nucleotide. Such a strong involvement of the first transcribed nucleotide in the cap recognition is a unique feature of CBC compared with the other known cap-binding proteins, e.g., eIF4E (Marcotrigiano et al. 1997; Matsuo et al. 1997; Niedzwiecka et al. 2002a; Tomoo et al. 2002) or viral methyltransferase VP39 (Hu et al. 1999). Comparison of strength of the CBC binding to the mono- and dinucleotide 5′ cap analogs as well as to the dinucleotide analogs and the tetramer clearly shows that the protein specifically recognizes the first two nucleotides in capped RNA. The presence of the second nucleotide is indispensable for formation of the tightly bound complex, even if the stacking of the second base with Tyr138 is missing. The dinucleotide analogs have greater affinity than do the mononucleotide analogs, three- to eightfold for the wild-type CBC, depending on the stacking ability of the second base, and approximately threefold for the Y138A CBC mutant. Therefore, in addition to the stacking with Tyr138, the second nucleoside of the cap must be stabilized by other nonspecific van der Waals contacts in the protein-binding center. The presence of the second nucleotide may also help to constrain the position of the third phosphate.
Stabilization of the CBC–cap complex by interactions involving the N2 amino group of m7G (Scheme 2) is crucial for the anchoring step (Mazza et al. 2002a), as suggested by the crystal structure. Indeed, even single methylation of the N2-amino group of m7G in the dinucleotide analogs precludes efficient binding to both the wild-type and mutated CBC due to disruption of the hydrogen bonds with Trp115 and Asp116, and substitution of the second methyl is of no further effect. In contrast, methylation of the N2-amino group in mononucleotide cap analogs has no effect on the binding showing that the mononucleotide caps, both methylated and unmethylated at N2, are bound in a different way in comparison with the dinucleotide analogs and longer capped-RNA fragments. Hence, the spectroscopic results reveal a significant cooperativity of the two parts of the CBC cap-binding center. The two nucleosides forming the cap are not bound independently, and it is only when the second nucleoside of the cap is stabilized by folding of CBP20 into the closed form that full discrimination of m7G against say m32,2,7 GTP is fully achieved.
The binding specificity of the cap analogs for CBC correlates with their ability to inhibit adenoviral pre-mRNA splicing in vitro and in vivo (Izaurralde et al. 1994). As determined qualitatively from gel electrophoresis, both m7GpppG and et7GpppG competed for CBC with the capped RNA ~1000-fold more efficiently than m22,7GpppG and m32,2,7GpppG, and ~30-fold more efficiently than mononucleotide cap analogs, m7GMP, m7GDP, and m7GTP. The results depend on the experimental conditions, e.g., the concentrations of the interacting species. The spectroscopically measured decrease of the affinity of N2-methylated analogs and mononucleotide analogs, expressed by Kas, is 100-fold and three- to eightfold, respectively (Table 1).
An important aspect of the interaction with the second nucleoside concerns the analogs with a methyl group at the 2′-hydroxyl of the second nucleoside, m7GpppGm2′ and m7GpppAm2′. Despite disruption of the putative hydrogen bond between the 2′-hydroxyl and the ring oxygen of Tyr138 (Scheme 2) the affinity of both analogs for CBC is not lower than that of the parent m7GpppG and m7GpppA (Table 1). Thus 2′-O-methylation in naturally occurring mRNA 5′ cap (cap1) does not lead to tighter binding to CBC.
In summary, the most important factor that contributes to specific recognition of cap by several cap-binding proteins, including eIF4E and VP39 (Quiocho et al. 2000), i.e., the stacking of the sandwiched m7G moiety with two aromatic side-chains, also plays a key role for CBC. In the CBC– cap complexes, the interaction with the second nucleotide is an additional key element of the specific recognition, and the two sites of the CBP20 cap-binding center, for m7G and for the second nucleoside, are highly cooperative.
The affinity of the dinucleotide analogs for CBC is threeto eightfold greater than the affinity of mononucleotides, contrary to eIF4E that binds m7GTP with the highest Kas of 108 × 106 M−1, compared with Kas of 2 × 106 M−1 to 8 × 106 M−1 for the dinucleotide analogs (Niedzwiecka et al. 2002a). The translation factor eIF4E binds to cap less tightly than CBC does (Fig. 2) but exhibits greater discrimination between the nucleotides possessing a substituent at N7 and those without such substituent. The corresponding Kas values for the binding to eIF4E differ 106-fold, whereas for CBC the difference is <103-fold. Biologically important methylation of the cap N2 amino group has differing effects on the binding to CBC versus eIF4E. The decrease of Kas is ~800-fold in the case of eIF4E and ~100-fold in the case of CBC. The substitution of one amino proton in m7G by a methyl group leads to as great a decrease of the affinity for CBC as the substitution of both protons by CH3, whereas single methylation leads to only slight decrease of the cap affinity for eIF4E.
Comparison of the cap-binding modes of CBC and eIF4E (Fig. 2) may have important biological consequences for exchange of CBC by the translation complex eIF4F at the mRNA 5′ terminus. CBC binds the dinucleotide cap analogs 40-fold stronger (~2 kcal/mol) and the tetranucleotide threefold stronger (~0.6 kcal/mol) than eIF4E (Fig. 2). Although the titrations of CBC and eIF4E (Niedzwiecka et al. 2002a; Zuberek et al. 2003) were performed at different salt concentrations, 200 mM NaCl and 100 mM NaCl, respectively (Fig. 2), to ensure optimal binding of the ligands, the elevated ionic strength was shown to decrease the eIF4E–cap association constants (Niedzwiecka et al. 2002a, Zuberek et al. 2004). Hence, the difference in affinity of eIF4E and CBC for capped-mRNA is even greater in the same buffer conditions. During the CBC-to-eIF4E exchange, the cap transfer occurs from the more to the less tightly binding protein. This process could be mediated by specific interactions between CBP80 and eIF4G (Fortes et al. 2000, McKendrick et al. 2001), although it was shown to be dispensable for translation in yeast (Baron-Benhamou et al. 2003). The latter large, adaptor protein eIF4G is bound to eIF4E within the eIF4F complex that is essential for both in vivo and in vitro translation (for review, see von der Haar et al. 2004). The role of a transient eIF4E–eIF4G– CBP80 interaction might be to decrease the affinity of CBC for cap through destablization of the CBC–cap complex (Fortes et al. 2000). In contrast, the interaction between eIF4E and eIF4G was found to increase the cap affinity for eIF4E (Haghighat and Sonenberg 1997; Gross et al. 2003). Thus carefully balanced modulation of cap-binding affinities by eIF4G may facilitate the replacement of CBC by eIF4E. However some results indicate that CBC may be the capbinding protein that initiates the first “pioneer” round of translation, where nonsense-mediated decay is triggered in the case of ribosome stalling at a premature stop codon (Ishigaki et al. 2001). The pioneer translation initiation complex is functionally distinct from but structurally overlaps with the steady-state translation initiation complex (Chiu et al. 2004). There are some suggestions that the replacement of CBC to eIF4E can occur in the mammalian nucleus (Wilkinson and Shyu 2002), but the exact mechanism and location of the process remains to be elucidated.
MATERIALS AND METHODS
Synthesis of cap analogs
The mono- and dinucleotide cap analogs (Scheme 1) were synthesized as reported previously (Darzynkiewicz et al. 1985, 1990; Jankowska et al. 1993, 1996; Stepinski et al. 1995). The tetranucleotide, m7GpppAm2′pUm2′pAm2′, was prepared fromtwo batches of the 5′-phosphorylated pAm2′pUm2′pAm2′ trimer, one synthesized according to the method of Zuberek et al. (2003), and the other purchased from TriLink BioTechnologies. A mixture of ammonium salt of the trimer (1.1 mg, 1 μmol), sodiumsalt of P2-imidazolide 7-methylguanosine 5′-diphosphate (2.7 mg, 5 μmol) (Sawai et al. 1999), andZnCl2 (14mg, 0.1 mMol) in dimethylformamide (0.3mL) was stirred for 24 h at room temperature. The reaction mixture was diluted with 1.5 mL of EDTA (3.7 mg, disodium salt) solution in water. The capped product was purified by HPLC (Spectra-Physics SP8800) using a reverse phase Supelcosil LC-18-T column (25 cm+2 cm guard) and eluted with a linear gradient ofmethanol, 0%–25% in 0.05 M ammonium acetate (pH 5.9) for 15 min, followed by 15-min elution keeping the final concentration of methanol. The final product m7GpppAm2′pUm2′pAm2′ was obtained as the ammonium salt (0.73 mg, 0.48 μmol, yield 48%). The predicted molecular mass for the free acid is 1463.9 Da, and the measured mass by ESI-MS was 1462.1 Da.
Expression and purification of wild-type and mutant CBC
Expression, purification, and assembly of wild-type CBC was performed as described earlier (Mazza et al. 2002b). Alanine mutagenesis of CBP20 (Y138A) was carried out according to the method of Mazza et al. (2002a).
Spectroscopic measurements and data analysis
Fluorescence titrations were performed on an LS-50B spectrofluorometer (Perkin Elmer Co.) in 50 mM Hepes/NaOH (pH 7.5), 200 mM NaCl, 0.2 mM EDTA, and 10 mM DTT, at 20 ± 0.2°C as described previously (Niedzwiecka et al. 2002a). Aliquots of 1 μL of increasing concentrations of the cap analog solutions were added to 1000 μL of 0.05 μM CBC. Fluorescence intensities (excitation at 275 nm, detection at 336 nm for most of the cap analogs; 290 nm/330 nm for hypermethylated dinucleotide cap analogs) were integrated over 30 sec, with a break of 30 sec for the ligand addition. The data were corrected for the sample dilution (<4.5%) and for the inner filter effect (Lakowicz 1999). Equilibrium association constants (Kas) were determined by fitting of the theoretical dependence of the fluorescence intensity (F) on the total concentration of the cap analog ([L]) to the experimental data points according to the equation:
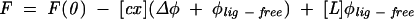 |
where F(0) is the initial fluorescence intensity; Δφ, difference between the fluorescence efficiencies of the apo protein and the complex; φlig-free, the fluorescence efficiency of the free cap analog; and [cx], the equilibrium concentration of the cap–CBC complex is given by:
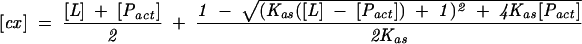 |
where [Pact] is the concentration of the active protein.
The numerical least-squares nonlinear regression analysis was performed by using ORIGIN 6.0 (Microcal Software Inc.). The final Kas values were calculated as weighted averages (Brandt 1999) of ln(Kasi), where Kasiis the association constant from a single titration i. The experimental error of the averaged Kas value was calculated from the Student t-distribution (Brandt 1999):
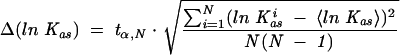 |
where N is the number of independent Kas values and tα,N is the factor of the Student’s t-distribution corresponding to the confidence interval α = 0.6826 (Brandt 1999), i.e., 1 SD. This procedure of averaging was chosen to avoid biasing of the results toward lower values, which would occur for the simple weighted averages of the Kas values. The statistically estimated errors of lnKas allow for a reliable comparison of the individual results.
The Gibbs free energy of binding ΔG° was calculated as ΔG° = −RT·lnKas. For symmetrical dinucleotide cap analogs, the microscopic association constant, 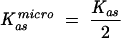 , has been used due to the entropic effects.
, has been used due to the entropic effects.
Total quenching (Q) was calculated from the fitting curves according to:
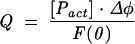 |
Acknowledgments
We are indebted to Teija Nittymaki and Harri Lonnberg for cooperation in preparing the tetranucleotide. This work was supported by the State Committee for Scientific Research KBN 3 P04A 021 25, KBN P04 A 006 28, PBZ/KBN/059/T09/2001, and BST 833/BF.
Abbreviations
CBC, cap-binding complex
eIF4E, eukaryotic initiation factor 4E
Gm2′, 2′-O-methylguanosine
Am2′, 2′-O-methyladenosine
Um2′, 2′-O-methyluridine
m6A, N6-methyladenosine
m7G, 7-methylguanosine
m22,7 G, N2,7-dimethyl guanosine
m32,2,7 G, N2,N2,7-trimethylguanosine
m7GTP, 7-methylguanosine triphosphate and similarly other nucleoside triphosphates
m22,7 GTP, N2,7-dimethylguanosine triphosphate
m7GpppG, P1-7-methylguanosine-5′ P3-guanosine-5′ triphosphate, and similarly other dinucleoside triphosphates
EDTA, ethylenediaminetetraacetic acid
DTT, dithiothreitol
HEPES, 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid
wt, wild type.
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.2850705.
REFERENCES
- Baron-Benhamou, J., Fortes, P., Inada, T., Preiss, T., and Hentze, M.W. 2003. The interaction of the cap-binding complex (CBC) with eIF4G is dispensable for translation in yeast. RNA 9: 654–662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blachut-Okrasinska, E., Bojarska, E., Niedzwiecka, A., Chlebicka, L., Darzynkiewicz, E., Stolarski, R., Stepinski, J., and Antosiewicz, A. 2000. Stopped-flow and Brownian dynamics studies of electrostatic effects in the kinetics of binding of 7-methyl-GpppG to the protein eIF4E. Eur. Biophys. J. 29: 487–498. [DOI] [PubMed] [Google Scholar]
- Brandt, S. 1999. Data analysis: Statistical and computational methods for scientists and engineers, 3d ed. Springer Verlag, New York.
- Cai, A., Jankowska-Anyszka, M., Centers, A., Chlebicka, L., Stepinski, J., Stolarski, R., Darzynkiewicz, E., and Rhoads, R.E. 1999. Quantitative assessment of mRNA cap analogues as inhibitors of in vitro translation. Biochemistry 38: 8538–8547. [DOI] [PubMed] [Google Scholar]
- Calero, G., Wilson, K., Ly, T., Rios-Steiner, J.R., Clardy, J., and Cerione, R.A. 2002. Structural basis of m(7)GpppG binding to the nuclear cap- binding protein complex. Nat. Struct. Biol. 9: 912–917. [DOI] [PubMed] [Google Scholar]
- Chiu, S.Y., Lejeune, F., Ranganathan, A.C., and Maquat, L.E. 2004. The pioneer translation initiation complex is functionally distinct from but structurally overlaps with the steady-state translation initiation complex. Genes & Dev. 18: 745–754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darzynkiewicz, E., Ekiel, I., Tahara, S.M., Seliger, L.S., and Shatkin, A.J. 1985. Chemical synthesis and characterization of 7-methylguanosine cap analogues. Biochemistry 24: 1701–1707. [Google Scholar]
- Darzynkiewicz, E., Stepinski, J., Tahara, S.M., Ekiel, I., Haber, D., Neuvonen, K., Lehikoinen, P., Labadi, I., and Lonnberg, H. 1990. Synthesis, conformation and hydrolytic stability of P1, P3-dinucleoside triphosphates related to mRNA 5′-cap, and comparative kinetic studies on their nucleoside and nucleoside monophosphate analogues. Nucleosides Nucleotides 9: 599–618. [Google Scholar]
- Flaherty, S.M., Fortes, P., Izaurralde, E., Mattaj, I.W., and Gilmartin, G.M. 1997. Participation of the nuclear cap binding complex in pre-mRNA 3′ processing. Proc. Natl. Acad. Sci. 94: 11893–11898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fortes, P., Inada, T., Preiss, T., Hentze, M.W., Mattaj, I.A., and Sachs, A.B. 2000. The yeast nuclear cap binding complex can interact with translation factor eIF4G and mediate translation initiation. Mol. Cell 6: 191–196. [PubMed] [Google Scholar]
- Gorlich, D., Kraft, R., Kostka, S., Vogel, F., Hartmann, E., Laskey, R.A., Mattaj, I.W., and Izaurralde, E. 1996. Importin provides a link between nuclear protein import and U snRNA export. Cell 87: 21–32. [DOI] [PubMed] [Google Scholar]
- Gross, J.D., Moerke, N.J., der Haar, T., Lugovskoy, A.A., Sachs, A.B., McCarthy, J.E., and Wagner, G. 2003. Ribosome loading onto the mRNA cap is driven by conformational coupling between eIF4G and eIF4E. Cell 115: 739–750. [DOI] [PubMed] [Google Scholar]
- Haghighat, A. and Sonenberg, N. 1997. Additions and corrections to eIF4G dramatically enhances the binding of eIF4E to the mRNA 5′-cap structure. J. Biol. Chem. 272: 21677–21680. [DOI] [PubMed] [Google Scholar]
- Hamm, J. and Mattaj, I.W. 1990. Monomethylated cap structures facilitate RNA export from the nucleus. Cell 63: 109–118. [DOI] [PubMed] [Google Scholar]
- Hu, G., Gershon, P.D., Hodel, A.E., and Quiocho, F.A. 1999. mRNA cap recognition: Dominant role of enhanced stacking interactions between methylated bases and protein aromatic side chains. Proc. Natl. Acad. Sci. 96: 7149–7154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishigaki, Y., Li, X., Serin, G., and Maquat, L.E. 2001. Evidence for a pioneer round of mRNA translation: mRNAs subject to nonsensemediated decay in mammalian cells are bound by CBP80 and CBP20. Cell 106: 607–617 [DOI] [PubMed] [Google Scholar]
- Izaurralde, E., Lewis, J., McGuigan, C., Jankowska, M., Darzynkiewicz, E., and Mattaj, I.W. 1994. A nuclear cap binding protein complex involved in pre-mRNA splicing. Cell 78: 657–668. [DOI] [PubMed] [Google Scholar]
- Izaurralde, E., Lewis, J., Gamberi, C., Jarmolowski, A., McGuigan, C., and Mattaj, I.W. 1995. A cap-binding protein complex mediating U snRNA export. Nature 376: 709–712 [DOI] [PubMed] [Google Scholar]
- Jankowska, M., Stepinski, J., Stolarski, R., Temeriusz, A., and Darzynkiewicz, E. 1993. Synthesis and properties of new NH2 and N7 substituted GMP and GTP 5′-mRNA cap analogues. Collect. Czech. Chem. Commun. 58: 138–141. [Google Scholar]
- Jankowska, M., Stepinski, J., Stolarski, R., Wieczorek, Z., Temeriusz, A., and Darzynkiewicz, E. 1996. H-1 NMR and fluorescence studies of new mRNA 5′-cap analogues. Collect. Czech. Chem. Commun. 61: 197–202. [Google Scholar]
- Lakowicz, J.R. 1999. Principles of fluorescence spectroscopy, 2d ed. Kluwer Academic/Plenum Publishers, New York.
- Lewis, J.D. and Izaurralde, E. 1997. The role of the cap structure in RNA processing and nuclear export. Eur. J. Biochem. 247: 461–469. [DOI] [PubMed] [Google Scholar]
- Lewis, J.D., Gorlich, D., and Mattaj, I.W. 1996. A yeast cap binding protein complex (yCBC) acts at an early step in pre-mRNA splicing. Nucleic Acids Res. 24: 3332–3336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcotrigiano, J., Gingras, A.-C., Sonenberg, N., and Burley, S.K. 1997. Cocrystal structure of the messenger RNA 5′ cap-binding protein (eIF4E) bound to 7-methyl-GDP. Cell 951: 951–961. [DOI] [PubMed] [Google Scholar]
- Matsuo, H., Li, H., McGuire, A.M., Fletcher, C.M., Gingras, A.-C., Sonenberg, N., and Wagner, G. 1997. Structure of translation factor eIF4E bound to m7GDP and interaction with 4E-binding protein. Nat. Struct. Biol. 4: 717–724. [DOI] [PubMed] [Google Scholar]
- Mazza, C., Ohno, M., Segref, A., Mattaj, I.W., and Cusack, S. 2001. Crystal structure of the human nuclear cap binding complex Mol. Cell 8: 383–396. [DOI] [PubMed] [Google Scholar]
- Mazza, C., Segref, A., Mattaj, I.W., and Cusack, S. 2002a. Large-scale induced fit recognition of an m(7)GpppG cap analogue by the human nuclear cap-binding complex. EMBO J. 21: 5548–5557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ———. 2002b. Co-crystallization of the human nuclear cap-binding complex with a m7GpppG cap analogue using protein engineering. Acta Cryst. D 58: 2194–2197. [DOI] [PubMed] [Google Scholar]
- McKendrick, L., Thompson, E., Ferreira, J., Morley, S.J., and Lewis, J.D. 2001. Interaction of eukaryotic translation initiation factor 4G with the nuclear cap-binding complex provides a link between nuclear and cytoplasmic functions of the m7 guanosine cap. Mol. Cell. Biol. 21: 3632–3641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niedzwiecka, A., Marcotrigiano, J, Stepinski, J., Jankowska-Anyszka, M., Wyslouch-Cieszynska, A., Dadlez, M., Gingras, A.-C., Mak, P., Darzynkiewicz, E., Sonenberg, N., et al. 2002a. Biophysical studies of eIF4E cap-binding protein: Recognition of mRNA 5′ cap structure and synthetic fragments of eIF4G and 4E-BP1 proteins. J. Mol. Biol. 319: 615–635. [DOI] [PubMed] [Google Scholar]
- Niedzwiecka, A., Stepinski, J., Darzynkiewicz, E., and Stolarski, R. 2002b. Positive heat capacity change upon specific binding of translation initiation factor eIF4E to mRNA 5′ cap. Biochemistry 41: 12140–12148. [DOI] [PubMed] [Google Scholar]
- Niedzwiecka, A., Darzynkiewicz, E., and Stolarski, R. 2004. Thermodynamics of mRNA 5′ cap binding by eukaryotic translation initiation factor eIF4E. Biochemistry 43: 13305–13317. [DOI] [PubMed] [Google Scholar]
- Ohno, M., Segref, A., Bachi, A., Wilm, M., and Mattaj, I.W. 2000. PHAX, a mediator of U snRNA nuclear export whose activity is regulated by phosphorylation. Cell 101: 187–198. [DOI] [PubMed] [Google Scholar]
- Quiocho, F.A., Guanghui, H., and Gershon, P.D. 2000. Structural basis of mRNAcap recognition by proteins, Curr. Opin. Struct. Biol. 10: 78–86. [DOI] [PubMed] [Google Scholar]
- Sawai, H., Wakai, H., and Nakamura-Ozaki, A. 1999. Synthesis and reactions of nucleoside 5′-diphosphate imidazolide: A nonenzymatic capping agent for 5′-monophosphorylated oligoribonucleotides in aqueous solution. J. Org. Chem. 64: 5836–5840. [Google Scholar]
- Shen, E.C., Stage-Zimmermann, T., Chui, P., and Silver, P.A. 2000. The yeast mRNA-binding protein Npl3p interacts with the Capbinding complex. J. Biol. Chem. 275: 23718–23724. [DOI] [PubMed] [Google Scholar]
- Stepinski, J., Bretner, J., Jankowska, M., Felczak, K., Stolarski, R., Wieczorek, Z., Cai, A.-L., Rhoads, R.E., Haber, D., and Darzynkiewicz, E. 1995. Synthesis and properties of P1, P2-, P1, P3- and P1, P4- dinucleotide di-, tri- and tetraphosphate mRNA 5′-cap analogues. Nucleosides Nucleotides 14: 717–721. [Google Scholar]
- Tomoo, K., Shen, X., Okabe, K., Nozoe, Y., Fukuhara, S., Morino, S., Ishida, T., Taniguhi, T., Hasegawa, H., Terashima, A., et al. 2002. Crystal structures of 7-methylguanosine 5′-triphosphate (m(7)GTP)- and P(1)-7-methylguanosine-P(3)-adenosine-5′,5′- triphosphate (m(7)GpppA)-bound human full-length eukaryotic initiation factor 4E: Biological importance of the C-terminal flexible region. Biochem. J. 362: 539–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visa, N., Izaurralde, E., Ferreira, J., Daneholt, D., and Mattaj, I.W. 1996. A nuclear cap-binding complex binds Balbiani ring premRNA cotranscriptionally and accompanies the ribonucleoprotein particle during nuclear export. J. Cell Biol. 133: 5–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von der Haar, T., Gross, J.D., Wagner, G., and McCarthy J.E.G. 2004. The mRNA cap-binding protein eIF4E in post-transcriptional gene expression. Nat. Struct. Biol. 11: 503–511. [DOI] [PubMed] [Google Scholar]
- Wallace, A.C., Laskowski, R.A., and Thornton, J.M. 1995. LIGPLOT: A program to generate schematic diagrams of protein–ligand interactions. Protein Eng. 8: 127–134. [DOI] [PubMed] [Google Scholar]
- Wilkinson, M.F. and Shyu, A.-B. 2002. RNA surveillance by nuclear scanning, Nat. Cell Biol. 4: E144–E147. [DOI] [PubMed] [Google Scholar]
- Zuberek, J., Wyslouch-Cieszynska, A., Niedzwiecka, A., Dadlez, M., Stepinski, J., Augustyniak, W., Gingras, A.-C, Zhang, Z., Burley, S.K., Sonenberg, N., et al. 2003. Phosphorylation of eIF4E attenuates its interaction with mRNA 5′ cap analogs by electrostatic repulsion: Intein-mediated protein ligation strategy to obtain phosphorylated protein. RNA 9: 52–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuberek, J., Jemielity, J., Jablonowska, A., Stepinski, J., Dadlez, M., Stolarski, R., and Darzynkiewicz, E. 2004. Influence of electric charge variation at residues 209 and 159 on the interaction of eIF4E with the mRNA 5′ terminus, Biochemistry 43: 5370–5379. [DOI] [PubMed] [Google Scholar]