Abstract
Ribosomal variants carrying mutations in active site nucleotides are severely compromised in their ability to catalyze peptide bond formation (PT) with minimal aminoacyl tRNA substrates such as puromycin. However, catalysis of PT by these same ribosomes with intact aminoacyl tRNA substrates is uncompromised. These data suggest that these active site nucleotides play an important role in the positioning of minimal aminoacyl tRNA substrates but are not essential for catalysis per se when aminoacyl tRNAs are positioned by more remote interactions with the ribosome. Previously reported biochemical studies and atomic resolution X-ray structures identified a direct Watson–Crick interaction between C75 of the A-site substrate and G2553 of the 23S rRNA. Here we show that the addition of this single cytidine residue (the C75 equivalent) to puromycin is sufficient to suppress the deficiencies of active site ribosomal variants, thus restoring “tRNA-like” behavior to this minimal substrate. Studies of the binding parameters and the pH-dependence of catalysis with this minimal substrate indicate that the interaction between C75 and the ribosomal A loop is an essential feature for robust catalysis and further suggest that the observed effects of C75 on peptidyl transfer activity reflect previously reported conformational rearrangements in this active site.
Keywords: ribosome, peptidyl transfer, A loop, tRNA, pH-rate profile
INTRODUCTION
Peptide bond formation takes place in the RNA-only active site of the large ribosomal subunit where universally conserved nucleotides surround the CCA ends of the peptidyl (P) and aminoacyl (A) tRNA substrates. While this chemically facile reaction between a nucleophilic amine and an electron-deficient carbon center likely requires little more than positioning by the active site, more “active” modes of catalysis including mechanisms that invoke general-acid–base assisted, substrate-assisted, or metal ion-assisted catalysis continue to be evaluated. Recent thermodynamic studies argued that catalysis of peptide bond formation was primarily driven by entropy (Sievers et al. 2004) whereas other studies argued for a strong enthalpic contribution deriving from potential direct involvement of the 2′-OH of the peptidyl-tRNA substrate (Weinger et al. 2004). Our previous mutational analysis of the active site of the ribosome led us to surprising conclusions about the role of active site nucleotides in the catalysis of peptide bond formation (Youngman et al. 2004). While mutation of the active site nucleotides A2451, U2506, U2585, or A2602 resulted in drastic deficiencies in peptidyl transfer with the minimal aminoacyl substrate puromycin (composed essentially of the terminal A76 of tRNA and its attached aminoacyl moiety), there were no discernible effects on catalysis when intact aminoacyl-tRNA substrates were used. These data suggested that when the more remote positioning elements of an intact tRNA substrate are in place, the nucleotides surrounding the active site do not contribute significantly to catalysis. In contrast, when remote positioning elements are not present, the local packing interactions of an active site that has coevolved with the universal A76 of tRNA become essential to catalysis.
We have been interested in understanding the molecular basis for the strikingly different behavior of the intact aminoacyl-tRNA substrate and puromycin in peptidyl transfer. Specifically, which interaction(s) between the intact tRNA and the ribosome are sufficient to make peptidyl transferase activity independent of the identity of core conserved nucleotides? While intact aminoacyl tRNA substrates clearly make a number of favorable interactions with both the large and small subunits of the ribosome (Cate et al. 1999), we wondered if the well-described Watson–Crick interaction between C75 of the A-site substrate and G2553 of 23S rRNA might be sufficient to restore tRNA-like behavior to puromycin in peptidyl transfer. Here, we test this hypothesis by comparing the peptidyl transferase activity of CPm with Pm on a collection of active site rRNA variants and then examine the binding parameters of its interaction with the ribosome and the pH dependence of catalysis with this substrate. These data clearly establish that robust peptidyl transferase activity derives from the precise positioning that results from the C75:G2553 tRNA:rRNA interaction. Based on earlier observations (Miskin et al. 1968; Bayfield et al. 2001), we further speculate that conformational rearrangements in the active site induced by this interaction may underlie the observed effects on peptidyl transfer activity.
RESULTS
Determination of the K1/2 for CPm in the tripeptide assay
The goal of these studies was to compare the activity of Pm and CPm on active site variant ribosomes in previously described dipeptide and tripeptide assays for peptide bond formation (Fig. 1 ▶). We first established the K1/2 for this compound in the tripeptide assay by measuring the concentration dependence of the reaction (Fig. 2 ▶). The observed rates of peptide bond formation in the tripeptide (fMetPheCpm) assay were measured in duplicate at 13 concentrations of CPm. The resulting curve was fit to a hyperbolic equation to establish a K1/2 value of 1.7 ± 1.6 mM for CPm on wild-type ribosomes (MRE600), similar to that previously measured for Pm (K1/2 = 3.8 ± 0.6 mM) (Youngman et al. 2004; Table 1 ▶). A saturating concentration (10 mM) was used for both Pm and CPm in the rapid quench experiments described below.
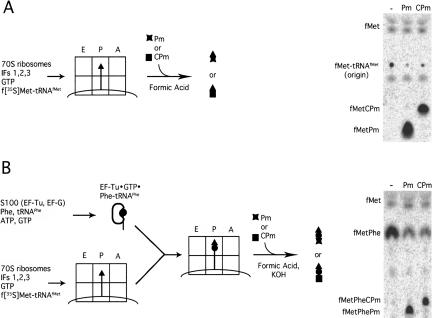
FIGURE 1.
Kinetic assays for peptide bond formation. Schematic representation of the assembly of ribosome complexes and the assays used to measure the kinetics of PT in dipeptide (A) and tripeptide (B) formation. Representative electrophoretic TLCs showing resolution of reactants and products in these reactions are shown at the right.
FIGURE 2.
Concentration dependence of PT kinetics with Pm and CPm. Observed rate constants for peptide bond formation were measured in the tripeptide assay at concentrations between 0.125 and 17 mM Pm (closed circles) and CPm (open circles) at pH 7.5 and 20° C. Error bars represent the standard deviation from at least two independent measurements. Fitting of these data to a hyperbolic (Michaelis–Menten) equation yielded a K1/2 of 3.8 mM for Pm and 1.7 mM for CPm, and maximum rate constants of 15 sec−1 for Pm and 18 sec−1 for CPm.
TABLE 1.
K1/2 and Kd values for Pm and CPm
| K1/2 (mM) | Kd (mM) | |
| Pm | 3.8 ± 1.6a,b | 1.8 ± 0.5c |
| CPm | 1.7 ± 0.6a | 0.014 ± 0.002d |
aTo measure K1/2, PT rate constants were measured at least twice at 8 to 13 concentrations of Pm or CPm. The average rate constants were plotted against [Pm] or [CPm] and the resulting curves fit to the Michaelis–Menten equation. Reported deviations are the error in the fit to the averaged data points.
bData from Youngman et al. (2004).
cBased on protection of G2505 of 23S rRNA from kethoxal modification by Pm.
dBased on protection of G2553 of 23S rRNA from kethoxal modification by CPm.
Peptidyl transferase activity of wild-type ribosomes with Pm and CPm as A-site substrates
We first measured the rate of peptide bond formation with wild-type ribosomes using Pm and CPm as A-site substrates in two different pre-steady-state assays (Youngman et al. 2004). In the first assay (Fig. 1A ▶), f-[35S]-Met-tRNAfMet was provided as a peptidyl-tRNA substrate, while in a second assay (Fig. 1B ▶), the length of the peptidyl moiety on the P-site substrate was increased to a dipeptide, f-[35S]-Met-Phe-tRNAPhe. The data are summarized in Table 2 ▶. These two assays are expected to report on similar catalytic properties and the results are generally comparable, though there are some differences. For example, the rate constants for reaction of Pm and CPm with the dipeptidyl tRNA substrate (f-[35S]-Met-Phe-tRNAPhe) are very similar (9.4 sec−1 vs. 11.9 sec−1 for Pm and CPm, respectively) and yet differ substantially with the simpler peptidyl substrate f-[35S]-Met-tRNAfMet (1.2 sec−1 vs. 18.9 sec−1 with Pm and CPm, respectively). Previous studies reported similar rate increases with puromycin as the peptidyl moiety in the P site was lengthened up to a pentamer (Katunin et al. 2002). These observations are consistent with the idea that the active site can assume alternative conformations and either the peptidyl moiety of the P-site tRNA or C75 of the A-site tRNA can stabilize the more catalytically active state.
TABLE 2.
Rate constants for peptidyl transfer of active site mutants
| fMetPm (sec−1) | fMetCPm (sec−1) | fMetPhePm (sec−1)a | fMetPheCPm (sec−1) | |
| Wild-type | 1.2 ± 0.2a | 18.9 ± 0.7 | 9.4 ± 2.5 | 11.9 ± 2.9 |
| A2451U | 0.07 ± 0.02a | 2.6 ± 0.6 | 0.05 ± 0.009b | 3.3 ± 0.6b |
| A2451C | 0.04 ± 0.02 | 1.0 ± 0.3 | 2.8 ± 0.04b,c 0.04 ± 0.007b,c |
0.6 ± 0.5b |
| A2451G | 0.01 ± 0.002 | 0.2 ± 0.006 | 0.02 ± 0.004b | 0.7 ± 0.8b |
| U2506C | 0.003 ± 0.0002a | 0.2 ± 0.1 | 0.002 ± 0.0003b | 0.4 ± 0.2 |
| U2506G | 0.005 ± 0.001 | 0.04 ± 0.01 | 0.002 ± 0.0003b,d | 0.08 ± 0.02b |
| U2506A | 0.009 ± 0.005 | 0.4 ± 0.01 | 0.001 ± 0.0004b | 0.4 ± 0.01b |
| U2585C | 0.02 ± 0.00a | 1.2 ± 0.1 | 0.02 ± 0.007 | 2.0 ± 0.8 |
| U2585G | 0.3 ± 0.04 | 2.0 ± 0.05 | 1.3 ± 0.07 | 8.6 ± 2.8 |
| U2585A | 0.05 ± 0.003 | 1.6 ± 0.04 | 0.02 ± 0.009 | 5.3 ± 2.5 |
| A2602C | 0.005 ± 0.0006a | 0.01 ± 0.006 | 0.03 ± 0.009 | 0.8 ± 0.3 |
| A2602U | 0.04 ± 0.02 | 0.07 ± 0.002 | 0.3 ± 0.1 | 1.9 ± 0.4 |
| A2602G | 0.01 ± 0.004 | 0.03 ± 0.01 | 0.07 ± 0.02 | 0.9 ± 0.7 |
PT reactions were carried out in buffer A at pH 7.5 and 10 mM Pm or CPm and, unless otherwise indicated. 7 mM Mg2+ and 20° C. The error listed is the standard deviation in the rate constants from at least two experiments.
aData from Youngman et al. (2004).
bRate constant measured at 15 mM Mg2+.
cTime courses for the fMetPhePm reaction on A2451C ribosomes consistently fit to double exponential curves. The rate constants for the first and second phases are reported where the first phase typically proceeds to an amplitude of 0.2 and the second phase to 0.5.
dRate constant measured at 37° C.
As a logical extension of these results, we were interested in knowing whether the addition of C74 (or more nucleotides) to CPm would further enhance rates of peptidyl transfer with f-[35S]-Met-tRNAfMet. However, the high K1/2 value for Pm and CPm (and presumably CCPm) and the large sample volumes required for rapid quench kinetics precluded us from performing these experiments under substrate saturating (kcat) conditions (since solution synthesis of large quantities of CCPm would be prohibitively expensive). When we compared CPm with CCPm at the subsaturating concentration of 5 μM (kcat/Km conditions), we saw no differences in the rate of peptidyl transfer (kobs = 0.01 sec−1) as might be detected if there were gross changes in the rates of catalysis or in K1/2 values (data not shown). The substrate CACCPm supplied at 5 μM did not react with the f-[35S]-Met-tRNAfMet substrate during the 2-min time course (data not shown), consistent with previous studies using puromycin derivatives of increasing length (Starck and Roberts 2002).
Peptidyl transferase activity of active site variant ribosomes with Pm and CPm as A-site substrates
We next measured the rate of peptide bond formation on active site variant ribosomes using Pm and CPm as A-site substrates. Our previous studies established that the variant ribosomes were unlikely to exhibit gross binding defects, and so the experiments were conducted with the same concentrations of Pm and CPm (10 mM) (Youngman et al. 2004). The general observation from these experiments is that defects associated with variant ribosomes when Pm is the A-site substrate are substantially recovered when CPm is used (Table 2 ▶, cf. columns 1 and 2, and cf. columns 3 and 4). For example, U2585A is quite deficient in peptidyl transfer with Pm as a substrate in both the dipeptide and tripeptide assays (0.05 sec−1 and 0.02 sec−1, respectively), whereas with CPm as a substrate, the rate constants are restored to 1.6 sec−1 and 5.3 sec−1. In some cases, the amount of activity restored is as much as 400-fold (for U2506A in the tripeptide assay). These data indicate that positioning by C75 of tRNA plays an important role in catalysis. There again appear to be some contributions made by the peptidyl moiety to active site function, as the recovery of activity by CPm is in a few cases more complete when a dipeptidyl f-[35S]-Met-Phe-tRNAPhe substrate is supplied in the P site as opposed to the simpler f-Met-tRNAfMet (see, for example, the A2602 mutants).
Determination of the equilibrium binding constant (Kd) for CPm and Pm by chemical modification analysis
Because of the additional hydrogen bonding interactions that are utilized by CPm in interacting with the ribosome, we had anticipated that its K1/2 (1.7 mM) might be substantially lower than that observed for Pm (3.8 mM). However, since the K1/2 value is minimally composed of three different rate constants (an on- and off-rate for the sub-strate [k1 and k−1] and the rate constant for peptidyl transfer [k2], it is not possible to extract a simple equilibrium binding constant (Kd) from these experiments. To further probe this phenomenon, we used a chemical footprinting procedure to establish the Kd of CPm and Pm for wild-type ribosomes. In this approach, increasing amounts of the puromycin substrates were added to 70S ribosomes carrying a model mRNA (gene 32) and the deacylated P-site substrate tRNAfMet. It was not possible to evaluate the properties of ribosome complexes carrying authentic peptidyl P-site substrates, as they would react with Pm and CPm and the measured Kd would represent that of a functionally (and perhaps structurally) distinct post-peptidyl transfer complex. The guanosine-specific reagent kethoxal was used to probe the accessibility of G2553 and G2505, two nucleotides known to be protected by bound Pm (Rodriguez-Fonseca et al. 2000). G2553 is an obvious candidate for such a protection experiment since this nucleotide directly interacts with C75 of the aminoacyl tRNA (Kim and Green 1999; Nissen et al. 2000). Primer extension analysis was used to visualize the extent of protection of these nucleotides at various Pm and CPm concentrations (Moazed and Noller 1986). Quantitation of such binding experiments established that the Kd of CPm for the ribosomal complexes is 14 μM whereas the Kd for Pm is 1.8 mM (Table 1 ▶). The relatively high Kd value for puromycin was independently confirmed using DMS modification enhancements at positions A508 and A1579 (Rodriguez-Fonseca et al. 2000; data not shown).
It is interesting to note the substantial discrepancy between the K1/2 and the Kd for CPm, in contrast to the similarity of these values for Pm. K1/2 and Kd values are generally the same for a given substrate (as is the case for puromycin) when the kcat of the reaction is slow relative to substrate dissociation. Conversely, differences between K1/2 and Kd values are generally seen when the kcat of the reaction is relatively fast (i.e., when compared to k−1), thus making the enzymatic reaction require more substrate to reach saturation than would be required to reach saturation for binding. In this case, we have information indicating that the kcat and K1/2 values for CPm and Pm are equivalent and yet that their Kd values differ by two orders of magnitude. Since the kcat values are the same for the two substrates, a relatively simple explanation for the similarities in K1/2 value would be a smaller value for k−1 for CPm that results in a stronger equilibrium binding constant (Kd) but has little overall effect on the K1/2 of the reaction. This seems probable, as the two orders of magnitude difference in Kd (~3 kcal/mol of energy) is in a range consistent with the three additional hydrogen bonds associated with the interaction between C75 and G2553 of the A loop of the ribosome. Although a more compelling argument would require direct measurement of k−1, this experiment is not trivial and was thus not pursued.
pH-dependence of peptidyl transfer with Pm and CPm substrates
The ability of CPm to mask the deficiencies of active site mutations seen with Pm suggested the interesting possibility that this additional nucleotide might somehow activate the peptidyl transferase site for catalysis. Such an idea would be consistent with earlier reports that the trinucleotide CCA can stimulate the related reaction of peptide release from peptidyl-tRNA bound in the P site (Caskey et al. 1971). Moreover, there are recent reports of conformational rearrangements in the active site of the ribosome dependent on the presence of C74 and C75 on tRNA in the A site (Schmeing et al. 2005). Here we observed differences in the rate of catalysis with CPm and Pm on certain substrates (f-Met-tRNAfMet) but not with others (the dipeptidyl f-Met-Phe-tRNAPhe) and wondered whether under the latter conditions catalytic differences might also exist but be masked by other rate-limiting steps. We reasoned that if CPm functions in some way to activate catalysis (for example, by promoting productive conformational rearrangements), then catalysis might be more rapid with CPm than with Pm at low pH where chemistry is more likely to be rate limiting for both reactions.
To address this possibility, we examined the rates of peptidyl transfer with Pm and CPm in the tripeptide assay over a range of pH values. These experiments highlight overall substantial differences in the response of these two peptidyl transfer reactions to pH (Fig. 3 ▶). Although both reactions are dependent on pH, the shapes of the curves suggest that different protonation events might be involved in the two reactions. The pH-rate profile that we observe for Pm is similar to that previously reported by Rodnina and colleagues where the plot of log(kpep) versus pH yields a slope of ~1.3 and kpep plateaus at 12 sec−1 at pH values > 7.5 (Katunin et al. 2002). The pH-rate profile for CPm has a much shallower slope of 0.6 but plateaus at a similar kpep of 12 sec−1 at pH values > 7. Further, as proposed above, at low pH values, CPm is a more reactive substrate than Pm (about 20-fold faster). These overall catalytic differences revealed by pH titrations are consistent with the different rates of catalysis by Pm and CPm in the dipeptide reaction presented above (Table 2 ▶).
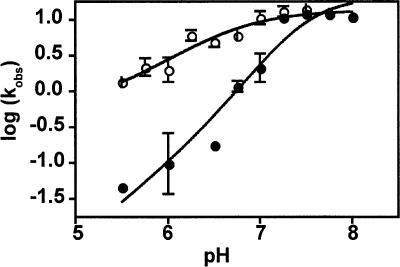
FIGURE 3.
pH-dependence of PT kinetics with Pm and CPm. Observed rate constants for peptide bond formation were measured in the tripeptide assay at 20° C and saturating (10 mM) Pm (closed circles) or CPm (open circles) at pH values between 5.5 and 8. Error bars represent the standard deviation from at least two independent measurements. For Pm, the curve shown is the fit to a model with two ionizing groups whose ionization state contributes to catalysis; for CPm, the curve is the fit to a model with one ionizing group (see Materials and Methods). The slopes of the linear portions of the curves are 1.3 ± 0.1 for Pm and 0.6 ± 0.07 for CPm.
As previously discussed (Katunin et al. 2002), the data for Pm are most simply explained by a model involving two critical protonation events. Fitting of the earlier data to the simplest potential model yielded two distinct pKa values of 6.9 and 7.5. These studies argued that pKa1 (the lower value) corresponds to protonation of the primary amine of puromycin leaving pKa2 to represent an unidentified ribosomal moiety whose protonation state is relevant to catalysis. This apparent pKa value may correspond to a general base involved in the reaction, a pH-dependent conformational change (Bayfield et al. 2001; Muth et al. 2001) or even to an aggregate pKa value representing a larger number of titratable groups of pKa far removed from this value (Knitt and Herschlag 1996). Subsequent studies have focused on trying to identify which nucleotide(s) might function as a general base or as the pH-dependent conformational switch critical for the peptidyl transfer reaction, though no definitive answer has emerged (Katunin et al. 2002; Beringer et al. 2003; Hesslein et al. 2004; Youngman et al. 2004; Okuda et al. 2005).
Importantly, the pH-rate profile for CPm described here is strikingly different from that observed for Pm. As for Pm, the highest relevant pKa can be directly taken from the log(kpep) versus pH plot as 7.0, but in this case the slope is substantially more shallow (0.6 ± 0.07). In contrast to the Pm reaction, these data cannot be fit to a model involving two protonation events that affect catalysis, but instead are well fit to a model with a single relevant pKa. The simplest interpretation of these data is that only a single protonation event is critical to peptidyl transfer with this substrate. We suggest that this single catalytically relevant protonation event is equivalent to pKa1 (6.9) observed with Pm and that it corresponds to deprotonation of the attacking nucleophilic amine group on these related compounds. Direct titrations of CPm with KOH were consistent with this proposal (data not shown).
While it is likely that the chemistry of peptidyl transfer is rate limiting for the CPm reaction at low pH, our data cannot establish that this is true over the entire range of pH, as has been shown likely for Pm (Katunin et al. 2002; Seila et al. 2005). Consequently, we cannot exclude the possibility that the shallower pH profile observed for the reaction with CPm reflects a change in rate-limiting step rather than the loss of a catalytically important pKa. While such alternative explanations cannot be excluded, the simplest view of these data is that the second catalytically relevant protonation event identified for the Pm reaction is not relevant for the reaction with CPm as substrate. In molecular terms, C75 interactions with the A loop promote changes (presumably conformational) in the active site that obviate the need for deprotonation to activate catalysis.
DISCUSSION
These studies were undertaken to understand at a molecular level the very different behaviors of active site ribosomal variants with the minimal substrate puromycin and with intact aminoacyl-tRNAs (Youngman et al. 2004). Earlier studies had suggested that the CCA end of the tRNA could activate the large subunit active site to catalyze the related peptidyl hydrolysis reaction (Caskey et al. 1971). We reasoned that known interactions between C75 of aminoacyl-tRNA and the A loop of the ribosome (Kim and Green 1999; Nissen et al. 2000) might be responsible for activation of peptidyl hydrolysis as well as peptidyl transfer. In the series of experiments described here, we provide critical support to indicate that interactions between C75 of aminoacyl-tRNA and the ribosome are important for promoting catalysis in the large subunit active site.
First, we show that catalytic deficiencies associated with active site ribosomal variants are substantially reversed when a single C75 equivalent is appended to the minimal puromycin substrate. From these results we conclude that restoration of activity by aminoacyl-tRNA substrates as reported (Youngman et al. 2004) must be, to a large extent, a consequence of interactions between C75 of tRNA and the A loop of the ribosome. A second important observation is that the pH dependence of the reaction changes substantially with the addition of the C75 equivalent to the minimal puromycin substrate. These data reveal a shallower pH-dependence for the CPm peptidyl transfer reaction and further suggest that a single proton is important for optimal catalysis, rather than the two protons reported for the puromycin reaction (Katunin et al. 2002). A potential explanation for this effect might be that CPm induces productive conformational changes in the active site that in the context of puromycin are accomplished by the titration of specific protons on the ribosome itself. Such conformational rearrangements in the active site, some dependent on pH, have been previously described (Miskin et al. 1968; Bayfield et al. 2001; Muth et al. 2001). In an attempt to correlate these earlier observations with our CPm data, we asked whether peptidyl transfer activity inactivated by dialysis against low monovalent ions (Miskin et al. 1968) was recovered by the presence of C75 on puromycin. These experiments were not successful (data not shown) and, in turn, suggest that the “Zamir-Elson” conformational activation is not equivalent to the one probed by our CPm data.
We suggest that conformational changes occur as a consequence of the interaction of C75 with the A loop of the ribosome and, as such, that tRNA itself would be predicted to induce similar structural rearrangements. In other recent studies using CPm as a ribosomal substrate (Okuda et al. 2005), the pH dependence of the reaction exhibited a slope of 2.0, consistent with the involvement of two distinct deprotonation events for optimal catalysis. Interestingly, in that study, a modified “fragment reaction” was used that depends on catalysis by the 50S subunit alone. This reaction involving only the large subunit is apparently not activated in the same fashion by the C75 equivalent in tRNA. Such differences may, in part, explain the relatively slow rates (0.06 sec−1) of catalysis in such minimal reaction systems (Schmeing et al. 2002).
The CPm data presented here provide a potential kinetic correlate for a number of structural observations (Schmeing et al. 2005; Bayfield et al. 2001; Muth et al. 2001). In particular, recent structural studies suggest that substantial conformational rearrangements are observed in this active site in the presence of both the C74 and the C75 nucleotides of the tRNA (Schmeing et al. 2005). Our data, however, indicate that the active site is substantially dependent on the C75 moiety of the aminoacyl substrate for optimal catalysis. Perhaps the C74 moiety makes important stacking interactions within the substrate that favor stabilization of the structure trapped by X-ray crystallography. It is not surprising that reactivity in this biologically central process is strongly buffered, here by a somewhat remote and stable interaction, so that it is generally insensitive to a variety of environmental perturbations.
MATERIALS AND METHODS
Peptidyl transferase assays
MRE wild-type ribosomes, MS2-tagged wild-type and variant ribosomes, gene 32 mRNA, f[35S]Met-tRNAfMet, Phe-tRNAPhe, EF-Tu, and EF-G were prepared as previously described (Youngman et al. 2004; Youngman and Green 2005). Initiation factors 1 and 3 were overexpressed (Spurio et al. 1991) and purified as previously described (Soffientini 1994; Dahlquist and Puglisi 2000; Dallas and Noller 2001). 6XHis-tagged IF2 was overexpressed and purified on Qiagen Ni-NTA resin by standard procedures. Cytidine-(2′-5′)-[α-Amino-p-methoxyhydrocinnamamido)]-3′-deoxy-N,N-dimethyl-adenosine (CPm) was synthesized by solution coupling reactions by TriLink BioTechnologies Inc. CPm was resuspended in water at a concentration of 30–50 mM and stored at −80° C. For use in kinetic assays, it was diluted in buffer A (50 mM Tris-HCl at pH 7.5, 70 mM NH4Cl, 30 mM KCl, 7 mM MgCl2, 1 mM DTT) and neutralized with ammonium hydroxide.
For dipeptide reactions, initiation complexes were formed and reacted directly with Pm or CPm as previously described (Youngman et al. 2004). Reactions were quenched with 25% formic acid and spotted on TLC-cellulose plates for electrophoresis since the previously used EDTA quench was insufficient for quenching the earliest time points in the CPm reactions. No specific hydrolytic step was used in this procedure since the dipeptidyl product falls off the ribosome upon reaction with Pm or CPm, and the unreacted f-[35S]-Met-tRNAfMet can be accounted for by simply including counts remaining at the origin.
For tripeptide reactions, reaction complexes were prepared and tested as described (Youngman et al. 2004) with the following modifications for reactions with CPm. Reaction complexes were mixed with CPm in the quench-flow machine and quenched with 25% formic acid (20° C). Reactions were hydrolyzed with 10 N KOH on ice for 10 min and spotted on TLC-cellulose plates for electrophoresis as described. These modifications were necessary to efficiently stop the CPm reaction and were sufficient to hydrolyze the product from the tRNA without extraneous hydrolysis products observed at higher temperatures. As previously reported (Youngman et al. 2004), some mutant ribosomes required additional Mg+2 to avoid endpoint deficiencies. To measure K1/2, the rates of tripeptide formation were measured at 13 different concentrations of CPm (ranging from 0.125 mM to 15 mM) and the resulting plots of rate versus [CPm] were fit to a hyperbolic (Michaelis–Menten) equation. The reported deviations are the error in the fit to the averaged data points from at least two experiments.
For determining the pH dependence of tripeptide formation, initiation and ternary complexes were formed generally as described above but with the following modifications. Initiation complexes were formed in buffer B (20 mM Bis-Tris, 50 mM Tris, 30 mM KCl, 70 mM NH4Cl, 7 mM MgCl2 at pH 5.5–8.0). After mixing the initiation and ternary complexes to form f-[35S]-Met-Phe tRNAPhe, the pH was measured with a microelectrode (Calomel MicroCombination Electrode, VWR) and adjusted to the desired pH. Reaction complexes were mixed in the quench-flow machine with Pm or CPm, diluted in buffer B at the appropriate pH, and quenched with 25% formic acid (20° C). Reactions were hydrolyzed with 10 N KOH on ice for 10 min and spotted on TLC-cellulose plates for electrophoresis as above.
Plots of log kobs versus pH were then fit to equations modeling a system where catalysis is dependent on either one or two ionizing groups (Fersht 1984). For a single ionizing group, the equation is log(kobs) = log(10− [H] Vmin + VmaxKa) – log(Ka + 10− [H]). For two ionizing groups, the equation is log(kobs) = log[(10− [H] K1VHA− ) + (K1K2Vmax)]− log[ K1K2 + 10− [H] K1 + (10− 2[H])], where K1 is the first ionization constant, K2 is the second ionization constant, and VHA− is the rate constant for the singly deprotonated species. The data for CPm could only be fit to the equation for the single Ka equation, whereas the data for Pm could be fit only to the 2 Ka equation.
Chemical footprinting
Ribosome complexes were formed by incubating 15 pmol 70S MRE600 ribosomes, 60 pmol gene 32 mRNA, and 45 pmol of deacylated tRNAfmet in 12.5 μL of buffer C (80 mM K-HEPES at pH 7.8, 15 mM MgCl2, 100 mM NH4Cl) at 37° C for 20 min. These reaction complexes were then mixed with an equal volume of either CPm (final concentration ranging from 5 μM to 1 mM) or Pm (final concentration ranging from 0.1 to 10 mM) prepared in buffer C, and the complete mixture was incubated at 37° C for 5 min. The ribosome complexes were then modified with kethoxal (from MP Biomedicals) as described (Merryman et al. 1999), with the following changes. The modification reactions were carried out at 37° C for 10 min in the presence of 25 mM kethoxal (by adding to the ribosomal complexes 1 μL of 0.625 M solution of kethoxal prepared by diluting a 2.5 M stock solution of kethoxal in ethanol with 3 volumes of water). The RNA was isolated and analyzed by primer extension essentially as described (Merryman et al. 1999). Primer extension gels were analyzed using a Molecular Dynamics Phosphorimager. The density of the protected band was normalized to a fixed band in the same primer extension lane and corrected by subtracting the normalized density of the corresponding band in the control lane modified with kethoxal in the absence of CPm and Pm. These values were then plotted against the CPm or Pm concentration, and the plots were fit to a hyperbolic equation to obtain the respective Kds.
Acknowledgments
We thank A. Kochaniak for early experiments and J. Lorsch, C. Merryman, and other members of the Green laboratory for helpful discussions and critical review of the manuscript. The work was supported by NIH grant R01GM059425 and salary support for R.G. was from HHMI.
Article and publication are at http://www.rnajournal.org/cgi/doi/10.1261/rna.2256706.
REFERENCES
- Bayfield, M.A., Dahlberg, A.E., Schulmeister, U., Dorner, S., and Barta, A. 2001. A conformational change in the ribosomal peptidyl transferase center upon active/inactive transition. Proc. Natl. Acad. Sci. 98: 10096–10101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beringer, M., Adio, S., Wintermeyer, W., and Rodnina, M. 2003. The G2447A mutation does not affect ionization of a ribosomal group taking part in peptide bond formation. RNA 9: 919–922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caskey, C.T., Beaudet, A.L., Scolnick, E.M., and Rosman, M. 1971. Hydrolysis of fMet-tRNA by peptidyl transferase. Proc. Natl. Acad. Sci. 68: 3163–3167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cate, J.H., Yusupov, M.M., Yusupova, G.Z., Earnest, T.N., and Noller, H.F. 1999. X-ray crystal structures of 70S ribosome functional complexes. Science 285: 2095–2104. [DOI] [PubMed] [Google Scholar]
- Dahlquist, K.D. and Puglisi, J.D. 2000. Interaction of translation initiation factor IF1 with the E. coli ribosomal A site. J. Mol. Biol. 299: 1–15. [DOI] [PubMed] [Google Scholar]
- Dallas, A. and Noller, H.F. 2001. Interaction of translation initiation factor 3 with the 30S ribosomal subunit. Mol. Cell 8: 855–864. [DOI] [PubMed] [Google Scholar]
- Fersht, A. 1984. Enzyme structure and mechanism. W.H. Freeman and Company, New York.
- Hesslein, A.E., Katunin, V.I., Beringer, M., Kosek, A.B., Rodnina, M.V., and Strobel, S.A. 2004. Exploration of the conserved A + C wobble pair within the ribosomal peptidyl transferase center using affinity purified mutant ribosomes. Nucleic Acids Res. 32: 3760–3770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katunin, V.I., Muth, G.W., Strobel, S.A., Wintermeyer, W., and Rodnina, M.V. 2002. Important contribution to catalysis of peptide bond formation by a single ionizing group within the ribosome. Mol. Cell 10: 1–20. [DOI] [PubMed] [Google Scholar]
- Kim, D.F. and Green, R. 1999. Base-pairing between 23S rRNA and tRNA in the ribosomal A site. Mol. Cell 4: 859–864. [DOI] [PubMed] [Google Scholar]
- Knitt, D.S. and Herschlag, D. 1996. pH dependencies of the Tetrahymena ribozyme reveal an unconventional origin of an apparent pKa. Biochemistry 35: 1560–1570. [DOI] [PubMed] [Google Scholar]
- Merryman, C., Moazed, D., McWhirter, J., and Noller, H.F. 1999. Nucleotides in 16S rRNA protected by the association of 30S and 50S ribosomal subunits. J. Mol. Biol. 285: 97–105. [DOI] [PubMed] [Google Scholar]
- Miskin, R., Zamir, A., and Elson, D. 1968. The inactivation and reactivation of ribosomal-peptidyl transferase of E. coli. Biochem. Biophys. Res. Commun. 33: 551–557. [DOI] [PubMed] [Google Scholar]
- Moazed, D. and Noller, H.F. 1986. Transfer RNA shields specific nucleotides in 16S ribosomal RNA from attack by chemical probes. Cell 47: 985–994. [DOI] [PubMed] [Google Scholar]
- Muth, G.W., Chen, L., Kosek, A.B., and Strobel, S.A. 2001. pH-dependent conformational flexibility within the ribosomal peptidyl transferase center. RNA 7: 1403–1415. [PMC free article] [PubMed] [Google Scholar]
- Nissen, P., Hansen, J., Ban, N., Moore, P.B., and Steitz, T.A. 2000. The structural basis of ribosome activity in peptide bond synthesis. [See comments.] Science 289: 920–930. [DOI] [PubMed] [Google Scholar]
- Okuda, K., Seila, A.C., and Strobel, S.A. 2005. Uncovering the enzymatic pKa of the ribosomal peptidyl transferase reaction utilizing a fluorinated puromycin derivative. Biochemistry 44: 6675–6684. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Fonseca, C., Phan, H., Long, K.S., Porse, B.T., Kirillov, S.V., Amils, R., and Garrett, R.A. 2000. Puromycin–rRNA interaction sites at the peptidyl transferase center. RNA 6: 744–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmeing, T.M., Seila, A.C., Hansen, J.L., Freeborn, B., Soukup, J.K., Scaringe, S.A., Strobel, S.A., Moore, P.B., and Steitz, T.A. 2002. A pre-translocational intermediate in protein synthesis observed in crystals of enzymatically active 50S subunits. Nat. Struct. Biol. 9: 225–230. [DOI] [PubMed] [Google Scholar]
- Schmeing, T.M., Huang, K., Strobel, S.A., and Steitz, T.A. 2005. An induced-fit mechanism to promote peptide bond formation and exclude hydrolysis of peptidyl-tRNA. Nature (in press). [DOI] [PubMed]
- Seila, A.C., Okuda, K., Nunez, S., Seila, A.F., and Strobel, S.A. 2005. Kinetic isotope effect analysis of the ribosomal peptidyl transferase reaction. Biochemistry 44: 4018–4027. [DOI] [PubMed] [Google Scholar]
- Sievers, A., Beringer, M., Rodnina, M.V., and Wolfenden, R. 2004. The ribosome as an entropy trap. Proc. Natl. Acad. Sci. 101: 7897–7901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soffientini A.L.R., GastaldoL., Parlett, J.H., Spurio, R., Lateana, A., and Islam, K. 1994. Purification procedure for bacterial translational initiation factors IF2 and IF3. Protein Expr. Purif. 5: 118– 124. [DOI] [PubMed] [Google Scholar]
- Spurio, R., Paci, M., Pawlik, R.T., La Teana, A., DiGiacco, B.V., Pon, C.L., and Gualerzi, C.O. 1991. Site-directed mutagenesis and NMR spectroscopic approaches to the elucidation of the structure–function relationships in translation initiation factors IF1 and IF3. Biochimie 73: 1001–1006. [DOI] [PubMed] [Google Scholar]
- Starck, S.R. and Roberts, R.W. 2002. Puromycin oligonucleotides reveal steric restrictions for ribosome entry and multiple modes of translation inhibition. RNA 8: 890–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinger, J.S., Parnell, K.M., Dorner, S., Green, R., and Strobel, S.A. 2004. Substrate-assisted catalysis of peptide bond formation by the ribosome. Nat. Struct. Mol. Biol. 11: 1101–1106. [DOI] [PubMed] [Google Scholar]
- Youngman, E.M. and Green, R. 2005. Affinity purification of in vivo-assembled ribosomes for in vitro biochemical analysis. Methods 36: 305–312. [DOI] [PubMed] [Google Scholar]
- Youngman, E.M., Brunelle, J.L., Kochaniak, A.B., and Green, R. 2004. The active site of the ribosome is composed of two layers of conserved nucleotides with distinct roles in peptide bond formation and peptide release. Cell 117: 589–599. [DOI] [PubMed] [Google Scholar]