Abstract
Distinct isoforms of spliceosomal RNAs may be involved in regulating pre-messenger RNA splicing in eukaryotic cells. During a large-scale effort to identify small noncoding RNAs in Drosophila, we isolated a U5 snRNA-like molecule containing a 5′ segment identical to that of the canonical (major) U5 snRNA but with a variant Sm binding site and a distinct 3′ hairpin sequence. Based on this finding, another six similar U5 snRNA-like sequences were identified within the Drosophila genome by sequence similarity to the invariant loop in the 5′ half of U5. Interestingly, although all of these variants are expressed in vivo, each shows a distinct temporal expression profile during Drosophila development, and one is expressed primarily in fly heads. The presence of these U5 snRNA variants within RNP particles suggests their role in splicing and implies a possible connection to regulation of developmental and tissue-specific gene expression.
Keywords: U5 snRNA, noncoding RNA, Drosophila
INTRODUCTION
The abundance of genomic, genetic, and biochemical tools available for Drosophila make the fruit fly an attractive system in which to investigate small noncoding RNAs (ncRNAs) that regulate post-transcriptional gene expression. Several previous studies have identified new ncRNAs in Drosophila, though in many cases the functions of these transcripts remain to be determined (Misra et al. 2002; Yuan et al. 2003; Tupy et al. 2005). In addition to novel ncRNAs, variant forms of small nuclear RNAs (snRNAs) and small nucleolar RNAs (snoRNAs) may play important roles in tissue-specific pre-messenger RNA (pre-mRNA) splicing and post-transcriptional RNA modifications, respectively (for reviews, see Bachellerie et al. 2002; Kiss 2002; Villa et al. 2002). Both snRNAs and snoRNAs function as part of ribonucleoprotein (RNP) complexes, and both have signature secondary structural motifs that enable identification of additional family members through bioinformatic methods (Lowe and Eddy 1999; Omer et al. 2000). The snRNAs, possibly remnants of an RNA-catalyzed machinery for intron removal, are responsible for targeting and aligning splice sites in pre-mRNAs through base-pairing interactions (Staley and Guthrie 1998; Collins and Guthrie 2000). For this reason, they could potentially regulate splice site choice and hence participate directly in increasing the size of the proteome through alternative splicing (Black 2000; Graveley 2001).
In an effort to identify small ncRNAs that might have new and possibly tissue-specific functions within Drosophila, we cloned and sequenced nonmessenger RNAs isolated from total fly RNA. The most interesting of the molecules discovered in this study is a class of RNAs with striking similarity to the spliceosomal U5 snRNA (Myslinski et al. 1984). Expression analysis showed that each of the seven U5-like snRNAs found within the Drosophila genome is expressed with a distinct developmental pattern, and one is specific to fly heads. The presence of some of these RNAs within large RNP complexes suggests their role in pre-mRNA splicing, perhaps as regulators of alternative or development-specific splicing.
RESULTS AND DISCUSSION
To identify and investigate novel ncRNAs that might have tissue-specific functions, a cDNA library was constructed using RNA isolated from Drosophila melanogaster. Briefly, total RNA was isolated from flies and hybridized to DNA oligonucleotides complementary to mRNA, 5S, 5.8S, U1, U2, U4, U5, and U6 RNA sequences in an effort to selectively remove these molecules prior to cloning. Surprisingly, several of the resulting clones encoded a noncanonical U5 snRNA-like RNA, called variant 63BC (accession number DQ-136000), which had previously been identified computationally based on sequence similarity to the canonical U5 snRNA sequence (Adams et al. 2000). The presence of this apparent U5 snRNA isoform within a sample from which snRNAs had been depleted suggested the possibility that this variant sequence might differ in structure or binding partners from the canonical U5 snRNA. Examination of snRNAs annotated in Flybase (http://www.flybase.org) revealed a total of seven nonredundant U5-like RNA sequences in addition to the canonical U5 snRNA. These eight RNAs are all ~125 nucleotides (nt) in length and consist of an identical 5′ 85-nt sequence followed by a variable 3′ sequence that in each case has the potential to form a single-hairpin secondary structure preceded by a short pyrimidine-rich region resembling the Sm protein binding site of canonical snRNAs (Wooley et al. 1982; Riedel et al. 1987; Fig. 1 ▶). The Sm protein binding motif is a hallmark of all snRNAs and indicates the site of assembly of the seven Sm proteins that form the common core of each snRNP particle within the spliceosome (for reviews, see Will et al. 1993; Kramer 1996; Will and Luhrmann 2001).
FIGURE 1.
Secondary structures of U5 snRNA isoforms. The secondary structures were generated based on the canonical human U5 snRNA secondary structure (Branlant et al. 1983). (Lowercase letters) The conserved 5′ region found in all U5 snRNAs, (boxed nucleotides) the conserved 11-nt region, (gray underlined letters) putative Sm binding sites, (bold capital letters) the unique 3′ variable regions that differ in length and structure. The asterisk indicates where the conserved 5′ region would be found in each U5 RNA variant. The name of each variant refers to its chromosomal location: U5 canonical is located in K03096; 23D, in AE003581; 34A, in AE003639; 63BC, in AE003477; 35D, in AE003648; 14B, in AE003501; 38ABa and 38ABb, in AE003664.
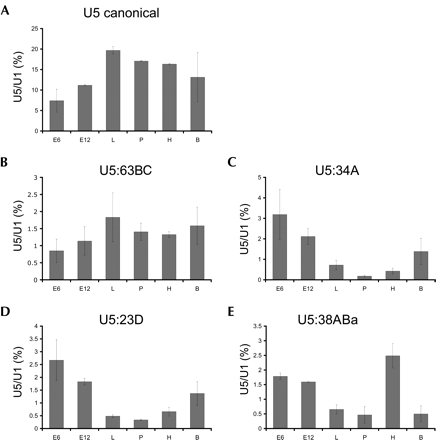
RNA blot hybridization verified expression of all variants except for U5 38ABb, though at apparently lower abundance than the canonical U5 snRNA. Variant U5 38ABa was detected in total RNA isolated from Drosophila heads but not bodies, suggesting that there may be tissue-specific expression of some of these U5 snRNA isoforms. To explore this idea further, RNA blot hybridization was performed on total RNA isolated from Drosophila harvested at discrete developmental stages including embryos, larvae, pupae, and adults, as well as from the commonly used Drosophila S2 and Kc cell lines. Expression levels were normalized by comparison to those for U1 snRNA in each sample; U1 snRNA levels remained relatively constant in all developmental stages and tissues tested (data not shown). In this analysis, the canonical U5 snRNA was found to have the highest level of expression, with the other well-expressed variants ~10-fold lower. The canonical U5 snRNA has a distinct developmental expression profile, with the lowest level of expression in 6-h embryos (Fig. 2A ▶). This level increases by about twofold in the larval state, at which point expression levels off. Variant U5 snRNA 63BC shares a similar profile (Fig. 2B ▶), with a twofold increase in expression between 6-h embryos and larvae and then leveling off. In contrast, U5 variant RNA species 34A, 23D, and 38ABa (Fig. 2C–E ▶) have the reverse expression pattern, with decreasing levels of expression throughout the developmental stages tested. Most remarkable is U5 variant 34A, which steadily decreases ~17-fold from 6-h embryos to the pupal stage, before rising again in adult flies. U5 variant 23D shares a similar profile in that it shows high expression levels in early embryos that decrease in later stages, but the decrease is smaller with only an eightfold difference. Finally, U5 variant U5 38ABa shares the overall pattern of decreasing expression throughout development. Unlike the other U5 variants, however, U5 38ABa exhibits a unique expression profile in adults that shows an approximately sixfold higher RNA level in heads versus bodies; in contrast, both U5 23D and U5 34A have higher expression in bodies relative to heads. U5 variants 14B, 35D, and 38ABb have lower expression levels than the other U5 isoforms examined, which made quantification of the transcripts difficult. However, all variants, including 38ABb, show low-level expression at many of the developmental stages, which was not observed in the initial head and body RNA blot hybridization (data not shown).
FIGURE 2.
Developmental profiles of U5 snRNAs. RNA blot analysis was performed using α-32P-dATP-labeled anti-sense probes complementary to the variable 3′ end of each U5 RNA; a U1 snRNA probe was used as a loading control. For each U5 snRNA, signal is quantified as the percent of U5 snRNA detected relative to U1 snRNA; four 20-min washes, including a final high-stringency 0.1X SSC wash, removed partially hybridized probes to avoid cross-reactivity of probes among the variant U5 species. RNA was prepared from the following sources: E6, 6-h-old embryos; E12, 12-h-old embryos; L, third instar larvae; P, pupae; H, adult heads; B, adult bodies. (A) Developmental profile for the canonical U5 snRNA, (B) U5:63BC, (C) U5:34A, (D) U5:23D, and (E) U5:38ABa.
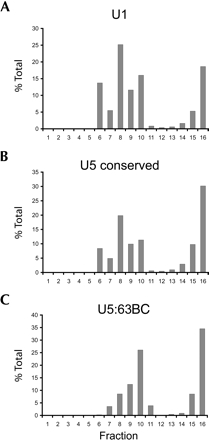
In cell nuclei, U5 snRNA is incorporated into snRNP particles composed of seven Sm proteins as well as numerous U5-specific proteins (for reviews, see Will et al. 1993; Will and Luhrmann 2001; Turner et al. 2004). If the U5 variants identified in Drosophila are involved in splicing, they should likewise be incorporated into similar large snRNP complexes. To determine whether this is the case, Kc nuclear extract was fractionated by glycerol gradient sedimentation, and fractions were probed for the presence of various U5-like RNA sequences by RNA blot hybridization. Probes for U1 snRNA and the conserved region of U5 snRNA, used as controls, hybridize to material in fractions 6–10 as well as near the bottom of the gradient (Fig. 3A,B ▶). A probe targeting the unique region of U5 snRNA variant 63BC showed that like U1 and bulk U5 snRNAs, U5 63BC is found primarily in fractions 7–10 and 15–16 (Fig. 3C ▶). RNA blot hybridization was also performed on U5 variants 34A, 23D, and 38ABa (data not shown). For variant 34A, signal was only seen in fractions nine and 10. The other two variants had no detectable signal in any fractions, perhaps due to low inherent expression levels.
FIGURE 3.
Glycerol gradient analyses of U5 snRNAs. Glycerol gradient sedimentation of Drosophila nuclear extract was performed on nuclear extract from Kc cells using 10%–30% linear glycerol gradients as previously described (Makarova et al. 2002). Fractions were analyzed by RNA blot hybridization using probes complementary to U1 snRNA (A), the conserved 5′ region of all U5 snRNAs (B), and U5 63BC (C). The Y-axis is the percent signal observed in each fraction over the sum of signal detected in all fractions.
The differences in expression levels of the U5 snRNA variants at distinct developmental stages, and their incorporation into complexes with density similar to those of canonical U1 and U5 snRNP particles, suggests possible roles in stage-specific pre-mRNA splicing events. One possibility is that these variants contribute to splicing by the minor spliceosome, which catalyzes splicing of the subset of introns that contain AT/AC splice junctions (Wu and Krainer 1999; Patel and Steitz 2003). Variants of the U1, U2, U4, and U6 snRNAs have been identified in the minor spliceosome, but U5 has been assumed to be the same in both the minor and major spliceosomal complexes. Interestingly, purification and analysis of the U5 snRNP by mass spectrometry showed that the particle undergoes a dynamic rearrangement of proteins during assembly of active spliceosomes; the fate of the RNA within the U5 snRNP was not investigated (Makarov et al. 2002).
Another possibility is that the U5 snRNA variants are involved in alternative splicing events. All known U5 snRNA sequences share nine phylogenetically conserved nucleotides within an 11-nt region in the 5′ hairpin secondary structure of the RNA (Frank et al. 1994; see Fig. 1 ▶). This invariant sequence was shown by cross-linking analysis to contact the 5′ splice site of a pre-mRNA prior to the first transesterification reaction (Wyatt et al. 1992). After cleavage at the 5′ exon–intron junction, the U5 snRNA invariant stem loop is thought to help align the 5′ and 3′ splice sites by direct base pairing prior to the exon-joining step (Newman and Norman 1992; Wyatt et al. 1992; Cortes et al. 1993; Sontheimer and Steitz 1993; Newman et al. 1995). The variant U5s are expected to be fully capable of these interactions due to conservation of the 5′ sequence, but their 3′ sequence differences relative to each other and the “major” U5 snRNA could lead to distinct protein composition of variant U5 snRNP particles. This might enable interaction with alternative splicing factors during assembly of spliceosomes on target pre-mRNAs, potentially allowing recognition of weak or noncanonical splice sites (Newman and Norman 1991). The observation that one of the U5 snRNA variants (U5 38ABa) is significantly more abundant in adult fly heads versus bodies suggests a possible role of this RNA in neuronal gene regulation.
Notably, alternative isoforms of U5 snRNA have been found previously in human cells, where they are incorporated into spliceosomal particles (Sontheimer and Steitz 1992). Differences in abundance of these U5 variants in different human cell lines suggested a possible role in alternative splicing that may be conserved among higher eukaryotes. Furthermore, the spliced leader (SL1 and SL2) RNAs of Caenorhabditis elegans have a secondary structure similar to that of U5 snRNA, and mutations in the 3′ sequence of SL2 alter or prevent its ability to function in trans-splicing (Evans and Blumenthal 2000). It is intriguing to speculate that the variant forms of U5 found in organisms that do not carry out trans-splicing represent a distantly related mechanism for recognition and splicing of noncanonical splice sites in pre-mRNAs.
Acknowledgments
We thank K. Wan and J. Carlson of the Berkeley Drosophila Genome Project for sequencing and data processing, respectively. This work was supported by NIH grants R01HG002673 (S.E.C.) and R01GM61987 (D.R.) and by the Howard Hughes Medical Institute.
Article and publication are at http://www.rnajournal.org/cgi/doi/10.1261/rna.2141505.
REFERENCES
- Adams, M.D., Celniker, S.E., Holt, R.A., Evans, C.A., Gocayne, J.D., Amanatides, P.G., Scherer, S.E., Li, P.W., Hoskins, R.A., Galle, R.F., et al. 2000. The genome sequence of Drosophila melanogaster. Science 287: 2185–2195. [DOI] [PubMed] [Google Scholar]
- Bachellerie, J.P., Cavaille, J., and Huttenhofer, A. 2002. The expanding snoRNA world. Biochimie 84: 775–790. [DOI] [PubMed] [Google Scholar]
- Black, D.L. 2000. Protein diversity from alternative splicing: A challenge for bioinformatics and post-genome biology. Cell 103: 367– 370. [DOI] [PubMed] [Google Scholar]
- Branlant, C., Krol, A., Lazar, E., Haendler, B., Jacob, M., Galego-Dias, L., and Pousada, C. 1983. High evolutionary conservation of the secondary structure and of certain nucleotide sequences of U5 RNA. Nucleic Acids Res. 11: 8359–8367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins, C.A. and Guthrie, C. 2000. The question remains: Is the spliceosome a ribozyme? Nat. Struct. Biol. 7: 850–854. [DOI] [PubMed] [Google Scholar]
- Cortes, J.J., Sontheimer, E.J., Seiwert, S.D., and Steitz, J.A. 1993. Mutations in the conserved loop of human U5 snRNA generate use of novel cryptic 5′ splice sites in vivo. EMBO J. 12: 5181– 5189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans, D. and Blumenthal, T. 2000. trans splicing of polycistronic Caenorhabditis elegans pre-mRNAs: Analysis of the SL2 RNA. Mol. Cell. Biol. 20: 6659–6667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank, D.N., Roiha, H., and Guthrie, C. 1994. Architecture of the U5 small nuclear RNA. Mol. Cell. Biol. 14: 2180–2190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graveley, B.R. 2001. Alternative splicing: Increasing diversity in the proteomic world. Trends Genet. 17: 100–107. [DOI] [PubMed] [Google Scholar]
- Kiss, T. 2002. Small nucleolar RNAs: An abundant group of noncoding RNAs with diverse cellular functions. Cell 109: 145–148. [DOI] [PubMed] [Google Scholar]
- Kramer, A. 1996. The structure and function of proteins involved in mammalian pre-mRNA splicing. Annu. Rev. Biochem. 65: 367–409. [DOI] [PubMed] [Google Scholar]
- Lowe, T.M. and Eddy, S.R. 1999. A computational screen for methylation guide snoRNAs in yeast. Science 283: 1168–1171. [DOI] [PubMed] [Google Scholar]
- Makarov, E.M., Makarova, O.V., Urlaub, H., Gentzel, M., Will, C.L., Wilm, M., and Luhrmann, R. 2002. Small nuclear ribonucleoprotein remodeling during catalytic activation of the spliceosome. Science 298: 2205–2208. [DOI] [PubMed] [Google Scholar]
- Makarova, O.V., Makarov, E.M., Liu, S., Vornlocher, H.P., and Luhrmann, R. 2002. Protein 61K, encoded by a gene (PRPF31) linked to autosomal dominant retinitis pigmentosa, is required for U4/U6*U5 tri-snRNP formation and pre-mRNA splicing. EMBO J. 21: 1148–1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misra, S., Crosby, M.A., Mungall, C.J., Matthews, B.B., Campbell, K.S., Hradecky, P., Huang, Y., Kaminker, J.S., Millburn, G.H., Prochnik, S.E., et al. 2002. Annotation of the Drosophila melanogaster euchromatic genome: A systematic review. Genome Biol. 3: http://genomebiology.com. [DOI] [PMC free article] [PubMed]
- Myslinski, E., Branlant, C., Wieben, E.D., and Pederson, T. 1984. The small nuclear RNAs of Drosophila. J. Mol. Biol. 180: 927–945. [DOI] [PubMed] [Google Scholar]
- Newman, A. and Norman, C. 1991. Mutations in yeast U5 snRNA alter the specificity of 5′ splice-site cleavage. Cell 65: 115–123. [DOI] [PubMed] [Google Scholar]
- ———. 1992. U5 snRNA interacts with exon sequences at 5′ and 3′ splice sites. Cell 68: 743–754. [DOI] [PubMed] [Google Scholar]
- Newman, A.J., Teigelkamp, S., and Beggs, J.D. 1995. snRNA interactions at 5′ and 3′ splice sites monitored by photoactivated cross-linking in yeast spliceosomes. RNA 1: 968–980. [PMC free article] [PubMed] [Google Scholar]
- Omer, A.D., Lowe, T.M., Russell, A.G., Ebhardt, H., Eddy, S.R., and Dennis, P.P. 2000. Homologs of small nucleolar RNAs in Archaea. Science 288: 517–522. [DOI] [PubMed] [Google Scholar]
- Patel, A.A. and Steitz, J.A. 2003. Splicing double: Insights from the second spliceosome. Nat. Rev. Mol. Cell. Biol. 4: 960–970. [DOI] [PubMed] [Google Scholar]
- Riedel, N., Wolin, S., and Guthrie, C. 1987. A subset of yeast snRNAs contains functional binding sites for the highly conserved Sm antigen. Science 235: 328–331. [DOI] [PubMed] [Google Scholar]
- Sontheimer, E.J. and Steitz, J.A. 1992. Three novel functional variants of human U5 small nuclear RNA. Mol. Cell. Biol. 12: 734–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ———. 1993. The U5 and U6 small nuclear RNAs as active site components of the spliceosome. Science 262: 1989–1996. [DOI] [PubMed] [Google Scholar]
- Staley, J.P. and Guthrie, C. 1998. Mechanical devices of the spliceosome: Motors, clocks, springs, and things. Cell 92: 315–326. [DOI] [PubMed] [Google Scholar]
- Tupy, J.L., Bailey, A.M., Dailey, G., Evans-Holm, M., Siebel, C.W., Misra, S., Celniker, S.E., and Rubin, G.M. 2005. Identification of putative noncoding polyadenylated transcripts in Drosophila melanogaster. Proc. Natl. Acad. Sci. 102: 5495–5500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner, I.A., Norman, C.M., Churcher, M.J., and Newman, A.J. 2004. Roles of the U5 snRNP in spliceosome dynamics and catalysis. Biochem. Soc. Trans. 32: 928–931. [DOI] [PubMed] [Google Scholar]
- Villa, T., Pleiss, J.A., and Guthrie, C. 2002. Spliceosomal snRNAs: Mg(2+)-dependent chemistry at the catalytic core? Cell 109: 149–152. [DOI] [PubMed] [Google Scholar]
- Will, C.L. and Luhrmann, R. 2001. Spliceosomal UsnRNP biogenesis, structure and function. Curr. Opin. Cell Biol. 13: 290–301. [DOI] [PubMed] [Google Scholar]
- Will, C.L., Behrens, S.E., and Luhrmann, R. 1993. Protein composition of mammalian spliceosomal snRNPs. Mol. Biol. Rep. 18: 121–126. [DOI] [PubMed] [Google Scholar]
- Wooley, J.C., Cone, R.D., Tartof, D., and Chung, S.Y. 1982. Small nuclear ribonucleoprotein complexes of Drosophila melanogaster. Proc. Natl. Acad. Sci. 79: 6762–6766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, Q. and Krainer, A.R. 1999. AT–AC pre-mRNA splicing mechanisms and conservation of minor introns in voltage-gated ion channel genes. Mol. Cell. Biol. 19: 3225–3236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyatt, J.R., Sontheimer, E.J., and Steitz, J.A. 1992. Site-specific cross-linking of mammalian U5 snRNP to the 5′ splice site before the first step of pre-mRNA splicing. Genes & Dev. 6: 2542–2553. [DOI] [PubMed] [Google Scholar]
- Yuan, G., Klambt, C., Bachellerie, J.P., Brosius, J., and Huttenhofer, A. 2003. RNomics in Drosophila melanogaster: Identification of 66 candidates for novel non-messenger RNAs. Nucleic Acids Res. 31: 2495–2507. [DOI] [PMC free article] [PubMed] [Google Scholar]