Abstract
Most of the 400 RNA editing sites in flowering plant mitochondria are found in mRNAs. Consequently, the sequence vicinities of homologous sites are highly conserved between different species and are presumably recognized by likewise conserved trans-factors. To investigate the evolutionary adaptation to sequence variation, we have now analyzed the recognition elements of an editing site with divergent upstream sequences in the two species pea and cauliflower. This variation is tolerated at the site selected, because the upstream cis-elements reach into the 5′-UTR of the mRNA. To compare cis-recognition features in pea and cauliflower mitochondria, we developed a new in vitro RNA editing system for cauliflower. In vitro editing assays with deleted and mutated template RNAs show that the major recognition elements for both species are located within the conserved sequence. In cauliflower, however, the essential upstream nucleotides extend further upstream than they do in pea. In-depth analysis of single-nucleotide mutations reveals critical spacing of the editing site and the specific recognition elements, and shows that the +1 nucleotide identity is important in cauliflower, but not in pea.
Keywords: RNA editing, plant mitochondria, cauliflower in vitro editing, atp9
INTRODUCTION
In the 15 years since RNA editing was first recognized in plant mitochondria and chloroplast as a post-transcriptional process that alters mostly C-to-U nucleotide identities in mRNAs and tRNAs, progress toward elucidating the enzymes and the specificity recognition has been restricted mostly by the lack of efficient in vitro systems. In vivo analysis of transgenic chloroplasts has brought important insights into the structure and extension of cis-elements, but this approach is difficult to extent toward a biochemical characterization and the identification of the corresponding trans-factors (Chaudhuri et al. 1995; Bock et al. 1996, 1997; Chaudhuri and Maliga 1996; Reed et al. 2001; Chateigner-Boutin and Hanson 2002).
With the development of reliable in vitro activities for chloroplasts (Hirose and Sugiura 2001; Miyamoto et al. 2002, 2004) and also for pea mitochondria (Takenaka and Brennicke 2003; Takenaka et al. 2004) in the past few years, characterization of the cis-requirements at individual sites has accelerated considerably.
For plant mitochondria, the in vitro RNA editing system from the pea has shown that for recognition by the RNA editing activity, only ~20 nucleotides are essential upstream, 40 are optimal, and basically none is necessary downstream of the first editing site in the atp9 mRNA. Analysis of the cis-requirements by targeted mutations of the template and competition experiments have narrowed the sequence requirements for the site specificity to the region 5–20 nucleotides upstream of this site in the atp9 mRNA (Takenaka et al. 2004).
Transfections of isolated wheat mitochondria with cox2 mRNA (Farré and Araya 2001; Farré et al. 2001; Staudinger and Kempken 2003) and mutational analysis of two sites in this transcript showed that similarly 16–20 nucleotides upstream are required to define these sites (Choury et al. 2004). However, in addition to these upstream elements, one or more nucleotide positions downstream were found to be crucial for efficient editing. These sequence requirements suggest that individual recognition elements vary between different editing sites. Extensive mutational analysis revealed that the important nucleotide identities are different for the two sites, confirming the variation of cis-elements between individual RNA editing sites (Choury et al. 2004).
These two experimental approaches with a dicot, the pea, and a monocot, wheat, thus suggest that between plants the recognition parameters of RNA editing sites are similar but can vary between individual sites. To investigate this inference in more detail, we have now assayed the cis-elements determining a given RNA editing site in two different plant species, cauliflower and pea, and compared the recognition parameters. Furthermore, we analyze the spacing requirements of the recognition elements and the edited nucleotides by specific insertion/deletion mutations.
RESULTS
The cauliflower in vitro mitochondrial extract
For a convenient source for mitochondria, we selected cauliflower inflorescences, since these offer several advantages to obtain large amounts of comparatively clean mitochondria from plants. These tissues contain few secondary plant compounds, which notoriously make biochemical and molecular analyses difficult. Furthermore, no chloroplasts differentiate in these pale white tissues, and few proplastids contaminate purification schemes of other organelles. Yet a third advantage is the economic source of material. Last not least, cauliflower (Brassica oleracea) is closely related to Arabidopsis thaliana, the model plant for which the complete genomic sequence and countless other data are readily available.
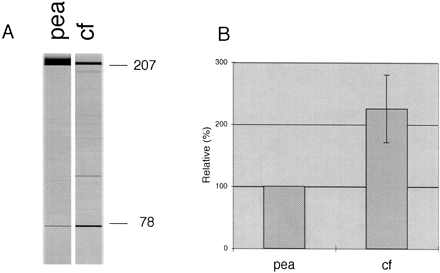
As detailed in the Materials and Methods section, we prepared an S-60 lysate from mitochondria purified from cauliflower inflorescences along the procedure adapted from the original protocol for tobacco chloroplasts (Hirose and Sugiura 2001). The cauliflower mitochondrial extract proved to be more active than the previously developed pea in vitro system from elongated hypocotyls (Fig. 1; Takenaka and Brennicke 2003). On average, 4%–7% of the template molecules are edited by the cauliflower system in comparison to 1.5%–3% in the pea lysate (Fig. 1). This may be due to the higher concentration of mitochondrial proteins consistently achieved with lysates from the cauliflower inflorescences (8.3 μg/μL on average) than in the pea (Pisum sativum), with ~1.9 μg/μL. The relationship is not linear, since the about five times higher protein content only yields a twofold increase in the editing activity.
FIGURE 1.
Comparison of the RNA editing lysates prepared from cauliflower and pea mitochondria. (A) Sample gel images of in vitro editing assays in the pea and cauliflower lysates. Numbers give the sizes of the full-length (207 nucleotides) and edited (78 nucleotides) RT-PCR-TDG products. (B) RNA editing activities in the two lysates as determined toward the pea atp9 −40/+49 template. The average of three experiments is given with the respective standard error shown for the cauliflower activity (cf) relative to the pea, the latter taken as 100% in each set of experiments.
The cauliflower and pea mitochondrial atp9 sequences diverge beyond 23 nucleotides upstream of the first RNA editing site
A disadvantage of any new system is the need to identify native sequences and confirm experimentally the presence of postulated RNA editing sites. This we did for the complete atp9 gene in cauliflower and determined the RNA editing sites by genomic and cDNA analysis (data not shown). The complete sequence information and editing sites have been deposited in the European Molecular Biology Laboratory (EMBL)/GenBank databases (accession no. DQ102391). The sequence comparison with the previously analyzed pea atp9 template (Takenaka et al. 2004) shows that the first RNA editing site in the open reading frame is conserved between these two species in the seventh codon from the pea AUG (Fig. 2). Upstream of this editing site, sequences diverge beyond nucleotide position −23 relative to the edited nucleotide. This sequence variation is possible because the editing site is close to the conserved AUG and the 5′-leader is not conserved between the two plants. Coding sequences in plant mitochondria are usually highly conserved between different species and thus do not allow much variation. Therefore, only an editing site located at the 5′-extremity of an open reading frame will display such natural sequence divergence in its upstream region and offers a choice of variable genuine wild-type templates from different plant species.
FIGURE 2.
Sequence comparison between pea (Pisum sativum) and cauliflower (Brassica oleracea) mitochondria surrounding the first RNA editing site in the atp9 coding region. Numbering is centered around this editing site. The native cauliflower and pea sequences deviate upstream of nucleotide −23, which is located in the 5′-untranslated leader region. This sequence variation between the two species allows a cross-wise comparison of the nucleotides necessary for site recognition. The AUG of the translational start in pea is boxed, the edited C is in large type, and identical nucleotides are indicated by the dashes between the two sequences. In cauliflower another AUG occurs in frame further upstream, which could theoretically be used (data not shown). The sequence between nucleotides +10 and +45 is not shown but is identical in the two plants. The complete sequence of the atp9 gene in cauliflower is deposited in the databases (accession no. DQ102391).
Both cauliflower and pea templates are recognized in the cauliflower lysate
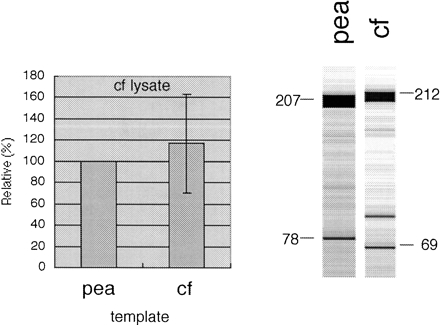
Despite this sequence variation, both the homologous cauliflower and the heterologous pea templates are correctly recognized and edited in the mitochondrial lysate from cauliflower (Fig. 3). Little difference is seen in the efficiencies of the in vitro modification between the homologous and heterologous templates. This result suggests that for cauliflower, all essential upstream cis-recognition elements are contained within the 23 nucleotides conserved between both templates, with the potential participation of the scattered further upstream positions identical between pea and cauliflower (Fig. 2).
FIGURE 3.
Pea and cauliflower templates are processed with similar efficiency in the cauliflower lysate. This observation suggests that in both plant species, the cis-elements for recognition of this editing site reside within the conserved 23 nucleotides (and possibly the few positions conserved further upstream). The cauliflower lysate activity appears to be slightly higher toward its cognate sequence, but the differences between the two templates are within the experimental variation. The average of four independent assays is shown with the activity toward the pea template taken as 100% in each assay. On the right, gel images are shown for representative assays of pea and cauliflower (cf) templates. Different cloning sites result in different respective TDG product sizes in pea (78) and cauliflower (69).
Deletion templates show that the essential cis-recognition elements for cauliflower are located within the upstream 20 nucleotides, but efficient editing also requires specific downstream nucleotide(s)
To investigate the evolutionary conservation of cis-elements, and by extrapolation the trans-elements recognizing an editing site, we employed the pea mitochondrial atp9 sequence in all of the following experiments, since this has been extensively examined in the native pea mitochondrial in vitro system (Takenaka et al. 2004).
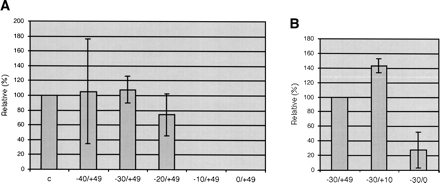
Successive deletions of the template sequences in steps of 10 nucleotides show that a template RNA with 20 upstream native nucleotides is correctly recognized (Fig. 4A). Further deletions up to −10 or right up to the edited nucleotide do not allow editing any more, suggesting that the 20 nucleotides directly upstream of the edited nucleotide are necessary as well as sufficient for recognition of the site.
FIGURE 4.
Delineation of the cis-recognition region by deletion clones in the cauliflower mitochondrial lysate. The sequences upstream and downstream of the monitored editing site were deleted in steps of 10 nucleotides; that is, they were in effect substituted by bacterial vector sequences. (A) Deletion of the 5′-sequence up to 20 upstream nucleotides (−20) still allows editing. Further deletion closer than −20 nucleotides from the editing site completely abolishes recognition of this editing site. Control was the pea template −40/+10; the deletions were all done+with the pea template to allow a direct comparison of the cis-elements. (B) Deleting the 3′-region has little effect up to +10 nucleotides, but the complete substitution up to the edited nucleotide (−30/0) shows substantial inhibition of the in vitro editing activity. The longest 3′-extension was used as standard, and the relative activities with the 3′-deleted templates are shown.
Conversely, substitution of all the downstream nucleotides by bacterial sequences still allows editing in vitro (Fig. 4B), but much of the activity is lost upon alteration of the 10 nucleotides immediately following the editing site. The lower editing activity in the complete 3′-substitution right up to the editing site is probably due to the effect of the identity of the adjacent nucleotide, which was investigated in detail through single nucleotide mutations (see below).
Scanning mutations and competitions suggest upstream and downstream extensions of the cis-recognition element in cauliflower versus pea
To investigate the necessary cis-sequences for recognition in more detail, the region surrounding the target editing site was mutagenized in steps of five nucleotides, which were exchanged for their respective complementary nucleotides. In the direct comparison of the relative activities of these templates in pea and cauliflower mitochondrial lysates (Fig. 5A), an overall similar pattern emerges, which corresponds to and extends the results of the deletion analysis.
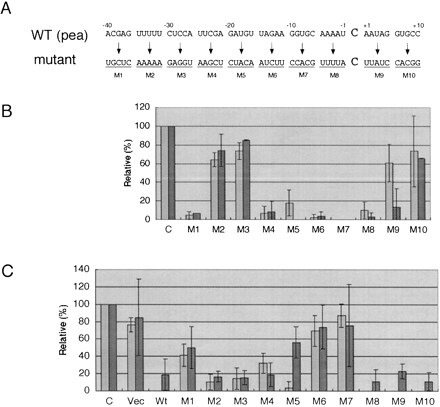
FIGURE 5.
The effects of scanning mutations around the first atp9 editing site as substrates and as competitors on in vitro RNA editing in pea and cauliflower mitochondrial lysates are compared. (A) The respective nucleotide quintet altered to its complementary sequence in each set of experiments for maximum effect and to maintain the G+C content is shown, and its designation is given beneath the mutated sequences. (B) The mutated pea templates are tested for their effectiveness in cauliflower (dark bars on the right) and pea (light bars on the left). Notable differences between the two species are observed toward mutants M5 and M9. (C) The wild-type pea template is competed with 1500-fold excess of the mutants M1–M10 from part A in the cauliflower lysate (dark bars; cf) and 1000-fold in the pea lysate assays, respectively (light bars; pea). Control template is the pea −40/+49 wild-type sequence without competitor. Vector sequences compete little, but the wild-type competitor suppresses recognition of the template completely in the pea lysate. Please note that this suppression is not complete in the more active cauliflower lysate, even though a 50% higher excess of competitor was used. The most striking difference between the lysates from these two plant species is seen with competitor M5. Further details are discussed in the text.
Surprisingly the upstream enhancer element between nucleotides −41 and −35 of the pea template active in its native in vitro lysate (Takenaka et al. 2004) appears to serve a similar positive function in the cauliflower lysate, because its alteration in construct M1 effects a similar decrease in the editing activity in both species. This is particularly intriguing, since in the native cauliflower mitochondrial atp9 sequence this element is not present (Fig. 2).
A second difference between the two plant species is the importance of the nucleotide identities between −21 and −15: While the pea lysate still shows ~20% activity with this mutant M5, the cauliflower lysate does not accept this modified template at all. This observation suggests that in cauliflower mitochondria, the major cis-recognition element extends further into the region upstream of nucleotide −15 than in pea mitochondria.
A third difference is observed with mutant M9 altered just downstream of the editing site between nucleotides 0 and +6: While this template loses little of its activity in the pea lysate, the cauliflower extract has difficulties recognizing this sequence at all and shows hardly any activity.
Competition of the wild-type pea template with itself and the various mutants reveals the higher capacity for RNA editing of the cauliflower lysate: Even with the wild-type sequence as competitor in 1000-fold excess, inhibition is not complete, but ~20% of residual editing activity remains (Fig. 5B). Therefore, all experiments with the mutated competitors must be accordingly interpreted; for example, the residual activities seen in the cauliflower lysate with the competing RNAs mutated between −6 and 0 (M8), 0 and +6 (M9), and +5 and +11 (M10) are comparable to the level of inhibition by the wild-type sequence. Their effects are thus identical to the observed effect in the pea lysate.
The differences between pea and cauliflower in the extent of the respective major cis-elements observed with the mutated templates are also reflected in the effects of the respective competitors: While in the pea the −20 to −16 mutant M5 as a competitor has a strong effect of ~90% reduction, in cauliflower only 40%–50% inhibition is seen. This difference reflects the greater extent of the essential cis-region, which, if covering a considerable part of these five nucleotides in the cauliflower, should inhibit less when mutated.
The absolute requirement of the nucleotides between five and 15 residues upstream of the editing site is confirmed by the absence of any significant effect on editing when the respectively mutated competitors are incubated with the reaction. The reduction observed is similar to the effect of added vector sequences in the control experiment (Fig. 5B). The relationship of the pea enhancer sequence between −41 and −35 to the major recognition element is similar in pea and cauliflower also in these competition experiments, their effect being considerably less than that of the next downstream mutants, which inhibit more strongly in both plants.
Mutants in the 3′ region of the editing site fully compete with the wild-type template in lysates from both plant species, showing that the upstream sequences are essential and limiting for RNA editing site recognition.
Point mutations reveal the importance of the +1 nucleotide in cauliflower
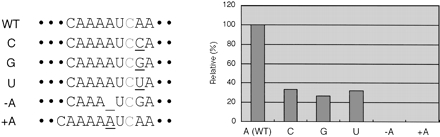
Since the exchange of the sequence downstream of the edited nucleotide by a bacterial sequence has a profound effect on the editing activity of the cauliflower lysate (Fig. 4B), we investigated the importance of the +1 nucleotide by mutating it through all four nucleotides (Fig. 6A). The effect of changing the wild-type adenosine to the other purine guanosine is similar to the effect of changing this nucleotide to any of the pyrimidines, showing that the adenosine identity is crucial at this position. This experiment thus focuses the effects of the respective substituted deletion template (Fig. 4) and the mutated pentanucleotide (Fig. 5) to this nucleotide position, which is changed in the former to a G and in the latter to a U.
FIGURE 6.
Importance of individual nucleotide alterations around the editing site. The influence of the identity of the first nucleotide downstream of the edited nucleotide was determined by mutating this position through all three alternative nucleotides. Any of these changes results in the loss of ~70% of the editing activity. All mutants were tested in four separate experiments, and the mean percentages of the wild-type editing activity were determined. The resulting standard error is indicated for each mutant. The tolerance of the in vitro editing activity for distance alterations between the upstream cis-recognition element and the edited nucleotide was investigated by deleting or inserting an adenosine nucleotide within the run of four As in the wild-type sequence. No activity was observed with either template, showing that the distance from the cis-element is crucial. Please note that in the deletion template a concomitant A-to-G change at the +1 position has occurred.
Single nucleotide insertion/deletion shows a low tolerance toward the spacing between the cis-element and the edited nucleotide
To examine the importance of the spacing between the major cis-recognition element covering nucleotides −20 and −5 to the editing site, we altered its distance to the edited C by one nucleotide in each direction (Fig. 6B). Both alterations, insertion or deletion of one nucleotide, completely block RNA editing in vitro. This result suggests zero tolerance for the editing complex presumably assembled at the conserved cis-elements to reach the nucleotide to be edited. The wild-type configuration clearly represents the only allowed distance between the cis-recognition region and this editing site.
DISCUSSION
RNA editing sites can vary between individual plant species. One species may require a given C-to-U alteration to specify a conserved and presumably functional open reading frame, but this same site can be genomically encoded as a T in another species and thus may not require editing. Particularly, third codon positions, which do not alter the amino acid specified, may be edited in one plant but may remain an unedited C nucleotide in another species. The editing specificities thus appear to be quite variable between different plants, suggesting a certain adaptive flexibility within rather short evolutionary distances. By extrapolation, the trans-factors recognizing a novel or altered editing site should have changed and adapted to (or away from) the concomitant novel or altered cis-elements.
To gain further information and insight about this dynamic potential for creating or altering specificities, we have compared the cis-elements at a homologous editing site in the atp9 mRNA in two different species of flowering plants, the pea and the cauliflower. We have chosen this site because the upstream sequences rapidly diverge between the two plants just upstream of the beginning of the respective coding regions, which is 23 nucleotides upstream of this editing site (Fig. 2).
The cis-recognition elements for an RNA editing site can vary between plant species
Deletion, substitutional mutation, and competition experiments suggest that the specific core nucleotide sequence necessary to address this first editing site in the atp9 mRNA in plant mitochondria is slightly different in the cauliflower in vitro system in comparison to the pea. The 5′-requirement of the core-recognition region (in pea, −15 to −5) extends in cauliflower further 5′ into the −20 to −15 region. The 3′-side of this core element, which is necessary and sufficient to specify this editing site in the pea, appears to be similarly delineated in both species around five nucleotides upstream of the edited nucleotide.
In cauliflower, however, effective in vitro editing depends furthermore on the identity of the nucleotide immediately downstream of the edited C nucleotide. This is different in the pea, where the identity of the adjacent nucleotide at +1 does not influence the editing efficiency. These results thus suggest that the cis-elements have evolved between the two plant species, and it thus can be assumed that also the respective trans-elements have changed and presently differ between the pea and the cauliflower. Nevertheless, the basic mode of editing site specification has of necessity been conserved in evolution between these two flowering plants, since both definitely require this C-to-U alteration for a functional mitochondrial ATPase (Hernould et al. 1993; Zabaleta et al. 1996).
The enhancer region of the pea is absent in cauliflower
Compensating adaptative mutation of the trans-factors may have been influenced by the nucleotide sequence changes further upstream. Here, in the region between −40 and −35, the pea template contains an enhancer element (Takenaka et al. 2004), which increases the in vitro editing activity in the homologous system. The cauliflower sequence, however, is—beyond the usual chance similarities—completely different in this region.
Comparing the in vitro editing activities of the cauliflower lysate toward the pea and the cauliflower templates, respectively, shows that both are recognized with about equal affinities (Fig. 1). Even without the pea enhancer sequence in the cauliflower template, the in vitro editing complex in cauliflower thus appears to concentrate sufficient activity at this site for efficient editing. This is confirmed by the in vivo situation, since in the analysis of the steady-state cDNA from cauliflower mitochondria, only molecules derived from mRNAs fully edited at this site are observed (data not shown).
The extension of the recognized core-region itself in cauliflower does not appear to compensate for the altered upstream sequence, since deletion of the pea enhancer element diminishes editing in this template (Fig. 4A). Likewise, the altered binding right at and around the edited nucleotide does not seem to be sufficient to enhance assembly of the trans-factors and to ensure very efficient editing. Although the identity of nucleotide +1 seems to play a crucial role for editing and the enhanced binding of the A nucleotide seems to consequently enhance attachment and assembly of the trans-factors, these parameters are all present in the template in which the 5′ pea enhancer has been deleted and is less efficient in the in vitro system.
The equally enhancing effect of the divergent upstream sequences in pea and in cauliflower is evidenced by the lower in vitro editing activity in the −20 deletion construct in comparison to the full-length templates from either species. Thus the different upstream sequence in cauliflower effectively substitutes the pea enhancer and now attracts a (presumably different) trans-factor able to similarly enhance the binding of the RNA editing complex to achieve rapid and faithful editing. To search for a candidate trans-factor recruited from another function and therefore potentially connecting to a similar sequence elsewhere, the mitochondrial genomes of Arabidopsis and rape seed were scanned with the cauliflower −40/−30 sequence. However, outside of the homologous atp9 upstream region, no overt similarities were detected that could donate an established trans-factor interaction to boost this RNA editing event.
The enhancer region from pea is addressed by a trans-factor conserved in cauliflower
It is surprising to find that deletion of the pea-specific enhancer element at −40/−35 also lowers the in vitro editing efficiency in the cauliflower mitochondrial lysate. This observation suggests that, even though cauliflower mitochondria do not encode this enhancer sequence of pea, they nevertheless still contain the potential to address this sequence motif through conserved trans-factor(s). Conservation of this trans-factor in cauliflower may be required if it has an additional other and essential function at another editing or processing site. A search of the genomic mitochondrial sequences from Arabidopsis and rape with the pea enhancer sequence did not reveal any striking similarity elsewhere in these genomes, but this search is difficult to conduct exhaustingly since the enhancer sequence itself is delimited to only about five nucleotides, too short to specify unique loci in the 350-kb genomes.
An analogous observation of a trans-factor conserved in evolution, although apparently not necessary any more, has been made in chloroplasts (Schmitz-Linneweber et al. 2001): Analysis of the allotetraploid tobacco Nicotiana tabacum has shown that the nuclear genome encodes a trans-factor necessary for recognition of an editing site that is not present in the N. tabacum chloroplast RNA. This chloroplast editing site is, however, found in spinach and in Nicotiana tomentosiformis, the paternal parent of N. tabacum (Schmitz-Linneweber et al. 2001). Again, one explanation could be a requirement of this trans-factor at another RNA processing event. More likely, this, in N. tabacum, superfluous factor just did not (yet) mutate in the relatively short evolutionary time between establishment of the line of N. tabacum from the cross between Nicotiana sylvestris and N. tomentosiformis.
Overall, the identification of RNA editing sites in plant mitochondria appears to be slowly adaptive, changing recognition parameters between different species and maintaining the capacity to recognize various elements. The trans-factors involved seem to act in a concert of several interconnecting proteins (and/or RNA?) that individually and together influence the overall efficiency at a given site. Meeting the challenge to identify and assign these trans-factors will be helped by the here-described novel in vitro system from cauliflower mitochondria.
MATERIALS AND METHODS
Preparation of plant mitochondrial extracts
RNA editing active mitochondrial lysates from pea seedlings (Pisum sativum L., var) were prepared as described (Takenaka and Brennicke 2003). Preparation of cauliflower lysates followed essentially the same protocol. About 2000 g fresh weight inflorescences from two to three heads of cauliflower (Brassica oleracea var. botrytis; purchased at local markets) were cut off from the stems. Cells were disrupted in a juice extractor, and the recovered 1 L of juice was diluted with 3× extraction buffer (3 × : 0.9 M mannitol, 90 mM Na-pyrophosphate, 6 mM EDTA, 2.4% PVP25 [w/v] with, added just before use, 0.9% BSA [w/v], 9 mM cysteine, 15 mM glycine, and 6 mM β-mercaptoethanol; pH 7.5 was adjusted with HCl). Mitochondria were enriched by several steps of differential centrifugation: Cell debris was pelleted by 10-min centrifugations each at increasing g-forces: 1100g, 2100g, and 3500g. Mitochondria were sedimented by 30 min at 11,300g and were resuspended in 1× extraction buffer. Subsequent purification on Percoll gradients was as described for the pea lysate. The mitochondrial fraction was diluted in 500 mL of wash buffer (1×: 0.3 M mannitol, 10 mM K-phosphate at pH 7.5, 1 mM EDTA with, added just before use, 0.1% BSA, 5 mM cysteine, and 15 mM glycine). Mitochondria were pelleted, resuspended in a small volume of wash buffer, and stored frozen. For lysate preparation 400 mg of isolated mitochondria were lysed in 1200 μL of lysis buffer (30 mM HEPES-KOH at pH 7.7, 10 mM Mg-acetate, 2 M KCl, 2 mM DTT, and 0.2% Triton X-100). After 30-min incubation on ice, the lysate was centrifuged at 60,000g for 20 min. The supernatant was recovered, and the KCl was removed by dialysis against 5 × 400 mL dialysis buffer (30 mM HEPES-KOH at pH 7.7, 3 mM Mg-acetate, 45 mM K-acetate, 30 mM ammonium acetate, and 10% glycerol) for a total of 5 h. All steps were carried out at 4° C. The resulting extract (~8 μg protein/μL) was rapidly frozen in liquid nitrogen.
Cloning and RNA substrates
DNA clones of the atp9 coding region and flanking sequences were constructed in an adapted pBluescript SK+ to allow run-off transcription of the editing template RNA as described (Takenaka and Brennicke 2003). Deletion clones were shortened by removing original mitochondrial sequences as indicated in the respective experiments. The outside bacterial anchors for PCR amplification accordingly moved closer into the editing sites. Coincidental nucleotide similarities between these and the substituted mitochondrial sequences were taken into consideration when evaluating nucleotide requirements for RNA editing. Cauliflower clones for sequence analyses were made from genomic mitochondrial DNA by PCR between primers derived from the respective Arabidopsis thaliana sequences and for cDNA analysis from mitochondrial RNA by RT-PCR, respectively.
In vitro RNA editing reactions
The in vitro RNA editing reactions were performed as described (Takenaka and Brennicke 2003). After incubation, template sequences were amplified by RT-PCR with one of the primers labeled with the Cy5 fluorophor. RNA editing activity was detected by mismatch analysis employing the TDG enzyme activity (thymine DNA glycosylase, Trevigen). The TDG treated fragments were separated, and the Cy5 fluorescence was scanned and displayed with an ALF express DNA sequencer (Amersham).
The efficiency of the in vitro RNA editing reaction was quantified by comparing the areas under the peaks of the cleaved and uncut DNA fragments. The ratio of cleaved (i.e., edited) fragment to uncut DNA was used to determine relative efficiencies of the investigated conditions in each experiment. To allow comparisons and to determine the variation between individual experiments, the ratios of cleaved to uncleaved fragments were displayed as percentages of the standard reaction conditions.
Generation of mutant substrates
The 5′-deletion mutants were constructed by inverted PCR from the cloned atp9 sequences with primers −40, −30, −20, −10, and −0, respectively, on the one side and primer invertion1 on the other. The resulting fragments were digested with EcoRI to generate sticky ends in the primer contained EcoRI recognition site and were self-ligated. The 3′-deletion mutants were constructed by inverted PCR from clone atp9–30 with primer invertion2 and primers +10 and +0, respectively. The PCR fragments were digested with XbaI and self-ligated.
The mutant templates with defined sequence regions exchanged to their opposite sequence were constructed by introducing the respective complement pentanucleotide in primers M1-M10. PCR was performed on deletion clone atp9–40 with primer invertion1 and primers M1, M2, M3, M4, and M5, respectively, and in the second series with primer invertion2 and primers M6, M7, M8, M9, and M10, respectively. The resulting fragments were digested with EcoRI or XbaI, respectively, and self-ligated. The point mutations were introduced by the same procedure using the respectively altered primers. All mutants were confirmed by sequence analysis.
Competition assays
Wild-type competitor RNA was synthesized from the PCR product amplified with primers T7 and +10 from clone atp9–40. An entirely plasmid derived control RNA was synthesized from the PCR product amplified from pBluescriptIISK+ with T7 and SK primers. The mutant competitors were synthesized from the PCR products amplified from clones M1–M5 with the T7 primer and primer +10, and from clones M6, M7, M8, M9, and M10 with T7 and the respective mutant primers. One hundred attomoles of substrate and 1000 times (100 fmol) for pea and 1500 times (150 fmol) for cauliflower competitor RNA were first mixed and then incubated with the mitochondrial in vitro assay as described above.
Acknowledgments
José Gualberto kindly introduced us to the juicer method for purification of mitochondria—many thanks. We thank Dagmar Pruchner for excellent experimental support. The technical help of Carmen Schilling-Kolle is gratefully acknowledged. We also thank Daniil Verbitskyi for many lively and constructive discussions. M.T. is a post-doctoral fellow of the JSPS. J.A.v.d.M. is a Ph.D. fellow of the DAAD. This work was supported by a grant from the DFG and by the Fonds der chemischen Industrie.
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.2740905.
REFERENCES
- Bock, R., Herrmann, M., and Kössel, H. 1996. In vivo dissection of cis-acting determinants for plastid RNA editing. EMBO J. 15: 5052–5059. [PMC free article] [PubMed] [Google Scholar]
- Bock, R., Herrmann, M., and Fuchs, M. 1997. Identification of critical nucleotide positions for plastid RNA editing-site recognition. RNA 3: 1194–1200. [PMC free article] [PubMed] [Google Scholar]
- Chateigner-Boutin, A.-L. and Hanson, M.R. 2002. Cross-competition in transgenic chloroplasts expressing single editing sites reveals shared cis elements. Mol. Cell. Biol. 22: 8448–8456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhuri, S. and Maliga, P. 1996. Sequences directing C to U editing of the plastid psbL mRNA are located within a 22 nucleotide segment spanning the editing site. EMBO J. 15: 5958–5964. [PMC free article] [PubMed] [Google Scholar]
- Chaudhuri, S., Carrer, H., and Maliga, P. 1995. Site-specific factor involved in the editing of the psbL mRNA in tobacco plastid. EMBO J. 14: 25951–25957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choury, D., Farré, J.-C., Jordana, X., and Araya, A. 2004. Different patterns in the recognition of editing sites in plant mitochondria. Nucleic Acids Res. 32: 6397–6406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farré, J.-C. and Araya, A. 2004. Gene expression in isolated plant mitochondria: High fidelity of transcription, splicing and editing of a transgene product in electroporated organelles. Nucleic Acids Res. 29: 2484–2491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farré, J.-C., Leon, G., Jordana, X., and Araya, A. 2001. Cis recognition elements in plant mitochondrion RNA editing. Mol. Cell. Biol. 21: 6731–6737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernould, M., Suharsono, S., Litvak, S., Araya, A., and Mouras, A. 1993. Male-sterility induction in transgenic tobacco plants with an unedited atp9 mitochondrial gene from wheat. Proc. Natl. Acad. Sci. 90: 2370–2374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirose, T. and Sugiura, M. 2001. Involvement of a site-specific trans-acting factor and a common RNA-binding protein in the editing of chloroplast RNA: Development of an in vitro RNA editing system. EMBO J. 20: 1144–1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyamoto, T., Obokata, J., and Sugiura, M. 2002. Recognition of RNA editing sites is directed by unique proteins in chloroplasts: Biochemical identification of cis-acting elements and trans-acting factors involved in RNA editing in tobacco and pea chloroplasts. Mol. Cell. Biol. 22: 6726–6734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ———. 2004. A site-specific factor interacts directly with its cognate RNA editing site in chloroplast transcripts. Proc. Natl. Acad. Sci. 101: 48–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed, M.L., Peeters, N.M., and Hanson, M.R. 2001. A single alteration 20 nt 5′ to an editing target inhibits chloroplast RNA editing in vivo. Nucleic Acids Res. 29: 1507–1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz-Linneweber, C., Tillich, M., Herrmann, R.G., and Maier, R.M. 2001. Heterologous, splicing-dependent RNA editing in chloroplasts: allotetraploidy provides trans-factors. EMBO J. 20: 4874–4883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staudinger, M. and Kempken, F. 2003. Electroporation of isolated higher-plant mitochondria: Transcripts of an introduced cox2 gene, but not an atp6 gene, are edited in organello. Mol. Gen. Genomics 269: 553–561. [DOI] [PubMed] [Google Scholar]
- Takenaka, M. and Brennicke, A. 2003. In vitro RNA editing in pea mitochondria requires NTP or dNTP, suggesting involvement of an RNA helicase. J. Biol. Chem. 278: 47526–47533. [DOI] [PubMed] [Google Scholar]
- Takenaka, M., Neuwirt, J., and Brennicke, A. 2004. Complex cis-elements determine an RNA editing site in pea mitochondria. Nucleic Acids Res. 32: 4137–4144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zabaleta, E., Mouras, A., Hernould, M., Suharsono, S., and Araya, A. 1996. Transgenic male-sterile plant induced by an unedited atp9 gene is restored to fertility by inhibiting its expression with antisense RNA. Proc. Natl. Acad. Sci. 93: 11259–11263. [DOI] [PMC free article] [PubMed] [Google Scholar]