Abstract
Protein biosynthesis requires numerous conformational rearrangements within the ribosome. The structural core of the ribosome is composed of RNA and is therefore dependent on counterions such as magnesium ions for function. Many steps of translation can be compromised or inhibited if the concentration of Mg2+ is too low or too high. Conditions previously used to probe the conformation of the mammalian ribosome in vitro used high Mg2+ concentrations that we find completely inhibit translation in vitro. We have therefore probed the conformation of the small ribosomal subunit in low concentrations of Mg2+ that support translation in vitro and compared it with the conformation of the 40S subunit at high Mg2+ concentrations. In low Mg2+ concentrations, we find significantly more changes in chemical probe accessibility in the 40S subunit due to subunit association or binding of the hepatitis C internal ribosomal entry site (HCV IRES) than had been observed before. These results suggest that the ribosome is more dynamic in its functional state than previously appreciated.
Keywords: ribosome dynamics, translation initiation, protein synthesis, chemical probing
INTRODUCTION
The ribosome is a dynamic molecular machine. In addition to association and dissociation of the two subunits, conformational changes occur within the ribosomal subunits at all stages of translation. The small ribosomal subunit in particular is known to undergo a number of conformational changes during the different stages of translation that are important for guiding mRNA decoding (Schilling-Bartetzko et al. 1992; Pape et al. 2000; Yusupov et al. 2001; Ogle et al. 2002; Stark et al. 2002; Savelsbergh et al. 2003; Valle et al. 2003a), the process of mRNA and tRNA translocation (Frank and Agrawal 2000; Stark et al. 2000; VanLoock et al. 2000; Valle et al. 2003b; Peske et al. 2004; Spahn et al. 2004a), termination (Klaholz et al. 2004; Gao et al. 2005), and possibly translation initiation (Spahn et al. 2001b, 2004b). The ribosome is composed primarily of RNA, and thus its structure and activity are extremely sensitive to the presence of divalent metal ions and polyamines (Zamir et al. 1974; Jelenc and Kurland 1979; Hoa et al. 1980; Sperrazza et al. 1980; Sperrazza and Spremulli 1983; Goss and Harrigan 1986; Moazed et al. 1986; Kozak 1990; Bayfield et al. 2001; Muth et al. 2001). Specific magnesium ion binding sites have been observed throughout the bacterial and archaeal ribosome in several different crystal structures (Wimberly et al. 2000; Klein et al. 2004; Schuwirth et al. 2005), suggesting that the correct folding of ribosomal RNA (rRNA) is dependent on these divalent cations (Sperrazza and Spremulli 1983; Cate et al. 1997).
A number of studies have examined the changes in conformation of rRNA in the ribosome upon binding of various ribosomal substrates and translation factors, as well as during different stages of translation. For example, changes in the chemical accessibility of rRNA nucleotides due to ribosomal subunit association have been measured using purified mammalian ribosomes (Holmberg et al. 1994). The effects of HCV IRES binding to purified 40S ribosomal subunits have also been examined (Otto et al. 2002). However, the magnesium ion dependence of these conformational changes has not been determined. Notably, we found that these experiments were carried out at concentrations of Mg2+ ions that completely inhibit translation in vitro, as determined from the Mg2+ ion concentration dependence of translation from a range of mRNAs. We therefore probed the effects of subunit association and HCV IRES binding on the conformation of the small ribosomal (40S) subunit at concentrations of Mg2+ that support translation. We find many more changes in the chemical accessibility of nucleotides in the 40S subunit than were seen in earlier studies at higher Mg2+ concentrations. These results suggest that the 40S subunit has a large degree of intrinsic flexibility in its functional state that had previously escaped detection.
RESULTS
Magnesium-dependence of translation in vitro
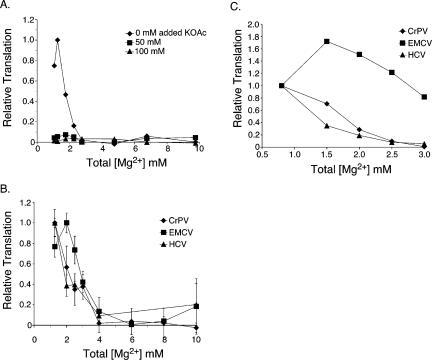
In order to study the effect of Mg2+ concentration on translation, several mRNAs were generated and tested for in vitro translation activity in rabbit reticulocyte lysate (RRL). The RRL used here had been treated to remove endogenous globin RNA (Pelham and Jackson 1976; Jackson and Hunt 1983), so that nearly all translation resulted from the exogenously added mRNA. The first mRNA tested (GLA mRNA) was a capped, polyadenylated mRNA that contained the globin 5′ nontranslated region (5′-NTR) and the luciferase open reading frame (ORF). Three other mRNAs contained the gene for enhanced green fluorescent protein (EGFP) in the ORF, downstream of either the Encephalo-myocarditis virus (EMCV) IRES, the Hepatitis C Virus (HCV) IRES, or the Cricket Paralysis Virus intergenic region (CrPV) IRES. For all of the above mRNAs, the level of translation was reduced to background levels at concentrations of Mg2+ >4 mM (Fig. 1). Optimal translation in terms of yield occurred at 1–2 mM total Mg2+ (Fig. 1).
FIGURE 1.
Effect of magnesium ion concentration on translation invitro. (A) GLA mRNA, which is capped and polyadenylated, was translated in RRL with the indicated total concentrations of Mg2+ and the indicated added KOAc. Relative levels of translation are shown. (B) The relative translation from CrPV-EGFP, HCV-EGFP, and EMCV-EGFP mRNAs shown at a range of Mg2+ concentrations. The reactions were carried out with no added KOAc. The yield of EGFP translation was determined by measuring the fluorescence of EGFP. The standard deviation of three experiments is shown. (C) The relative translation from the IRES mRNAs shown for translation at a lower range of total Mg2+. Translation was carried outin the presence of 35S-Methionine. The translation yield was determined by resolving the newly translated protein on SDS–PAGE gels and autoradiography.
Chemical probing of 40S subunits and 80S ribosomes
As noted above, chemical probing performed to date on the mammalian ribosome has been carried out at concentrations of Mg2+ much higher than the optimal levels for translation. The conformations of nonprogrammed and programmed mouse 80S ribosomes have been studied at 5 mM Mg2+ (Sloma and Nygard 2001). The effects of subunit association were studied for mammalian ribosomes at 7.5 mM Mg 2+ (Holmberg et al. 1994), and the effects of HCV IRES binding to the 40S ribosomal subunit were studied at 10 mM Mg2+ (Otto et al. 2002). The use of such high concentrations of magnesium ions may have masked conformational changes within the ribosome that are important for active translation. Therefore, we used chemical probing to investigate the effect of Mg2+ concentration on the conformation of the 40S subunit as well as the changes in conformation of the 40S subunit upon subunit association and upon HCV IRES binding.
Dimethyl sulfate (DMS) chemical probing was used to assess the chemical accessibility of specific nucleotides in 18S rRNA within the 40S subunit. This method allows changes in the environment of the rRNA due to conformational rearrangements and ligand or translation factor binding to be studied (Merryman and Noller 1998). DMS methylates exposed bases in the RNA by reacting with imino nitrogens. DMS reacts well with the N1 of adenosine and the N3 of cytidine bases (Merryman and Noller 1998). It has also been shown to react with the N7 atom of guanosine (Moine et al. 1998). In rare cases, DMS will react with the N3 position of uridines (Moazed and Noller 1986; Sloma and Nygard 2001), reactivity that would require a tautomeric form of uridine or a shift in its pKa. The effects of changes in magnesium ion concentration on the 40S subunit were first assessed in the absence of additional factors. Human 40S ribosomal subunits in buffer at 2.5 mM and 10 mM Mg2+ were subjected to chemical modification by DMS. At the higher concentration of Mg2+, many more nucleotides throughout the rRNA were protected from modification than at the lower concentration (data not shown). Sucrose gradients indicate that the 40S ribosomal subunits remain monomeric at 10 mM Mg2+, indicating that aggregation was not responsible for the changes (data not shown). The extensive protection at higher Mg2+ therefore suggests that the subunit becomes more conformationally “rigid” at the higher Mg2+ concentration, such that fewer nucleotides are exposed to the solvent.
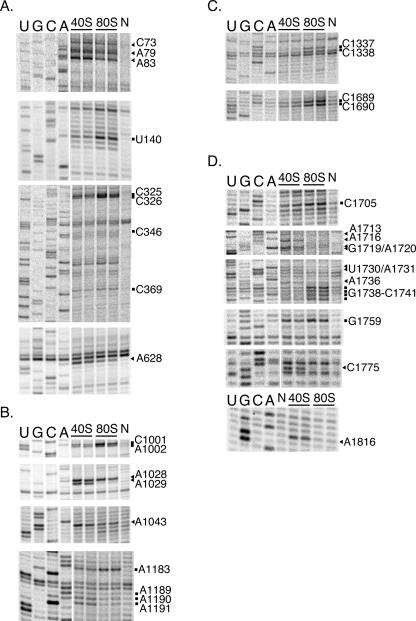
The interface between the two ribosomal subunits consists of a large number of RNA–RNA interactions and a small number of protein-based interactions at the periphery (Yusupov et al. 2001; Schuwirth et al. 2005). Therefore, it is likely that the concentration of magnesium ions could influence conformational changes induced upon subunit association. The effect of subunit association on the mouse 40S subunit has been studied previously at 7.5 mM (high) Mg2+. Upon subunit association, nine nucleotides were found to have altered reactivity to DMS, and five to CMCT, which modifies N3 of uridines (Holmberg et al. 1994). In order to determine whether subunit association leads to a different chemical reactivity profile of the 18S rRNA at low Mg2+, purified 40S subunits and 80S run-off ribosomes (monosomes) from HeLa cytoplasmic extract were subjected to modification by DMS at 2.5 mM Mg2+. At low Mg2+, 37 nucleotides displayed different DMS reactivity upon subunit association. Sequencing gels of probing reactions that revealed changes in 18S rRNA accessibility are shown in Figure 2. Table 1 lists the 37 nucleotides that have either reduced or enhanced reactivity with DMS in the 80S ribosome when compared with the 40S subunit. A secondary structure map of human 18S rRNA (Cannone et al. 2002) showing the differences in modification is shown in Figure 3.
FIGURE 2.
DMS modification of 40S subunits and 80S ribosomes. Sequencing lanes are shown to the left. Nucleotides with enhanced reactivity in 80S ribosomes are marked with a square; nucleotides protected in 80S ribosomes are marked with an arrow. Note that bands in the sample lanes appear one nucleotide below the corresponding sequencing band. Two 40S subunit reactions, two 80S ribosome reactions, and nonmodified (N) 18S rRNA are shown. Panels A–D are taken from representative gels.
TABLE 1.
Changes in DMS modification due to subunit association, 2.5 mM Mg2+
| Nucleotide, human | Domain | Changea | Nucleotide, E. colib |
| C73 | 5′ | − | NA |
| A79 | 5′ | − | NA |
| A83 | 5′ | − | NA |
| U140 | 5′ | ++ | NA |
| C326 | 5′ | + | NA |
| C346 | 5′ | + | U229 |
| C369 | 5′ | + | G251 |
| A628 | 5′ | − − | A532 |
| C1001 | Central | + | C732 |
| A1002 | Central | + | G733 |
| A1028 | Central | − − | A759 |
| A1029 | Central | − − − | G760 |
| A1043 | Central | − | G774 |
| A1183 | Central | + | A901 |
| A1189 | Central | − | A907 |
| A1190 | Central | − | A908 |
| A1191 | Central | − | A909 |
| C1337 | 3′ Major | − − | C1059 |
| C1338 | 3′ Major | − − | U1060 |
| C1689 | 3′ Major | ++ | G1387 |
| C1690 | 3′ Major | ++ | C1388 |
| C1705 | 3′ Minor | + | C1403 |
| A1713 | 3′ Minor | − | A1410 |
| A1716 | 3′ Minor | − − | A1413 |
| G1719 | 3′ Minor | − | G1417 |
| A1720 | 3′ Minor | − − | A1418 |
| U1730 | 3′ Minor | − | NA |
| A1731 | 3′ Minor | − | NA |
| A1736 | 3′ Minor | − | NA |
| C1739 | 3′ Minor | + | NA |
| C1740 | 3′ Minor | + | NA |
| C1741 | 3′ Minor | + | NA |
| G1738 | 3′ Minor | + | NA |
| G1759 | 3′ Minor | + | NA |
| C1775 | 3′ Minor | − − | NA |
| A1816 | 3′ Minor | − − | A1483 |
aDifferences in chemical accessibility of nucleotides in 18S rRNA, comparing 80S ribosomes with 40S subunits; − Protection; + Enhancement.
bThe equivalent E. coli nucleotide number is given where relevant.
FIGURE 3.
Secondary structure of the human 18S rRNA showing differences in DMS reactivity of nucleotides in 80S ribosomes compared with 40S subunits. Shaded regions where changes in chemical reactivity occur are enlarged below and to the right of the secondary structure. Squares show enhancements in reactivity; arrows, reduced reactivity. The secondary structure map is from Cannone et al. (2002). The domain organization of 18S rRNA is indicated on the secondary structure, as are helices in E. coli numbering relevant to the main text. The sequence register for human 18S rRNA is indicated by gray numbers for nucleotides divisible by 50, and gray dashes underlining nucleotides divisible by 10.
The fact that 37 differences were observed at low Mg2+, whereas only nine DMS-dependent differences were observed in the previous study performed under high Mg2+ conditions (cataloged in Table 2), supports the hypothesis that structural rearrangements in the 40S subunit are masked at the higher magnesium concentration. Of the nine DMS modification changes observed previously at high Mg2+, four were also observed here. All the nucleotides in which changes in modification were observed at either Mg2+ concentration are conserved in mouse and human ribosomes, ruling out differences in sequence as an explanation for differences in chemical probe accessibility.
TABLE 2.
Changes in DMS modification due to subunit association, 10 mM Mg2+
| Nucleotide, mouse | Changea | Nucleotide, human | Observed at 2.5 mM Mg 2+ |
| A888 | ++ | A886 | No |
| U941 | ++ | U939 | CMCT |
| U942 | ++ | U940 | CMCT |
| U971 | −−− | U970 | CMCT |
| A1003 | +++ | A1002 | Yes |
| U1023 | −−− | U1022 | CMCT |
| U1024 | −−− | U1023 | CMCT |
| A1027 | −− | A1026 | No |
| C1028 | −−− | C1027 | No |
| A1029 | −− | A1028 | Yes |
| A1030 | −−− | A1029 | Yes |
| A1072 | −− | A1071 | No |
| A1816 | −−− | A1816 | Yes |
| A1819 | −−− | A1819 | Stop |
aDifferences in chemical accessibility of nucleotides in 18S rRNA, comparing 80S ribosomes with 40S subunits probed with DMS; − Protection; + Enhancement. Modifications not assessed in the present experiment are marked CMCT (CMCT probing), and Stop, due to a reverse transcriptase stop (Holmberg et al. 1994).
The small ribosomal subunit is composed of four tertiary structural domains (Wimberly et al. 2000) that correlate well with secondary structural domains in 18S rRNA (Spahn et al. 2001a). The 5′ rRNA domain forms the body of the subunit, the central rRNA domain forms the platform, and the 3′ major domain in the rRNA forms the head of the small subunit. The 3′ minor domain in the rRNA includes the long penultimate stem that runs the length of the subunit body and platform (Wimberly et al. 2000). Subunit association induced changes in DMS reactivity in all four domains of the 40S subunit. In the body of the small subunit, four nucleotides were protected in the 80S ribosomes, and five became enhanced (Fig. 3; Table 1). This is in contrast to the data gathered at 7.5 mM Mg2+, in which no changes were observed in the entire body of the small subunit (Table 2). In the platform region of the subunit, nine changes were observed, three enhancements and six protections. In the head domain, four differences in reactivity were observed, whereas no changes were reported in this domain at the high magnesium concentration. The region in which the most changes were observed here was the penultimate stem in the 3′ minor domain. This long helix makes extensive contacts with the large subunit (Yusupov et al. 2001; Schuwirth et al. 2005). Therefore, it is not surprising that many nucleotides in this helix have altered reactivity upon subunit association.
Probing of the 40S:HCV IRES complex
The HCV and CrPV IRES RNAs are both able to bind directly to the 40S ribosomal subunit and position the start codon correctly for initiation. The effects of binding of these RESs to the 40S subunit have been investigated using cryo-EM. In both cases, conformational changes in the 40S ribosome were observed upon binding of the IRES RNA, and were proposed to play functional roles in initiation (Spahn et al. 2001b, 2004b). Notably, both of these studies were carried out in 2.5 mM Mg2+.
Chemical probing has previously been used to study the effect of HCV IRES binding on the conformation of the 40S subunit. Notably, the experiments were carried out at 10 mM Mg2+, and no changes in chemical accessibility were reported (Otto et al. 2002). In order to determine whether changes in the DMS modification pattern occur in the 40S subunit upon binding of the HCV IRES at a lower concentration of Mg2+ (2.5 mM), chemical probing was performed on the 40S subunit and the 40S:HCV IRES complex. The most prominent changes in DMS reactivity induced by HCV IRES binding to the 40S subunit that we have so far observed occurred in the base of helix 27 (Escherichia coli helix numbering) in the 18S rRNA (Fig. 4). We also tested the effect of a mutant form of the HCV IRES (termed H3V RNA) in which domain 2 has been deleted, which still allows 40S subunit binding but severely weakens IRES activity. Interestingly, the H3V RNA led to greater enhancements in reactivity in helix 27 than the full-length HCV IRES. This is surprising, given that the H3V RNA did not induce observable conformational changes in the cryo-EM reconstructions (Spahn et al. 2001b).
FIGURE 4.
Changes in DMS reactivity in 40S subunits when bound to HCV IRES or H3V RNAs. (A) DMS chemical probing results of 40S with the HCV IRES and H3V RNAs. Nucleotides with enhanced reactivity in the HCV IRES or H3V RNA complexes relative to the 40S subunits aloneare indicted by squares. (B) Changes in solvent accessibility upon binding of the HCV IRES to 40S ribosomal subunits in the central domain mapped onto the secondary structure of the 18S rRNA. Enhancements, protections, and secondary structural elements are marked as in Figure 3. Helix 27 (h27, E. coli numbering) is indicated.
DISCUSSION
Ribosomes are extremely sensitive to their ionic environment, especially to the concentration of divalent ions and polyamines (Jelenc and Kurland 1979; Bartetzko and Nierhaus 1988; Triana-Alonso et al. 1995; Gromadski and Rodnina 2004). For example, bacterial 30S subunits formed by dissociating 70S ribosomes at 1 mM Mg2+ are inactive. The binding of tRNA and other activities can be restored to these inactive subunits by heating them in an appropriate buffer at 10 mM Mg2+ (Zamir et al. 1969, 1971, 1974). The bacterial 50S subunit also assumes an inactive form under certain ionic conditions that cause conformational changes in the peptidyl transferase center (PTC) (Muth et al. 2000, 2001; Bayfield et al. 2001). Therefore, it is crucial that ionic conditions that maintain the ribosome in an active form are used in experiments to probe conformational changes that take place in the rRNA.
Chemical probing analyses have been performed previously on the bacterial ribosome. Changes in reactivity on subunit association were determined for reactions with the base-specific probes DMS, DEPC, CMCT, and kethoxal, as well as with hydroxyl radicals that cleave exposed regions of the RNA in a non-base-specific manner (Merryman et al. 1999). For bacterial subunit association, changes in reactivity were observed in the penultimate stem as well as in several of the helices in which changes were observed here. However, the extent of the changes observed in the bacterial study was less than that observed in the present study on human 40S subunits. This may be because the concentration of magnesium ions used in the earlier experiment was 10 mM (Merryman et al. 1999), whereas optimal translation with E. coli ribosomes occurs at 3.5 mM Mg2+ in the presence of polyamines (Jelenc and Kurland 1979; Bartetzko and Nierhaus 1988; Gromadski and Rodnina 2004).
In the present study, purified 40S subunits were compared with purified 80S monosomes. This differs slightly from the methods used in the experiments at higher Mg2+ concentrations, in which the two ribosomal subunits were purified separately, and the association was allowed to take place in vitro prior to chemical modification. The rationale in the earlier experiments was to ensure that no protein translation factors were present on the 80S ribosomes that could produce artifactual differences when compared with 40S subunits. We found that purified human 40S and 60S subunits did not readily associate at 2.5 mM Mg2+, and therefore decided to use purified 80S monosomes, which are stable at 2.5 mM Mg2+ and low monovalent salt concentrations (data not shown). Although we used purified 80S monosomes, other translation factors are unlikely to be present, as the ribosomes are not in mRNA-bound polysomes. In addition, the two subunits of the purified 80S ribosomes can be easily and entirely dissociated at 500 mM KCl at both 2.5 mM and 7.5 mM Mg2+ (data not shown). Their ease of dissociation suggests that the monosomes are indeed run-off ribosomes that do not contain any mRNA, tRNA, or additional protein factors (Falvey and Staehelin 1970b). Further support for the 80S ribosomes being run-off monosomes comes from the intermediate protection pattern of A628 seen in the present experiments (Fig. 2A), which is indistinguishable from the protection pattern of 80S run-off ribosomes used in previous experiments (Sloma and Nygard 2001). A628, which is positioned in the vicinity of mRNA in the aminoacyl-tRNA binding site (Yusupova et al. 2001; Ogle et al. 2002), is fully protected in native or salt-washed polysomes that contain mRNA (Sloma and Nygard 2001). The changes observed in the present experiments when compared with earlier ones at higher Mg2+ concentration are therefore most likely due to differences in the conformational dynamics of the 40S subunit at the two Mg2+ concentrations.
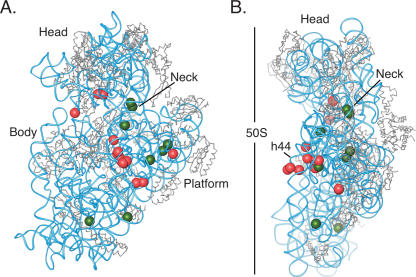
The positions of the changes in DMS modification that occurred in regions conserved in E. coli were mapped onto the crystal structure model of the E. coli ribosome (Cannone et al. 2002; Schuwirth et al. 2005). This allowed the three-dimensional positions of the changes in modification to be visualized (Fig. 5). Many nucleotides with changes in reactivity reside at the interface of the ribosome, suggesting that the protections occur due to direct binding of the large subunit. Other changes occur in interior regions of the 40S subunit, indicative of long-range conformational changes that take place upon binding of the 60S subunit.
FIGURE 5.
Changes observed in the human 40S subunit due to 60S subunit association, modeled onto the crystal structure of the E. coli ribosome. (A) The small subunit shown from the 60S subunit interface side. (B) The small subunit shown from the Exit-tRNA side, where the large subunit would be to the left. The 30S ribosomal subunit from the E. coli 70S ribosome is shown with RNA in blue and proteins in gray. Positions of nucleotides that become protected from DMS modification in 80S ribosome compared with 40S subunits are indicated by red spheres; nucleotides that have enhanced reactivity in 80S monosomes are indicated by green spheres. Domains in the subunit are marked: Head, Body, Platform, h44, penultimate stem in the 3′ minor domain.
A number of the protections induced upon subunit binding in this study have also been observed at equivalent nucleotide positions in bacterial ribosomes. Nucleotides A1190, A1191, A1716, A1720, and A1816 (E. coli nucleotides A908, A909, A1413, A1418, and A1483) are protected on subunit binding in both human and E. coli ribosomes (Merryman et al. 1999). Three of the nucleotides protected on 60S binding in the present study have equivalent nucleotides in E. coli that are also protected by tRNA binding. A1191 (A909) and A1716 (A1413) are among the “class III” sites in the 30S subunit that are protected by either binding of tRNA in an mRNA-independent manner, or by binding of the 50S subunit (Moazed and Noller 1986, 1987). In addition, A628 (A532) becomes protected in E. coli 30S ribosomal subunits upon binding of a P-site tRNA (Moazed and Noller 1986) and can be directly cross-linked to mRNA (Rinke-Appel et al. 1991; Dontsova et al. 1992).
Another modification of interest occurs at G1719 (G1417). This nucleotide, part of intersubunit bridge B3 (Yusupov et al. 2001; Schuwirth et al. 2005), is protected in the human 80S ribosome when compared with the 40S subunit. In the E. coli 70S ribosome, the equivalent nucleotide is involved in the inner-sphere coordination of a Mg2+ ion through the guanosine O6 atom (Schuwirth et al. 2005). Notably, in the crystal structure of the Thermus thermophilus 30S ribosome, the Mg2+ ion is fully hydrated, and involves only outer-sphere coordination with the rRNA (Wimberly et al. 2000). The rRNA sequence in this region is identical in T. thermophilus and E. coli, indicating that the difference in coordination of the Mg2+ is not due to differences in sequence. This change is the only clear example of a specific Mg2+ ion binding site that becomes occupied upon subunit association (Schuwirth et al. 2005). The change in coordination of the Mg2+ ion may account for the fact that the N7 of G1719 is reactive in the free 40S subunit and becomes protected in the 80S ribosome. Remarkably, the change in protection also suggests that the Mg2+ ion binding site is conserved from bacteria to mammals.
The dynamics of the ribosome have been studied during a number of stages of translation. The head of the small subunit in bacteria is known to close or “nod” relative to the body on subunit binding (Wimberly et al. 2000; Vila-Sanjurjo et al. 2003; Jenner et al. 2005). During translocation, the small subunit moves in a ratchet-like way relative to the large subunit (Frank and Agrawal 2000; Spahn et al. 2004a). In addition, the head has been observed in a swiveled orientation relative to the body in crystal structures of the bacterial ribosome and cryo-EM reconstructions of yeast ribosomes. The swiveling action was proposed to be involved in guiding the tRNAs during translocation (Spahn et al. 2004a; Schuwirth et al. 2005). The motion involves rotation about helix 28 (E. coli numbering), which is a GC-rich helix that is highly conserved (Cannone et al. 2002). Interestingly, in the experiments presented here, two of the cytidine residues in this helix are not reactive with DMS in the 40S subunit but become reactive in the 80S ribosome. The fact that DMS cannot react with the cytidines in this helix in the 40S subunits suggests that the nucleotides are base-paired as in the 30S subunit (Wimberly et al. 2000). Modification of helix 28 was not observed in chemical probing experiments performed on E. coli 30S subunits and 70S ribosomes (Merryman et al. 1999). The enhancement in reactivity of these bases due to 60S subunit binding therefore comes as a surprise. It suggests that in the human ribosome, the head of the small subunit may move upon subunit association such that the base pairing in the helix is disrupted or weakened, exposing the bases to DMS.
The enhanced reactivity in helix 28 may represent a real difference in the dynamics of eukaryotic and bacterial ribosomes. For example, the empty yeast 80S ribosome exists in the ratcheted state, whereas the vacant 70S ribosome exists in the nonratcheted state (Spahn et al. 2004a). The DMS reactivity pattern observed in helix 28 in the 80S ribosomes suggests that upon binding of the large subunit, some movement of the head of the 40S subunit relative to the body and platform takes place. However, it is not yet clear whether this movement involves a closing of the head or a swiveling movement. Higher resolution cryo-EM or X-ray crystal structures of the human ribosome may be able to discern the nature of the movements that take place in the small subunit upon subunit association.
Three prominent changes in chemical accessibility observed in the 18S rRNA upon HCV IRES binding are present in helix 27 (E. coli numbering). These occurred at nucleotides G1168–G1170 (G886–888 in E. coli), which reside near to the decoding center of the small subunit (Wimberly et al. 2000). Mutations in yeast ribosomes at nucleotides 886 and 888 (E. coli numbering) lead to phenotypes of increased accuracy (Velichutina et al. 2000). The fact that binding of the HCV IRES RNA results in changes in the chemical reactivity of these nucleotides suggests that the IRES could have an effect on the decoding center of the ribosome. Notably, the set of nucleotides with altered reactivity due to HCV IRES binding is different from the set altered by 60S subunit binding. This is especially true in the helix 27 region where nucleotides 1168–1170 become reactive in the 40S subunit complex with the HCV IRES, whereas nucleotides 1189–1191 remain unprotected (data not shown). Thus, the conformational change in the 40S subunit induced by binding of the HCV IRES differs from the conformation adopted by the 40S subunit within the 80S ribosome. Interestingly, the CrPV IRES, a different class of IRES that can bind directly to 40S subunits (Wilson et al. 2000), induces a conformational change in the 40S subunit that is not maintained in the CrPV IRES:80S ribosome complex (Spahn et al. 2001b).
The nucleotides in the central domain of the 40S subunit with altered chemical reactivity due to HCV IRES or H3V RNA binding were mapped onto the crystal structure of the E. coli ribosome (Fig. 6). By comparison with the cryo-EM structure of the 40S:HCV IRES complex, these nucleotides would not directly contact the IRES RNA (Cate et al. 1999; Wimberly et al. 2000; Spahn et al. 2001b; Yusupov et al. 2001). Therefore, the alterations must be due to long-range conformational changes induced in the 40S subunit by the IRES. The functional significance of these changes is not clear at present. More detailed analysis of initiation complexes with the HCV IRES will be needed to probe the role of conformational changes in HCV IRES-driven translation initiation.
FIGURE 6.
Changes observed upon binding of the HCV IRES or H3V RNA to 40S ribosomal subunits modeled onto the 30S subunit of the E. coli ribosome. Four enhancements in DMS reactivity (green spheres) were observed in the central domain of the 40S subunit upon binding of the HCV IRES or H3V RNA. Domains in the subunit are marked as in Figure 5. The binding site for the HCV IRES, as determined by cryo-EM, is also shown (Spahn et al. 2001b).
During translation the ribosome undergoes many conformational changes that are dependent on Mg2+ concentration. The ribosome requires Mg2+ ions in order to remain stably folded and to function, but at high concentrations of Mg2+, translation cannot take place. Chemical probing analyses of subunit association have been performed previously on the bacterial (Merryman et al. 1999) and eukaryotic ribosome (Holmberg et al. 1994) at prohibitively high Mg2+ ion concentrations. Although many of the changes observed upon subunit association in those prior studies were also observed here, many additional changes in chemical probe accessibility appeared at a lower Mg2+ ion concentration that supports in vitro translation. Similarly, probing of the 40S:HCV IRES complex revealed several changes induced in the 40S subunit upon IRES binding that were not observed previously in experiments performed at a high Mg2+ concentration (Otto et al. 2002). These results therefore suggest that in functionally relevant conditions, the ribosome has a greater range of flexibility than previously estimated from chemical probing experiments.
MATERIALS AND METHODS
Cloning
The pGLA plasmid encoding the GLA mRNA containing the globin 5′ NTR, luciferase open reading frame (ORF), and an encoded poly(A) tail was used without modification (Holden and Harris 2004). The IRES-EGFP plasmids were constructed from the pIRES2 plasmid (Clontech). The T7pI-EGFP plasmid, containing a T7 RNA polymerase promoter, the EMCV IRES, and the EGFP ORF, was made by inserting the T7 promoter between the SalI and the XmaI sites of the pIRES2. The CrPV IRES was amplified from the pCI-luc plasmid (Jan and Sarnow 2002) and cloned into the T7pI-EGFP plasmid between BamHI and BstXI sites, thereby removing the EMCV IRES. The HCV IRES RNA gene was amplified from the pHCV plasmid (Kieft et al. 1999) and cloned into the T7pI-EGFP plasmid similarly to the CrPV IRES using primers developed for the same two restriction sites. The H3V RNA was used from the pHCV del(1–119) plasmid with no alterations (Kieft et al. 1999).
In vitro transcription
All mRNAs were transcribed in vitro using T7 RNA polymerase, essentially as described (Doudna 1997). The RNAs were phenol/ chloroform extracted, LiCl salt precipitated, resuspended in water, and used without further purification. To ensure m7G cap incorporation, the concentration of GTP was lowered to 0.6 mM, and m7G cap analog (NEB) was included at 1.3 mM.
In vitro translation
In vitro translations (IVTs) were performed in Flexi Rabbit Reticulocyte Lysate (RRL) that contained 2.02 mM endogenous Mg2+. For the capped GLA mRNA, lysate was used at 40% v/v in 20-μL reactions supplemented with 87.5 mM KCl, 1.8 mM DTT, 17.5 mM HEPES at pH 7.5, 7.8 mM creatine phosphate, 25 μM amino acids except methionine, Rnasin RNase inhibitor at 0.6 U/μL, 15 μCi 35S-methionine (Promega; 1 μL of stock at >1000 Ci/mmol per 20 μL IVT reaction), and 10 μg/mL GLA mRNA. Translations also contained additional MgCl2 to bring the Mg 2+ion concentration to the indicated totals, and either 0, 50, or 100 mM additional KOAc. Translations were allowed to proceed for 60 min at 30 °C, and the degree of 35S incorporation was measured by nitrocellulose filter binding and scintillation counting. Counts bound to the filters correlated well with full-length translated protein, as determined by quantification of autoradiograms of SDS gels (not shown). Translation was fully inhibited by 1 mM m7G cap analog in the IVT reactions (not shown), demonstrating the cap-dependence of the reactions.
For the IRES-driven constructs in which translation was quantified by EGFP fluorescence, 10-μL reactions were made with 66% v/v Flexi Rabbit Reticulocyte Lysate, 0.1 μM mRNA, 22.3 mM added KCl, 35 μM complete amino acid mix, 0.36 U/μL Rnasin RNase inhibitor, protease inhibitor, DTT and HEPES buffer as above, and Mg(OAc)2 to give the indicated final concentration. Samples were incubated for 18 h at 30 °C. 100 μL of GFP dilution buffer (15 mM Tris at pH 7.5, 5 mM EDTA) was added to each sample. The samples were incubated for an additional 1 h at room temperature to ensure complete folding of the EGFP. 20 μL of each sample was used to measure fluorescence in a 1.5-mm path length quartz cuvette. Emission was measured at 507 nm with excitation at 488 nm in an ISA instruments Fluoromax 2 fluorimeter.
For the IRES-driven constructs in which translation was quantified by autoradiography, Flexi Rabbit Reticulocyte Lysate was used at 40% v/v in 20-μL reactions supplemented with 87.5 mM KOAc, 7.8 mM creatine phosphate, 25 μM all amino acids except methionine, 0.6 U/μL Rnasin RNase inhibitor, 15 μCi 35S-Methionine, and 10 μg/mL mRNA. MgCl 2 was added to bring the Mg 2+ ion concentration to the indicated totals. Translations were allowed to proceed for 60 min at 30 °C. The proteins were resolved by SDS–PAGE, and the degree of 35S-Methionine incorporation was measured by autoradiography on a STORM phosphorimager (Molecular Dynamics) and quantified using ImageQuant.
Ribosome purification
The following buffers were used during the purification. Buffer A: 15 mM Tris (pH 7.5), 300 mM NaCl, 6 mM MgCl2, 1% Triton X-100, 1mg/mL heparin, 1mM DTT. Buffer B: 20 mM Tris (pH 7.5), 6 mM Mg(OAc)2, 150 mM KCl, 2 mM DTT. Buffer C: 20 mM HEPES (pH 7.5), 6 mM MgOAc2, 0.5 M KCl, 2 mM DTT. Buffer P: 20 mM HEPES (pH 7.5), 100 mM KCl, 2.5 mM MgCl2, 2 mM DTT. Subunits Sucrose Cushion: 0.988 M Sucrose, 0.5 M KCl, 40 mM Tris (pH 7.5), 1 mM EDTA, 6 mM Mg(OAc)2, 2 mM DTT. 80S Sucrose Cushion: 0.988 M sucrose in buffer P.
The 40S subunits were purified under conditions of high salt (0.5 M KCl) in order to remove any bound initiation factors (Falvey and Staehelin 1970a,b; Sundkvist et al. 1974; Benne and Hershey 1976). HeLa cytoplasmic extract was obtained from the Robert Tjian lab (UC-Berkeley), with cells treated as described elsewhere (Dignam et al. 1983). Each purification was performed on either fresh lysate or lysate stored at −80°C from 10–15 L of cells. Mitochondria were pelleted by spinning for 10 min at 12,600 rpm in a JA-20 rotor. The supernatant was carefully removed and transferred to a new tube. The lysate was further cleared by spinning at 18,000 rpm in a JA-20 rotor for 100 min. The supernatant was decanted and made up to a total volume of 90 mL with buffer P.
15 mL of cleared lysate was loaded onto each of six sucrose cushions in Ti45 tubes containing 50 mL of chilled subunit sucrose cushion solution. Sucrose cushions were spun at 33,000 rpm for 22 h in a Beckman Ti45 rotor. The supernatants were discarded and the pellets washed briefly with 7 mL of buffer B and resuspended in ~2 mL buffer B per tube. Resuspensions were carried out at 4°C with rapid shaking for 6–8 h.
The resuspended pellets were pooled and loaded over six 0.3 M–1.0 M (10%–30% w/v) sucrose gradients in buffer C. Gradients were spun in a SW-32 or SW-28 rotor for 18 h at 22,000 rpm. The gradients were then resolved, and 40S and 60S fractions were collected at 4°C. Each set of fractions was pooled, made up to a total volume of 65 mL with buffer C, and spun in a Ti45 rotor for 4 h at 40,000 rpm. The resulting pellet was washed briefly with 7 mL of buffer P and resuspended in 1–1.5 mL of buffer P for several hours until fully resuspended. The concentration was measured using the absorbance at 260 nm and an absorbance coefficient of 57 μg/mL per OD260 unit.
80S ribosomes were purified in a similar manner except for the following modifications: The initial sucrose cushion was made with buffer P and 1.0 M sucrose, and the tubes were spun for 40,000 rpm at 4 h. The pellets were washed with and resuspended in buffer P. The subsequent sucrose gradients were made up in buffer P and were spun for 12 h at 22,000 rpm. 80S ribosome fractions from the gradients were collected and pooled. The fractions were made up to a total of 65 mL with buffer P, pelleted in a Ti45 rotor, and resuspended as described above.
Some preparations of human ribosomes were performed using a cell pellet from 10 L of cells grown in suspension, supplied by the National Cell Culture Center. In this case the cells were first resuspended in buffer A, which contains detergent to disrupt the cell membranes. The cells were incubated on ice for 30 min, and the resulting lysate wass cleared by centrifugation at 23,000g for 30 min. The cleared lysate was loaded onto sucrose cushions, and ribosomes were purified as described above.
The integrity, purity, and homogeneity of ribosome samples were evaluated using composite acrylamide:agarose native gels (Dahlberg 1974) and analytical sucrose gradients (not shown).
Chemical probing
Chemical probing was carried out essentially as described (Merryman and Noller 1998). Probing reactions were carried out in 20 mM HEPES at pH 7.5, 100 mM KCl, 2 mM DTT, and either 2.5 mM or 10 mM Mg2+, as indicated in the text and figures. Polyamines were not included in the reactions, in order to avoid nonspecific labeling of the polyamines and rRNA (S. Joseph, pers. comm.). Ribosomes were incubated with or without ligands for 10 min at 30°C, then placed on ice. A control sample was used to which no DMS was added. This constituted the “nonmodified” sample and was also used for sequencing reactions. Each DMS reaction contained 100 pmol of ribosomes in 100 μL. The non-modified reaction contained 200 pmol in 200 μL. For probing with the HCV IRES and H3V RNA, three equivalents of the HCV IRES or H3V RNA were added to the 40S sample prior to incubation with DMS. The probing and extraction were carried out as described (Merryman and Noller 1998), and the positions of the modifications were determined using reverse transcriptase with 30 different DNA 33P-labeled primers that allowed probing of nearly all of the 18S rRNA (1870 nucleotides). The samples, along with sequencing reactions for each primer, were resolved on 10% acrylamide sequencing gels. The gels were dried and exposed to a phosphorimager screen for at least 12 h, and the screen was visualized using a STORM phosphorimager (Molecular Dynamics). Weak enhancements or protections were quantified from at least three separate experiments using ImageJ (Abramoff et al. 2004), to confirm their validity.
Acknowledgments
We thank E. Jan, C. Fraser, J. Hershey, S. Lanham, N. Shenvi, and J. Doudna for assistance and helpful discussions. The pGLA, pCI-luc, and pHCV plasmids were kind gifts of the E. Harris, P. Sarnow, and J. Doudna labs, respectively. HeLa cells were supplied by the National Cell Culture Center (NCCC). This work was funded in part by a Howard Hughes Medical Institute predoctoral fellowship for C.L.S., National Institutes of Health (NIH) CMB training grant T32-GM07232-27 to E.M.F., a NIH NRSA training grant (1 T32 GMO66698-01A1) to K.C.D. and J.A.H., and NIH grant R01-GM65050 to J.H.D.C.
Article and publication are at http://www.rnajournal.org/cgi/doi/10.1261/rna.2192805.
REFERENCES
- Abramoff, M.D., Magelhaes, P.J., and Ram, S.J. 2004. Image processing with ImageJ. Biophotonics International 11: 36–42. [Google Scholar]
- Bartetzko, A. and Nierhaus, K.H. 1988. Mg2+/NH4 +/polyamine system for polyuridine-dependent polyphenylalanine synthesis with near in vivo characteristics. Methods Enzymol. 164: 650–658. [DOI] [PubMed] [Google Scholar]
- Bayfield, M.A., Dahlberg, A.E., Schulmeister, U., Dorner, S., and Barta, A. 2001. A conformational change in the ribosomal peptidyl transferase center upon active/inactivetransition. Proc. Natl. Acad. Sci. 98: 10096–10101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benne, R. and Hershey, J.W. 1976. Purification and characterization of initiation factor IF-E3 from rabbit reticulocytes. Proc. Natl. Acad. Sci. 73: 3005–3009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannone, J.J., Subramanian, S., Schnare, M.N., Collett, J.R., D’Souza, L.M., Du, Y., Feng, B., Lin, N., Madabusi, L.V., Muller, K.M., et al. 2002. The comparative RNA web (CRW) site: An online database of comparative sequence and structure information for ribosomal, intron, and other RNAs. BMC Bioinformatics 3: 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cate, J.H., Hanna, R.L., and Doudna, J.A. 1997. A magnesium ion core at the heart of a ribozyme domain. Nat. Struct. Biol. 4: 553– 558. [DOI] [PubMed] [Google Scholar]
- Cate, J.H., Yusupov, M.M., Yusupova, G.Z., Earnest, T.N., and Noller, H.F. 1999. X-ray crystal structures of 70S ribosome functional complexes. Science 285: 2095–2104. [DOI] [PubMed] [Google Scholar]
- Dahlberg, A.E. 1974. Two forms of the 30S ribosomal subunit of Escherichia coli. J. Biol. Chem. 249: 7673–7678. [PubMed] [Google Scholar]
- Dignam, J.D., Martin, P.L., Shastry, B.S., and Roeder, R.G. 1983. Eukaryotic gene transcription with purified components. Methods Enzymol. 101: 582–598. [DOI] [PubMed] [Google Scholar]
- Dontsova, O., Dokudovskaya, S., Kopylov, A., Bogdanov, A., Rinke- Appel, J., Junke, N., and Brimacombe, R. 1992. Three widely separated positions in the 16S RNA lie in or close to the ribosomal decoding region; a site-directed cross-linking study with mRNA analogues. EMBO J. 11: 3105–3116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doudna, J.A. 1997. Preparation of homogeneous ribozyme RNA for crystallization. Methods Mol. Biol. 74: 365–370. [DOI] [PubMed] [Google Scholar]
- Falvey, A.K. and Staehelin, T. 1970a. Structure and function of mammalian ribosomes. I. Isolation and characterization of active liver ribosomal subunits. J. Mol. Biol. 53: 1–19. [DOI] [PubMed] [Google Scholar]
- ———. 1970b. Structure and function of mammalian ribosomes. II. Exchange of ribosomal subunits at various stages of in vitro polypeptide synthesis. J. Mol. Biol. 53: 21–34. [DOI] [PubMed] [Google Scholar]
- Frank, J. and Agrawal, R.K. 2000. A ratchet-like inter-subunit reorganization of the ribosome during translocation. Nature 406: 318–322. [DOI] [PubMed] [Google Scholar]
- Gao, N., Zavialov, A.V., Li, W., Sengupta, J., Valle, M., Gursky, R.P., Ehrenberg, M., and Frank, J. 2005. Mechanism for the disassembly of the posttermination complex inferred from cryo-EM studies. Mol. Cell 18: 663–674. [DOI] [PubMed] [Google Scholar]
- Goss, D.J. and Harrigan, T. 1986. Magnesium ion dependent equilibria, kinetics, and thermodynamic parameters of Artemia ribosome dissociation and subunit association. Biochemistry 25: 3690–3695. [DOI] [PubMed] [Google Scholar]
- Gromadski, K.B. and Rodnina, M.V. 2004. Kinetic determinants of high-fidelity tRNA discrimination on the ribosome. Mol. Cell 13: 191–200. [DOI] [PubMed] [Google Scholar]
- Hoa, G.H.B., Begard, E., Beaudry, P., Maurel, P., Grunberg-Manago, M., and Douzou, P. 1980. Analysis of cosolvent and divalent cation effects on association equilibrium and activity of ribosomes. Biochemistry 19: 3080–3087. [DOI] [PubMed] [Google Scholar]
- Holden, K.L. and Harris, E. 2004. Enhancement of dengue virus translation: Role of the 3′ untranslated region and the terminal 3′ stem-loop domain. Virology 329: 119–133. [DOI] [PubMed] [Google Scholar]
- Holmberg, L., Melander, Y., and Nygard, O. 1994. Probing the conformational changes in 5.8S, 18S and 28S rRNA upon association of derived subunits into complete 80S ribosomes. Nucleic Acids Res. 22: 2776–2783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson, R.J. and Hunt, T. 1983. Preparation and use of nuclease-treated rabbit reticulocyte lysates for the translation of eukaryotic messenger RNA. Methods Enzymol. 96: 50–74. [DOI] [PubMed] [Google Scholar]
- Jan, E. and Sarnow, P. 2002. Factorless ribosome assembly on the internal ribosome entry site of cricket paralysis virus. J. Mol. Biol. 324: 889–902. [DOI] [PubMed] [Google Scholar]
- Jelenc, P.C. and Kurland, C.G. 1979. Nucleoside triphosphate regeneration decreases the frequency of translation errors. Proc. Natl. Acad. Sci. 76: 3174–3178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenner, L., Romby, P., Rees, B., Schulze-Briese, C., Springer, M., Ehresmann, C., Ehresmann, B., Moras, D., Yusupova, G., and Yusupov, M. 2005. Translational operator of mRNA on the ribosome: How repressor proteins exclude ribosome binding. Science 308: 120–123. [DOI] [PubMed] [Google Scholar]
- Kieft, J.S., Zhou, K., Jubin, R., Murray, M.G., Lau, J.Y., and Doudna, J.A. 1999. The hepatitis C virus internal ribosome entry site adopts an ion-dependent tertiary fold. J. Mol. Biol. 292: 513–529. [DOI] [PubMed] [Google Scholar]
- Klaholz, B.P., Myasnikov, A.G., and Van Heel, M. 2004. Visualization of release factor 3 on the ribosome during termination of protein synthesis. Nature 427: 862–865. [DOI] [PubMed] [Google Scholar]
- Klein, D.J., Moore, P.B., and Steitz, T.A. 2004. The contribution of metal ions to the structural stability of the large ribosomal subunit. RNA 10: 1366–1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak, M. 1990. Evaluation of the fidelity of initiation of translation in reticulocyte lysates from commercial sources. Nucleic Acids Res. 18: 2828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merryman, C. and Noller, H.F. 1998. Footprinting and modification-interference analysis of binding sites on RNA. In RNA: protein interactions—A practical approach (ed. SmithC.W.J. ), pp. 237– 253. Oxford University Press, Oxford, UK.
- Merryman, C., Moazed, D., McWhirter, J., and Noller, H.F. 1999. Nucleotides in 16S rRNA protected by the association of 30S and 50S ribosomal subunits. J. Mol. Biol. 285: 97–105. [DOI] [PubMed] [Google Scholar]
- Moazed, D. and Noller, H.F. 1986. Transfer RNA shields specific nucleotides in 16S ribosomal RNA from attack by chemical probes. Cell 47: 985–994. [DOI] [PubMed] [Google Scholar]
- ———. 1987. Interaction of antibiotics with functional sites in 16S ribosomal RNA. Nature 327: 389–394. [DOI] [PubMed] [Google Scholar]
- Moazed, D., Van Stolk, B.J., Douthwaite, S., and Noller, H.F. 1986. Interconversion of active and inactive 30 S ribosomal subunits is accompanied by a conformational change in the decoding region of 16 S rRNA. J. Mol. Biol. 191: 483–493. [DOI] [PubMed] [Google Scholar]
- Moine, H., Ehresmann, B., Ehresmann, C., and Romby, P. 1998. Probing RNA structure and function in solution. In RNA structure and function (eds. R.W. Simons and M. Grunberg-Manago), pp. 77– 116. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Muth, G.W., Ortoleva-Donnelly, L., and Strobel, S.A. 2000. A single adenosine with a neutral pKa in the ribosomal peptidyl transferase center. Science 289: 947–950. [DOI] [PubMed] [Google Scholar]
- Muth, G.W., Chen, L., Kosek, A.B., and Strobel, S.A. 2001. pH-dependent conformational flexibility within the ribosomal peptidyl transferase center. RNA 7: 1403–1415. [PMC free article] [PubMed] [Google Scholar]
- Ogle, J.M., Murphy, F.V., Tarry, M.J., and Ramakrishnan, V. 2002. Selection of tRNA by the ribosome requires a transition from an open to a closed form. Cell 111: 721–732. [DOI] [PubMed] [Google Scholar]
- Otto, G.A., Lukavsky, P.J., Lancaster, A.M., Sarnow, P., and Puglisi, J.D. 2002. Ribosomal proteins mediate the hepatitis C virus IRES-HeLa 40S interaction. Mol. Cell. Biol. 8: 913–923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pape, T., Wintermeyer, W., and Rodnina, M.V. 2000. Conformational switch in the decoding region of 16S rRNA during aminoacyl-tRNA selection on the ribosome. Nat. Struct. Biol. 7: 104–107. [DOI] [PubMed] [Google Scholar]
- Pelham, H.R.B. and Jackson, R.J. 1976. An efficient mRNA-dependent translation system from reticulocyte lysates. Eur. J. Biochem. 67: 247–256. [DOI] [PubMed] [Google Scholar]
- Peske, F., Savelsbergh, A., Katunin, V.I., Rodnina, M.V., and Wintermeyer, W. 2004. Conformational changes of the small ribosomal subunit during elongation factor G-dependent tRNA-mRNA translocation. J. Mol. Biol. 343: 1183–1194. [DOI] [PubMed] [Google Scholar]
- Rinke-Appel, J., Junke, N., Stade, K., and Brimacombe, R. 1991. The path of mRNA through the Escherichia coli ribosome; sitedirected cross-linking of mRNA analogues carrying a photoreactive label at various points 3′ to the decoding site. EMBO J. 10: 2195–2202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savelsbergh, A., Katunin, V.I., Mohr, D., Peske, F., Rodnina, M.V., and Wintermeyer, W. 2003. An elongation factor G-induced ribosome rearrangement precedes tRNA–mRNA translocation. Mol. Cell 11: 1517–1523. [DOI] [PubMed] [Google Scholar]
- Schilling-Bartetzko, S., Bartetzko, A., and Nierhaus, K.H. 1992. Kinetic and thermodynamic parameters for tRNA binding to the ribosome and for the translocation reaction. J. Biol. Chem. 267: 4703–4712. [PubMed] [Google Scholar]
- Schuwirth, B.S., Borovinskaya, M.A., Hau, C.W., Zhang, W., Vila- Sanjurjo, A., Holton, J.M., and Cate, J.H.D. 2005. Structures of the bacterial ribosome at 3.5 A° resolution. Science (in press). [DOI] [PubMed]
- Sloma, M.S. and Nygard, O. 2001. Chemical accessibility of 18S rRNA in native ribosomal complexes: Interaction sites of mRNA, tRNA and translation factors. Biol. Chem. 382: 661–668. [DOI] [PubMed] [Google Scholar]
- Spahn, C.M., Beckmann, R., Eswar, N., Penczek, P.A., Sali, A., Blobel, G., and Frank, J. 2001a. Structure of the 80S ribosome from Saccharomyces cerevisiae—tRNA–ribosome and subunit–subunit interactions. Cell 107: 373–386. [DOI] [PubMed] [Google Scholar]
- Spahn, C.M., Kieft, J.S., Grassucci, R.A., Penczek, P.A., Zhou, K., Doudna, J.A., and Frank, J. 2001b. Hepatitis C virus IRES RNA-induced changes in the conformation of the 40s ribosomal subunit. Science 291: 1959–1962. [DOI] [PubMed] [Google Scholar]
- Spahn, C.M., Gomez-Lorenzo, M.G., Grassucci, R.A., Jorgensen, R., Andersen, G.R., Beckmann, R., Penczek, P.A., Ballesta, J.P., and Frank, J. 2004a. Domain movements of elongation factor eEF2 and the eukaryotic 80S ribosome facilitate tRNA translocation. EMBO J. 23: 1008–1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spahn, C.M., Jan, E., Mulder, A., Grassucci, R.A., Sarnow, P., and Frank, J. 2004b. Cryo-EM visualization of a viral internal ribosome entry site bound to human ribosomes: The IRES functions as an RNA-based translation factor. Cell 118: 465–475. [DOI] [PubMed] [Google Scholar]
- Sperrazza, J.M. and Spremulli, L.L. 1983. Quantitation of cation binding to wheat germ ribosomes: Influences on subunit association equilibria and ribosome activity. Nucleic Acids Res. 11: 2665–2679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sperrazza, J.M., Russell, D.W., and Spremulli, L.L. 1980. Reversible dissociation of wheat germ ribosomal subunits:Cation-dependent equilibria and thermodynamic parameters. Biochemistry 19: 1053–1058. [DOI] [PubMed] [Google Scholar]
- Stark, H., Rodnina, M.V., Wieden, H.J., van Heel, M., and Wintermeyer, W. 2000. Large-scale movement of elongation factor G and extensive conformational change of the ribosome during translocation. Cell 100: 301–309. [DOI] [PubMed] [Google Scholar]
- Stark, H., Rodnina, M.V., Wieden, H.J., Zemlin, F., Wintermeyer, W., and van Heel, M. 2002. Ribosome interactions of aminoacyl-tRNA and elongation factor Tu in the codon-recognition complex. Nat. Struct. Biol. 9: 849–854. [DOI] [PubMed] [Google Scholar]
- Sundkvist, I.C., McKeehan, W.L., Schreier, M.H., and Staehelin, T. 1974. Initiation factor activity associated with free 40 S subunits from rat liver and rabbit reticulocytes. J. Biol. Chem. 249: 6512–6516. [PubMed] [Google Scholar]
- Triana-Alonso, F.J., Chakraburtty, K., and Nierhaus, K.H. 1995. The elongation factor 3 unique in higher fungi and essential for protein biosynthesis is an E site factor. J. Biol. Chem. 270: 20473–20478. [DOI] [PubMed] [Google Scholar]
- Valle, M., Zavialov, A., Li, W., Stagg, S.M., Sengupta, J., Nielsen, R.C., Nissen, P., Harvey, S.C., Ehrenberg, M., and Frank, J. 2003a. Incorporation of aminoacyl-tRNA into the ribosome as seen by cryo-electron microscopy. Nat. Struct. Biol. 10: 899–906. [DOI] [PubMed] [Google Scholar]
- Valle, M., Zavialov, A., Sengupta, J., Rawat, U., Ehrenberg, M., and Frank, J. 2003b. Locking and unlocking of ribosomal motions. Cell 114: 123–134. [DOI] [PubMed] [Google Scholar]
- VanLoock, M.S., Agrawal, R.K., Gabashvili, I.S., Qi, L., Frank, J., and Harvey, S.C. 2000. Movement of the decoding region of the 16 S ribosomal RNA accompanies tRNA translocation. J. Mol. Biol. 304: 507–515. [DOI] [PubMed] [Google Scholar]
- Velichutina, I.V., Dresios, J., Hong, J.Y., Li, C., Mankin, A., Synetos, D., and Liebman, S.W. 2000. Mutations in helix 27 of the yeast Saccharomyces cerevisiae 18S rRNA affect the function of the decoding center of the ribosome. RNA 6: 1174–1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vila-Sanjurjo, A., Ridgeway, W.K., Seymaner, V., Zhang, W., Santoso, S., Yu, K., and Cate, J.H. 2003. X-ray crystal structures of the WT and a hyper-accurate ribosome from Escherichia coli. Proc. Natl. Acad. Sci. 100: 8682–8687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson, J.E., Pestova, T.V., Hellen, C.U., and Sarnow, P. 2000. Initiation of protein synthesis from the A site of the ribosome. Cell 102: 511–520. [DOI] [PubMed] [Google Scholar]
- Wimberly, B.T., Brodersen, D.E., Clemons Jr., W.M., Morgan-Warren, R.J., Carter, A.P., Vonrhein, C., Hartsch, T., and Ramakrishnan, V. 2000. Structure of the 30S ribosomal subunit. Nature 407: 327– 339. [DOI] [PubMed] [Google Scholar]
- Yusupov, M.M., Yusupova, G.Z., Baucom, A., Lieberman, K., Earnest, T.N., Cate, J.H.D., and Noller, H.F. 2001. Crystal structure of the ribosome at 5.5A° resolution. Science 292: 883–896. [DOI] [PubMed] [Google Scholar]
- Yusupova, G.Z., Yusupov, M.M., Cate, J.H., and Noller, H.F. 2001. The path of messenger RNA through the ribosome. Cell 106: 233–241. [DOI] [PubMed] [Google Scholar]
- Zamir, A., Miskin, R., and Elson, D. 1969. Interconversions between inactive and active forms of ribosomal subunits. FEBS Lett. 3: 85–88. [DOI] [PubMed] [Google Scholar]
- ———. 1971. Inactivation and reactivation of ribosomal subunits: Amino acyl-transfer RNA binding activity of the 30 S subunit of Escherichia coli. J. Mol. Biol. 60: 347–364. [DOI] [PubMed] [Google Scholar]
- Zamir, A., Miskin, R., Vogel, Z., and Elson, D. 1974. The inactivation and reactivation of Escherichia coli ribosomes. Methods Enzymol. 30: 406–426. [DOI] [PubMed] [Google Scholar]