Abstract
Synthesis of wild-type levels of turnip crinkle virus (TCV)-associated satC complementary strands by purified, recombinant TCV RNA-dependent RNA polymerase (RdRp) in vitro was previously determined to require 3′ end pairing to the large symmetrical internal loop of a phylogenetically conserved hairpin (H5) located upstream from the hairpin core promoter. However, wild-type satC transcripts, which fold into a single detectable conformation in vitro as determined by temperature-gradient gel electrophoresis, do not contain either the phylogenetically inferred H5 structure or the 3′ end/H5 interaction. This implies that conformational changes are required to produce the phylogenetically inferred H5 structure for its pairing with the 3′ end, which takes place subsequent to the initial conformation assumed by the RNA and prior to transcription initiation. The DR region, located 140 nucleotides upstream from the 3′ end and previously determined to be important for transcription in vitro and replication in vivo, is proposed to have a role in the conformational switch, since stabilizing the phylogenetically inferred H5 structure decreases the negative effects of a DR mutation in vivo. In addition, high levels of aberrant transcription correlate with a specific conformational change in the Pr while maintaining the same conformation of the 3′ terminus. These results suggest that a series of events that promote conformational changes is needed to expose the 3′ terminus to the RdRp for accurate synthesis of wild-type levels of complementary strands in vitro.
Keywords: RNA-dependent RNA polymerase, RNA virus replication, satellite RNAs, temperature-gradient gel electrophoresis, RNA structure rearrangement, RNA switches
INTRODUCTION
Replication of (+)-strand RNA viruses requires reiterative copying of the infecting genome to generate complementary (−)-sense intermediates, followed by reiterative copying of the intermediates to generate progeny (+)-strand RNAs. The viral-encoded RNA-dependent RNA polymerase (RdRp) must associate specifically with its cognate RNA and interact with the 3′ end for de novo or primer-dependent initiation of RNA synthesis (Kao et al. 2001; van Dijk et al. 2004). All RdRps must generate copies of the infecting RNA using a similar mechanism of catalysis, reflected by conservation of basic structural features such as a right-hand-like form with fingers, palm, and thumb subdomains (van Dijk et al. 2004). While RdRp initiation complexes composed of template, RdRp, initiation NTP, and a second NTP have been well defined for several viruses, little is known about the events that lead up to the establishment of this complex.
Promoter elements that specifically interact with the polymerase prior to initiation of complementary strand synthesis are generally located proximal to the 3′ terminus and usually comprise one or more hairpins and adjoining single-stranded sequence (Dreher 1999). Using a reductionist approach, core promoters for (−)-strand synthesis have been identified for many (+)-strand viruses (Duggal et al. 1994; Buck 1996; Chapman and Kao 1999; Dreher 1999; Turner and Buck 1999). Core promoters generally are composed of one or a few 3′ proximal hairpins that contain multiple sequence and structural features required for efficient RdRp recognition.
While core promoters are capable of directing complementary strand synthesis without additional viral sequences, an increasing number of elements that positively or negatively influence replication have been found distal to the 3′ ends of both (+)- and (−)-strands. These include elements at the 5′ ends that may be required for genome circularization (Barton et al. 2001; Frolov et al. 2001; Herold and Andino 2001; Khromykh et al. 2001; You et al. 2001), and internal elements such as repressors, enhancers, and other replication-required elements, which function either in cis (Kim and Makino, 1995; Klovins et al. 1998; Nagy et al. 1999; Ray and White 1999 Ray and White 2003; Murray and Barton 2003; Panavas and Nagy 2003; Pogany et al. 2003; Zhang and Simon 2003; Zhang et al. 2004b) or in trans (Sit et al. 1998; Eckerle and Ball 2002). Enhancers are generally found on viral (−)-strands, need not be proximal to the core promoter, contain sequence and/or structural features of core promoters, and can promote transcription in the presence of sequences resembling the transcription initiation site (Nagy et al. 1999; Panavas and Nagy 2003; Ray and White 2003). Repressors (silencers) have been identified for the family Tombusviridae and are located on (+)-strands just upstream from the core promoter. The positioning of transcriptional enhancing and repressing elements on opposite strands has led to the suggestion that these elements function to regulate asymmetric levels of (+)- and (−)-strand synthesis (Pogany et al. 2003; Zhang et al. 2004b).
The discovery of replication-associated elements throughout viral genomes suggests that the generation of progeny (+)-strands, which requires the cessation of translation of the input template and usually the synthesis of asymmetric levels of (+)- and (−)-strands (Buck 1996), is a much more complicated process than previously thought. Recent evidence obtained using several unrelated viruses suggests that RNA conformational switches may be needed to hide and expose viral 3′ ends, which may temporally regulate (+)- and (−)-strand synthesis. Some viruses appear to activate these switches by changing the conformation of 3′-proximal structures, a process mediated by one or more unstable base pairs between complementary short sequences located within and outside of hairpins (Olsthoorn et al. 1999; Koev et al. 2002; Pogany et al. 2003; Zhang et al. 2004b). For example, Barley yellow dwarf virus is proposed to repress (−)-strand synthesis by embedding its 3′ end in a “pocket” structure, thus making it unavailable to the RdRp (Koev et al. 2002). A similar molecular switch in the Mouse hepatitis virus genome involves sequences within a stem–loop and pseudoknot in the 3′ untranslated region (UTR) (Goebel et al. 2004). In addition to cis-acting sequences, trans-acting cellular factors or virally encoded proteins may affect the balance between alternative structural conformations (Olsthoorn et al. 1999; Schuppli et al. 2000). For example, the 3′ terminal five bases of Qβ bacteriophage are involved in long-distance base-pairing that does not permit efficient access to the polymerase. Either the host protein Hfq or a series of mutations, including alterations to the 3′ end and interacting sequence, is needed for efficient replication to destabilize the secondary structure in the region and allow access of the polymerase to the 3′ end. These findings among unrelated viruses suggest that conformational switches preceding transcription initiation may be a general requirement for exposing the 3′ end to the RdRp and controlling both the timing and levels of viral RNA synthesis.
Turnip crinkle virus (TCV), a member of the genus Carmovirus in the family Tombusviridae, has a monopartite genome of 4054 nucleotides (nt) (Carrington et al. 1989; Oh et al. 1995). TCV genomic RNA is translated into p28 and the ribosomal read-through product p88, which are required for virus replication (Hacker et al. 1992). p88 contains the conserved polymerase active site motif GDD and alone is capable of directing complementary strand synthesis of exogenously added (+)- and (−)-strands of TCV subviral RNAs in vitro, producing double-stranded products that are not templates for further transcription (Rajendran et al. 2002). TCV is able to support the replication of several subviral RNAs, including satC (356 nt), a hybrid molecule composed of sequence unrelated to TCV at the 5′ end and two regions with 90% similarity to TCV at the 3′ end (Simon and Howell 1986).
Phylogenetic analysis of carmoviral 3′ UTR combined with computer mFold modeling (Zuker 2003) led to the proposal that the related 3′ regions of TCV and satC contain four hairpins (H4a, H4b, H5, Pr) (Zhang et al. 2004b). Using in vitro and in vivo approaches, the Pr was previously identified as the satC core promoter (Song and Simon 1995; Carpenter and Simon 1998). H5, also critical for satC and TCV replication (McCormack and Simon 2004; Zhang et al. 2004a; Zhang and Simon 2005), contains a large symmetrical internal loop (LSL) whose sequence is highly conserved among related carmoviruses (Zhang et al. 2004b). A combination of approaches including single-site mutagenesis, in vivo genetic selection (SELEX), and sequence replacements with analogous segments from the related carmovirus Cardamine chlorotic fleck virus (CCFV) indicates that both specific sequence and structural features throughout H5 are necessary for robust satC accumulation in plants and protoplasts (Zhang et al. 2004a; Zhang and Simon 2005).
Mutations in either the 3′ terminal cytidylates or the 3′ side of the H5 LSL led to enhanced transcription in vitro, and transcription was reduced to near wild-type levels when compensatory alterations restored possible interaction between the two sequences (Zhang et al. 2004b). SatC with a deletion of the 3′ terminal three cytidylates contained structural alterations in the H5 region and also produced high levels of transcription in vitro, with many products shorter than full-length. The suggestion was made that releasing the 3′ end from its partner sequence in the H5 LSL might lead to rearrangement of the local structure, an event required for activation of (−)-strand synthesis. However, wild-type satC transcripts fold into an initial conformation that does not contain the phylogenetically inferred H5 structure (Zhang et al. 2004b), bringing into question at what point the 3′ end interacts with H5 in the progression of events leading to transcription initiation in vitro. We now report that wild-type satC transcripts, which fold into a single detectable conformation in vitro as determined by temperature-gradient gel electrophoresis (TGGE), do not contain the 3′ end/H5 LSL interaction. This implies that a conformational switch is required to produce the phylogenetically inferred H5 structure for its pairing with the 3′ end, which takes place subsequent to the initial conformation assumed by the RNA and prior to transcription initiation. The DR region, which flanks the 5′ side of H4a and was previously determined to be important for transcription in vitro and replication in vivo (Sun et al. 2005), is proposed to have a role in the conformational switch, since stabilizing the phylogenetically inferred H5 structure decreases the negative effects of a DR mutation in vivo. In addition, high levels of aberrant transcription in vitro correlate with a specific conformational change in the Pr while maintaining the initial conformation of the 3′ terminus. These results suggest that conformational changes in the 3′ region are needed to expose the 3′ terminus to the RdRp for accurate synthesis of wild-type levels of complementary strands in vitro.
RESULTS
Transcripts of satC (+)-strands synthesized in vitro fold into one detectable conformation
Our previous results indicated that enhanced synthesis of satC complementary strands by the TCV RdRp in vitro, which occurred when templates were missing their 3′ terminal three cytidylates, correlated with substantial structural alterations in the H5 region (Zhang et al. 2004b). However, due to the methodology of the solution structure probing, we were unable to determine whether the H5 structural changes were accompanied by changes in the core Pr promoter located proximal to the 3′ end. For that study, transcripts generated by T7 RNA polymerase run-off transcription in vitro were prepared in an identical fashion for both solution structure analysis and RdRp assays. This procedure did not involve treatments subsequent to transcription that can eliminate or vastly reduce metastable misfolded intermediates that become kinetically trapped in the folding process of most in vitro synthesized RNAs (Emerick and Woodson 1993; Pan et al. 1997). Therefore, before continuing our structural analysis of satC, it was necessary to determine the structural homogeneity of satC transcripts synthesized in vitro.
To determine if wild-type satC transcripts contain stable and metastable forms following transcription in vitro, transcripts synthesized using T7 RNA polymerase were subjected to various treatments, followed by analysis of the resulting folded molecules using TGGE (Rosenbaum and Riesner 1987; Riesner et al. 1989). TGGE establishes a temperature gradient within the gel that allows RNAs to adopt conformations according to their stability at the underlying temperature and undergo transitions as the temperature increases. TGGE has been successfully employed to determine if different structures coexist in solution in equilibrium or if structural ensembles include metastable intermediates separated by an activation energy barrier that prevents conversion to the equilibrium structures (Baumstark and Riesner 1995; Baumstark et al. 1997; Schroeder et al. 1998). The separation and analysis of coexisting structures are possible since molecules of the same size, yet different conformation and stability, undergo local or global transitions at different temperatures and to different degrees, which can be detected by changes in hydrodynamic properties, i.e., shifted bands in the gel (Riesner and Steger 2005).
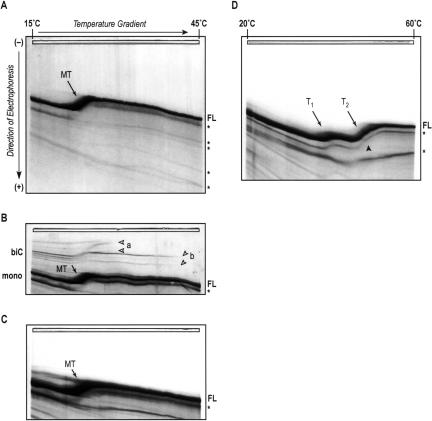
To determine the potential for wild-type satC to adopt different structures, we subjected product from T7 in vitro transcription to different structure-promoting pretreatments, followed by analysis of the resulting RNA structures using TGGE (Fig. 1 ▶). As a first approach, transcripts were denatured in low-salt TE buffer at 95°C and then rapidly renatured by transfer to ice. This procedure dissociates biologically irrelevant bimolecular complexes involving either full-length satC or shorter transcription products that frequently form during in vitro transcription reactions. Furthermore, snap-cooling in low salt promotes RNA folding into conformations that are kinetically favored (Baumstark and Riesner 1995). Under these conditions, satC adopts a single conformation characterized by one major band at the low-temperature side of the gradient gel (Fig. 1A ▶). This RNA structure undergoes a single transition at 23.7°C, and gel temperatures up to 60°C and inclusion of 4 M urea (not shown) did not provide evidence for any further conformational changes. The transition width of ~4°C is fairly narrow, implying that structural elements somewhat cooperatively dissociate in this transition. Typically, two types of transitions can be observed in a TGGE experiment: Reversible transitions appear as smooth curves, where the band thickness does not change before, during, or after the transition. Irreversible transitions display a stepwise behavior, characterized by a discontinuous mobility change at the transition temperature (Riesner and Steger 2005). For satC transcripts, the broadening of the band during the transition accompanied by a slightly fuzzy definition indicates a conformational change that is not completely reversible under the low ionic strength electrophoresis conditions of the TGGE. Shorter transcription products that were not removed for this analysis mirror the behavior of the full-length species down to a certain threshold length (Fig. 1A ▶, asterisks). While we cannot rule out that some secondary structure in the form of several small hairpins may still be present after the main observed transition, their independent denaturation would arguably not contribute to a mobility change large enough to visibly alter the migration of the 356-nt satC RNA in this type of electrophoretic separation. We therefore conclude that the majority, if not all, of the wild-type satC RNA structure assumed following heating and snap-cooling is disrupted in a single step in 0.2× TBE.
FIGURE 1.
TGGE analysis of structures formed by wild-type satC transcripts. Transcripts (500 ng) synthesized in vitro and not size-selected were treated as outlined in the text and below and then subjected to gel electrophoresis in 0.2× TBE with temperature gradients from 15°C–45°C (A–C), or in 0.2× TB buffer including 10 μM MgOAc with a gradient from 20°C–60°C (D). RNA bands were visualized by silver staining (Schumacher et al. 1986). (A) SatC transcripts in TE buffer were pretreated by denaturation and snap-cooling on ice. Full-length RNA (FL) denatures in a one-step main transition (MT). The arrow points to a midpoint of 23.7°C. Shorter RNA species generated by abortive T7 transcription events are marked by asterisks at the high-temperature side of the gel. (B) SatC transcripts were untreated prior to TGGE. The untreated RNA also folds into a single structural form with identical transition temperature as in (A). In vitro transcription reactions also yielded low levels of bimolecular complexes (biC) that denature into the single strands at higher transition temperatures (B, a,b). These complexes are formed by nascent RNA strands base-pairing with each other under the conditions of the T7 reaction mixture, and are disfavored by the low-salt snap-cooling treatment shown in (A). (C) SatC transcripts were denatured and slowly renatured in high-salt buffer and then dialyzed against 0.2× TBE buffer (see Materials and Methods). These conditions promote thermodynamic equilibrium and also disfavor formation of bimolecular complexes. A single conformation with identical transition temperature as found in (A) and (B) resulted from this pretreatment. (D) SatC transcripts in 0.2× TB were treated as in (A) (low-salt snap-cooling) and then adjusted to 10 μM MgOAc. The same concentration of Mg2+ ions was included in the electrophoresis buffer and gel. Under these conditions, a single RNA conformation was discernable that was more stable at higher temperatures, with two clear consecutive transitions (T1, T2) at 40°C and 48°C, respectively. The arrowhead designates a kink in the RNA band during the second transition that may indicate an overlap of two separate conformational changes too close in their transition temperature to resolve.
SatC transcripts were next analyzed without pretreatment prior to TGGE (Fig. 1B ▶). Apart from the occurrence of bimolecular complexes of two major stabilities (a,b), the dominating monomolecular structure had identical denaturation properties as that formed after low-salt snap-cooling, including the characteristic main transition at ~24°C. There was also no change in the transition pattern when transcripts were first snap-cooled in low-salt buffer to remove the bimolecular complexes and then subjected to thermal denaturation and slow renaturation in a buffer of high ionic strength (Fig. 1C ▶). This treatment allows for formation of the thermodynamically most stable RNA structure and possibly less stable structures coexisting in equilibrium with the optimal structure. The presence of a single, identical transition regardless of RNA pretreatment indicates an inherent propensity of T7 RNA polymerase-generated wild-type satC transcripts to adopt a single, stable conformation in the absence of metal ions, with no evidence for any kinetically trapped, metastable structures or any major population of suboptimal, alternative structures.
Formation of the functional form of RNA often requires divalent metal ions to stabilize interactions between distinct secondary structural elements, and between these elements and single-stranded linker sequences, to form tertiary structural elements (Draper 1996; Tinoco and Bustamante 1999). When wild-type satC transcripts were denatured and rapidly renatured in 0.2× TB buffer, adjusted to 10 μM MgOAc, and then analyzed in gradient gels containing the same Mg2+ concentration, a single dominant structural species was still observed (Fig. 1D ▶). However, in contrast to a main transition at 23.7°C–24°C in the absence of divalent ions (Fig. 1A–C ▶), the RNA structure was strongly stabilized by Mg2+, undergoing two consecutive transitions at 40°C and 48°C (Fig. 1D ▶, T1 and T2). This shift in transition temperatures represents a Mg2+-induced stabilization of the initial structure of 16°C. Both transitions appear wider, and the corresponding band more defined, than the single transition that occurs in the absence of Mg2+, indicating a loss of cooperativity and a more reversible nature of the underlying conformational changes. Thus, Mg2+ appears to allow individual regions of the RNA structure to denature independently from each other. Further evidence for this interpretation is indicated by the nature of the second transition, which contains two distinctly visible phases marked by a kink in the smooth transition curve (Fig. 1D ▶, closed arrowhead). We therefore conclude that wild-type satC RNA transcripts adopt a single conformation, either in the presence or absence of Mg2+, and that Mg2+ leads to significant stabilization of satC RNA structure most likely by enabling additional tertiary interactions.
Tertiary elements stabilized by Mg2+ are prevalent in the 3′ region of satC
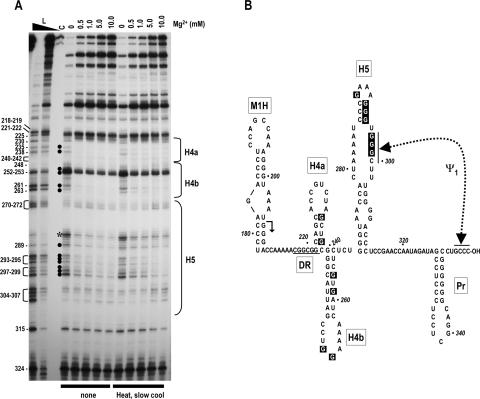
Since TGGE suggests the presence of elements stabilized by Mg2+ in the structure of satC transcripts, we were interested in determining if such elements were present in the 3′ terminal 140 nt that contain the four phylogenetically conserved hairpins H4a, H4b, H5, and Pr (Fig. 2B ▶). Therefore, full-length satC transcripts that were gel-purified for length to remove any bimolecular or truncated species were 3′ end-labeled with [32P]-pCp, subjected to heating/slow cooling or not further treated, and then partially digested with RNase T1 in the presence of increasing concentrations of Mg2+. RNase T1 cleaves specifically at single-stranded guanylates in a reaction that is not influenced by Mg2+ (Eun 1996); thus, cleavage differences due to the presence of Mg2+ reflect RNA structural changes and not alterations in enzymatic activity. All seven guanylates in the upper portion of H5 (positions 289–299) exhibited reduced susceptibility to RNase T1 as the Mg2+ concentration increased (Fig. 2A ▶), suggesting that these residues pair in a configuration stabilized by Mg2+. These guanylates include the three consecutive residues in positions 297–299, whose interaction with the 3′ terminal three cytidylates is necessary for normal (reduced) levels of transcription in vitro (Zhang et al. 2004b). Surprisingly, RNase T1 cleavage of three consecutive guanylates at positions 293–295 diminished with increasing concentrations of Mg2+, suggesting that the initial structure assumed by satC transcripts in vitro might not contain the short stem closed by a highly stable GAAA tetraloop that is evident in the phylogenetically inferred H5 structure. Six guanylates in the stems of H4a and H4b also displayed reduced susceptibility to RNase T1 in the presence of Mg2+, suggesting that the phylogenetically conserved H4a and H4b stem–loops may also not be present in the structure formed by the transcripts. Only two of 30 guanylates between positions 218 and 314 (G252, G253) were strongly susceptible to RNase T1 in the presence or absence of Mg2+, suggesting that the 3′ region of satC is highly structured. No differences in digestion patterns were discernable between untreated or heated/slow-cooled RNA transcripts, supporting the results obtained by TGGE that the population of satC transcripts does not contain detectable amounts of kinetically trapped intermediates (Pan et al. 1997).
FIGURE 2.
Mg2+ concentration dependence of RNase T1 cleavages in the 3′ region of satC. (A) 3′ end-labeled wild-type satC transcripts were either not treated or subjected to heating/slow cooling and then treated with identical concentrations of RNase T1 in increasing amounts of Mg2+. Mg2+ levels used are shown above each lane. L, RNase T1 ladder produced by cleavage of denatured transcripts. High and low concentrations of RNase T1 are indicated by the closed triangle above the lane. Position of guanylates are given at left. The location of the phylogenetically inferred 3′ hairpins is shown to the right. Filled circles indicate guanylates whose digestion level varies when the Mg2+ concentration increases. Asterisk denotes a nonspecific cleavage of U285. (B) Phylogenetically inferred structure of the 3′ region of satC. Arrow denotes sequence from that location to the 3′ end that is shared with TCV. The DR region (underlined) was previously determined to be important for satC accumulation in vivo and transcription in vitro. Dashed line denotes ψ1, an interaction previously determined to be important for wild-type levels of transcription by the TCV RdRp in vitro (Zhang et al. 2004b). The RNase T1 susceptibility of boxed guanylates varied with increasing concentration of Mg2+.
Releasing the 3′ end causes structural rearrangement of Pr
As described above, we previously demonstrated that the 3′ terminal GCCC-OH pairs with positions 297–300 in the H5 LSL at some point leading up to transcription initiation in vitro, using purified recombinant TCV RdRp expressed as a fusion with maltose-binding protein in Escherichia coli (Zhang et al. 2004b). Mutations at either C300 or G353 increased transcription of complementary strands, while the compensatory alterations together reduced transcription to near wild-type levels. The 3′ end/H5 interaction was confirmed for TCV genomic RNA by assaying mutants with single and compensatory mutations for accumulation in protoplasts (J. McCormack and A.E. Simon, in prep.). However, similar attempts to substantiate the interaction for satC in vivo were not successful, suggesting that the alterations disrupted other important functions in the sub-viral RNA. This 3′ end/H5 interaction, termed ψ1, is analogous to the 3′ end/SL3 pairing in tombusviruses (Pogany et al. 2003).
Solution structure analysis was used to investigate the conformation of the 3′ end of satC. Previous solution structure analysis of the satC Pr region was conducted using extension of primers to determine which nucleotides were susceptible to single-stranded-specific nucleotide-modifying chemicals and double-stranded-specific enzymatic cleavages (Song and Simon 1995). Since the Pr region comprises the 3′ terminal 28 nt, this analysis necessitated using satC transcripts containing an extended 3′ end composed of non-template residues to which a complementary primer could be annealed. Our reasoning for using these elongated transcripts was that they were efficient templates for transcription of satC-sized complementary products in vitro (using partially purified RdRp isolated from infected plants), and thus must contain an active Pr structure (Song and Simon 1994). However, more recent solution structure analysis of the 3′ region using wild-type satC transcripts and transcripts with a deletion of the 3′ terminal three cytidylates (CΔ3C) indicated that deletion of the terminal cytidylates caused significant structural changes to the H5 region, including enhanced susceptibility of guanylates at positions 297–299 to single-stranded-specific reagents. While this finding supports the existence of ψ1 in the initial structure of satC transcripts, it also implies that additional non-template 3′ nucleotides might disrupt ψ1 and possibly affect the structure of Pr and/or H5.
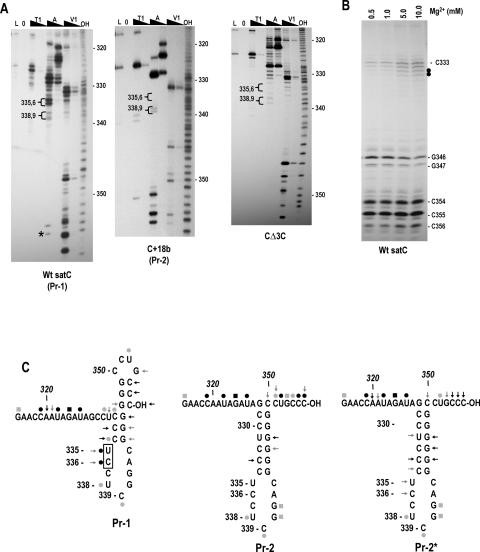
We therefore determined the structure of satC Pr with and without added 3′ terminal bases beyond the single cytidylate necessary to radioactively label the 3′ end. Transcripts of wild-type satC and satC with 18 plasmid-derived 3′ bases (C+18b) were radioactively end-labeled at their 3′ ends by addition of [32P]-pCp and then subjected to solution structure analysis in 10 mM Mg2+ using RNase T1, RNase A (specific for single-stranded pyrimidine nucleotides), and RNase V1 (specific for double-stranded or stacked nucleotides). A ladder of guanylate residues was prepared by thermally denaturing the transcripts prior to RNase T1 treatment. Although the RNA was presumably denatured, no digestion of any guanylates in the Pr region was detected using wild-type transcripts, although cuts at G340 and G341 were detected in the ladder lane using other templates (see below). A control reaction of RNA with no enzymatic treatment was also included to identify any bands produced by spurious RNA cleavages. The results indicate that very different patterns of RNase digestion are generated using transcripts with and without 18 added 3′ bases (Fig. 3 ▶). For wild-type satC, the reactivity of residues in the Pr region was consistently problematic, with a number of weak, nonspecific cleavages by RNase T1 and RNase A (Fig. 3A ▶, left). Nonspecific cleavages were not evident 5′ of position 315 (see Fig. 4A ▶, for example), with the exception of an inconsistent cleavage at U285 (see Fig. 2A ▶). The 3′ terminal cytidylates (positions 354–356) were strongly cleaved by RNase V1, as was G346. C333 and C334 also produced intense RNase V1 cleavages, with less intense cleavages at U335 and C336. These residues are likely involved in base-pairing or assume a stacked configuration.
FIGURE 3.
SatC with additional 18 plasmid-derived bases at the 3′ end has a structurally altered Pr region. (A) SatC, C+18b, and CΔ3C transcripts were subjected to partial treatment with two concentrations each of RNase T1 (T1), RNase A (A), or RNase V1 (V1). (L) RNase T1 ladder; (0) no added enzymes; (OH) transcripts were partially digested with alkaline buffer. U335/C336 and U338/C339 are in brackets since their susceptibility to RNase A was always clearly different in Pr-1 and Pr-2 forms of the Pr. Asterisk denotes cleavage was not consistent in wild-type satC transcripts. (B) Wild-type satC 3′ end-labeled transcripts were subjected to RNase V1 digestion with increasing concentrations of Mg2+. Closed circles denote residues whose cleavage was dependent on higher levels of Mg2+. (C) Possible secondary structures of Pr-1, Pr-2, and Pr-2*. (←) RNase V1; (•) RNase A; (▪) RNase T1. Intensity of the symbols implies relative intensity of the cleavages. Boxed residues are cleaved differentially by RNase V1 in the presence of different concentrations of Mg2+. The structure shown for Pr-2 and Pr-2* is the phylogenetically conserved structure for carmoviral Pr. Representative autoradiographs for Pr-2* are shown in Figure 5B ▶.
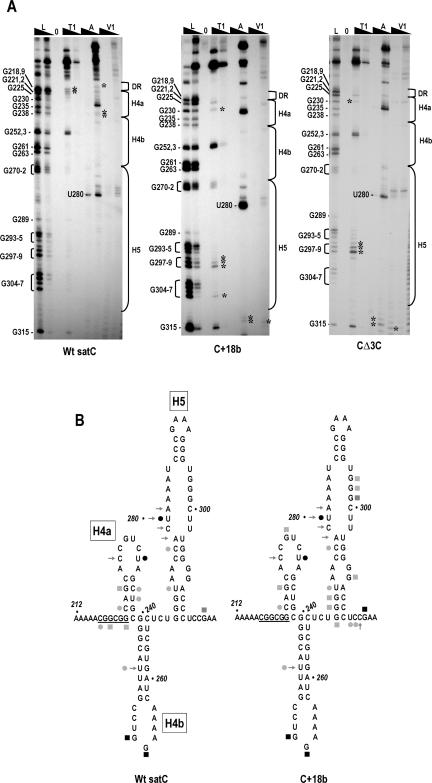
FIGURE 4.
Structural differences between wild-type satC, C+18b, and CΔ3C in the Dr/H4a and H5 regions. (A) SatC, C+18b, and CΔ3C transcripts were subjected to partial treatment with two concentrations of each enzyme. Abbreviations above each lane are as described in the legend to Figure 3 ▶. Guanylate residues in the RNase T1 ladder lane are identified by their positions, as is the prominent cleavage at U280. The boundaries of the H5, H4b, H4a, and DR regions are shown to the right. Asterisks denote cleavages exclusively in wild-type satC or the mutants. Cleavages at G270–272 that were visible using C+18b were also found in some, but not all, gels of wild-type satC. For this reason, they are not marked with asterisks. (B) Reactivity of residues in the DR/H4a/H4b and H5 regions. For convenience, cleavage locations are shown on the phylogenetically inferred satC structure. Symbols are as described in legend to Figure 3 ▶. The DR sequence is underlined.
To determine if any of the RNase V1 cleavages in the Pr region were due to possible tertiary interactions, the susceptibility of residues to RNase V1 was determined using different concentrations of Mg2+. Since RNase VI requires ≥0.3 mM Mg2+ for activity, the lowest concentration of Mg2+ used was 0.5 mM. As shown in Figure 3B ▶, RNase V1 digestion of the terminal cytidylates C354–356, as well as C333 and C334, did not vary substantially with increasing concentrations of Mg2+. This was surprising, because it suggested that the three terminal cytidylates might not be paired in a long-distance interaction with G297–299 in the H5 LSL in the initial structure of wild-type satC. In contrast, U335 and C336 were not detectably digested by RNase V1 at low Mg2+ concentrations, but were increasingly digested at higher concentrations. These two residues were also strongly digested by RNase A, suggesting that they exist in a tertiary conformation that may not be consistent in all structures. These results suggest an alternative folding of Pr compared with the phylogenetically inferred secondary structure. One possible structure is shown in Figure 3C ▶, where the 3′ terminal cytidylates form a small terminal hairpin by pairing with three of the six consecutive Pr guanylates, and the remaining three guanylates pairing with pyrimidine residues in positions 332–334.
In contrast, structure probing of C+18b revealed no nonspecific cleavages (Fig. 3A ▶, right). The extra 3′ residues caused the satC 3′ terminal five bases to now react strongly with RNase A (UGCCC-OH) and RNase T1 (UGCCC-OH), suggesting that the 3′ end is sterically hindered from pairing with its partner sequence or no longer assumes a stacked configuration. Besides these differences, G340 and G341 showed enhanced susceptibility to RNase T1; U335/C336 were not detectably cleaved by RNase A; and U335/C336, whose RNase V1 cleavage varied with Mg2+ concentration in wild-type satC transcripts, were no longer recognized by the enzyme. These differences denote that a structural rearrangement in the Pr region accompanies the addition of extra bases to wild-type satC and release of the 3′ end. This Pr rearrangement still results in an active template, with the RdRp initiating at the internally located satC 3′ end (Song and Simon 1994). As with wild-type satC, structure probing of C+18b did not indicate susceptibility of positions 328–333, which form the base of the phylogenetically inferred Pr structure, to RNase V1. Lack of RNase V1 digestion at these positions could reflect sequence/structure preferences of the enzyme, the inaccessibility of these bases to RNase digestion, or the non-existence of paired residues at this location. Comparative analysis of other carmovirus genomic RNAs, along with previous in vivo genetic selection data and site-specific mutagenesis of the satC Pr region, suggest that these residues are paired (Stupina and Simon 1997; Carpenter and Simon 1998). Combined with additional data described below suggesting that the C+18b Pr conformation reflects the active structure of Pr, we have mapped the new structural data for C+18b on the phylogenetically inferred structure of Pr (Fig. 3C ▶). For convenience, the putative initial Pr structure of wild-type satC transcripts will be referred to as “Pr-1,” and the phylogenetically inferred structure with the free 3′ end is called “Pr-2.”
Unpairing the satC 3′ end causes structural changes in the H5, H4a, and DR regions
To determine if freeing the 3′ end and consequential rearrangement of Pr caused by the extended sequence in C+18b had any effect on the structure of upstream regions, the enzymatic digestion patterns of upstream residues were anayzed using lower percentage polyacrylamide gels. As previously shown (Sun et al. 2005), the digestion pattern of wild-type satC in the H5 region was unusual, with a strong RNase A cleavage at U280 and weaker cleavages at C277 and U273 (Fig. 4A ▶). The strong RNase A cleavage at U280 was superimposed with one of four consecutive, weak RNase V1 cleavages (residues 278–281). No other consistent cuts were discernable in the H5 region using wild-type satC in the presence, but not the absence, of Mg2+ (see Fig. 2 ▶), suggesting that the H5 pyrimidines and guanylates are in a highly structured configuration with many tertiary contacts that are not susceptible or accessible to RNase V1. Solution structure analysis of C+18b transcripts in this same region revealed new RNase T1 cuts at G307 as well as G297–299, the residues previously determined to pair with the 3′ end. Interestingly, additional differences with wild-type satC were located in the sequence flanking H5. This sequence (5′ UCCG), which is critical for satC fitness in planta (Guan et al. 2000), forms a pseudoknot with the loop sequence of H4b that is important for satC accumulation in protoplasts and affects transcription by the RdRp in vitro (J. Zhang, G. Zhang, and A.E. Simon, in prep.). These results indicate that addition of sequences to the satC 3′ end caused structural alterations that included the H5 region of satC.
The H4b region did not vary detectably between wild-type satC and C+18b. However, several weak but consistent cleavages present in the DR/H4a region in wild-type satC were absent in C+18b. These included weak RNase A cuts at positions C217, C236, and U237, and weak RNase T1 cleavages at positions G219 and G222. In contrast, C+18b had a weak RNase T1 cleavage at G230 that was not detected in wild-type satC. These results suggest that structural rearrangements due to the presence of the 18 extra bases extended at least 140 bases from the 3′ end.
One possible explanation for the rearrangements in C+18b was an unintended effect of the additional residues at the 3′ end. To address this possibility, we determined the cleavage pattern of CΔ3C, since deleting the terminal cytidylates might have a similar consequence as “freeing” the 3′ end. Previous structural analysis of CΔ3C using chemical modifications and primer extension detected a significant structural rearrangement in the H5 region compared with wild-type satC (Zhang et al. 2004b). By subjecting CΔ3C to our current conditions, direct comparison of cleavage products could be made with those of wild-type satC and C+18b. As shown in Figure 4A ▶, three of the four new cleavage sites in the H5 region of C+18b were also present in CΔ3C, and the DR/H4a region of CΔ3C was identical to that of C+18b. These similarities included the moderate RNase T1 digestion at G297–299 in the H5 LSL region. This suggests that the structural changes in the DR/H4a/H5 regions of CΔ3C and C+18b were related, and thus unlikely to be dependent on the additional 3′ residues in C+18b. The sequence flanking the 3′ side of H5 also contained additional cleavages in CΔ3C transcripts not found in wild-type satC that were similar, but not identical, to C+18b. The structure of Pr in CΔ3C was also substantially different than wild-type satC, with fewer nonspecific cleavages and stronger RNase A cleavage of U338/C339 compared with U335/C336, similar to C+18b (Fig. 3A ▶). However, unlike C+18b, the five terminal bases reacted strongly with RNase V1, suggesting that they remain in a paired (or stacked) configuration. In addition, U335 and C336 remained weakly digested by RNase V1, similar to wild-type satC. These results indicate that the structural rearrangements evident in C+18b cannot be explained by the presence of the additional bases and therefore are likely the consequence of eliminating 3′ end base-pairing while retaining the satC terminal three cytidylates.
Mutations that cause a conformational change in the Pr region with no release of the 3′ end correlate with enhanced aberrant complementary strand transcription in vitro
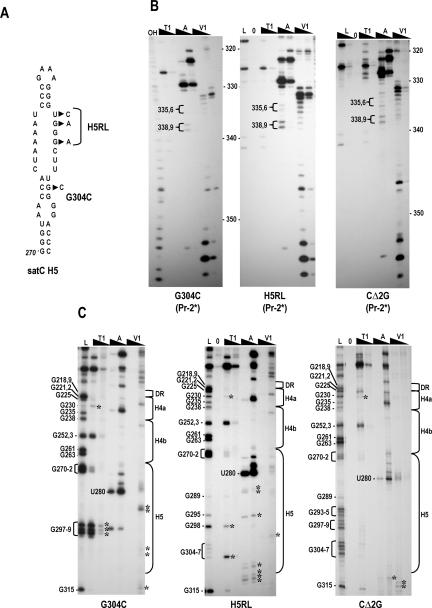
We previously reported that transcripts of CΔ3C and satC with mutations in H5 that disrupted either three residues in the H5 LSL (H5RL; Fig. 5A ▶) or a base pair in the H5 lower stem (G304C; Fig. 5A ▶) were identically transcribed by recombinant TCV RdRp purified from E. coli to levels much higher than that of wild-type satC in vitro (Zhang et al. 2004b; see Fig. 7A ▶, below). The transcriptional enhancement was unusual in that it was accompanied by increased levels of aberrant internal initiation (Guan and Simon 2000; Zhang et al. 2004b). Our explanation at the time was that enhanced levels of (−)-strand synthesis may have resulted from disrupting the 3′ end interaction with the H5 LSL, thus exposing the satC 3′ terminus to the RdRp. Our current results, that the 3′ end in CΔ3C remains strongly reactive to RNase V1 while the Pr is conformationally altered to a Pr-2-like structure, suggest an alternative explanation: Enhanced transcription is due to Pr rearrangement to a more active form, while aberrant internal initiation is the consequence of improper retention of a paired or stacked 3′ end.
FIGURE 5.
Structural differences in the 3′ region of G304C, H5RL, and CΔ2G. (A) Location of alterations in G304C and H5RL. CΔ2G contains a deletion of the 5′ terminal two guanylates (not shown). (B) Cleavage pattern in the Pr regions of G304C, H5RL, and CΔ2G. Note the distinctive RNase A cleavages of U338/C339 and RNase T1 cleavages of G340/G341 that are similar to those of C+18b (see Fig. 3A ▶). Abbreviations above each lane are as described in the legend to Figure 3 ▶. (C) Cleavage pattern in the DR/H4a/H4b and H5 regions of G304C, H5RL, and CΔ2G. Guanylate residues in the RNase T1 ladder lane are identified by their positions, as is the prominent cleavage at U280. The boundaries of the H5, H4b, H4a, and DR regions are shown to the right. Asterisks denote cleavages not found in wild-type satC.
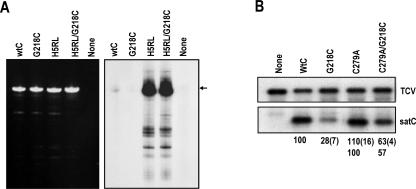
FIGURE 7.
Importance of the DR in the satC conformational switch. (A) In vitro transcription of satC transcripts containing alterations as indicated above each lane. Left panel shows the ethidium bromide-stained gel of templates. Autoradiograph of the gel is shown at the right. H5RL alterations are described in Figure 5A ▶. G218C is described in Figure 6A ▶. (wtC) wt satC; (none) no satRNA added. Arrow denotes the position of full-length satC. (B) RNA gel blot of total RNA isolated at 40 h post-inoculation of protoplasts with transcripts of TCV genomic RNA and satC variants. Values below each lane are the averages of at least two independent experiments. Standard deviation is in parentheses. Since C279A levels were greater than wild-type satC, accumulation of the double mutant (C279A/G218C) is compared with the C279A parental RNA (lower set of values). The presence of satC in the inoculum routinely decreases accumulation of TCV genomic RNA.
To explore this possibility, we examined the Pr structures of H5RL and G304C transcripts (Fig. 5B ▶). Transcripts of both mutants contained identical cleavages of Pr residues that were more similar to the Pr-2 form than the Pr-1 form, using the following criteria: there were no detectable non-specific cleavages; U338/C339 were digested more strongly by RNase A than U335/C336; and RNase T1 digested G340/G341. However, U335/C336 were cleaved by RNase V1 along with additional consecutive downstream residues, similar to wild-type satC and CΔ3C. In addition, the structure of the 3′ terminal 16 nt of H5RL and G304C did not vary reproducibly from wild-type satC, including strong RNase V1 cleavage of the three terminal cytidylates. This Pr-2-like structure with an undisrupted 3′ end was designated as Pr-2* (Fig. 3C ▶).
In addition to the Pr structural alterations, H5RL and G304C transcripts contained many unique cleavages in their H5 regions that did not extend into H4b (Fig. 5C ▶), indicating that their alterations mainly affected local H5 structure. Interestingly, the DR/H4a region of H5RL and G304C had identical cleavages as C+18b and CΔ3C (with the exception of reduced RNase T1 cleavage at G225), suggesting that structural differences in this region correlate with Pr-2/Pr-2* structures.
Since CΔ3C, H5RL, and G304C exhibited structural alterations in their H5 regions as well rearrangement of their Pr to Pr-2* (or Pr-2*-”like” for CΔ3C), we wanted to determine whether one or both of these structural changes were responsible for the enhanced, aberrant template activity in vitro. To address this question, the structure of satC with a deletion of the 5′ terminal two guanylates (CΔ2G) was examined, as this mutant also produces enhanced aberrant transcription in vitro, but without any nucleotide alterations in its 3′ region (Zhang et al. 2004b). Solution structure analysis of the Pr region of CΔ2G revealed a Pr-2*-like structure, with enhanced RNase A digestion of U338/C339 compared with U335/C336, detectable RNase T1 cleavage of G340/G341, and few nonspecific cleavages (Fig. 5B ▶). The DR/H4a region was also identical to that of the other Pr-2*-containing transcripts. This result indicates that the 5′ end of satC stabilizes the putative Pr-1 structure in satC transcripts, since lack of the 5′ terminal bases causes a rearrangement from Pr-1 to Pr-2*. In contrast, the H5 region was not altered from the cleavage pattern found for wild-type satC (Fig. 5C ▶), with the exception of positions flanking the 3′ side of H5. Altogether, these results suggest that enhanced transcription with aberrant initiation by the TCV RdRp in vitro is not due to structural differences in H5 or a disruption of ψ1, but rather is the consequence of a Pr-2* structure in the initial transcripts.
The DR is not an alternative binding site for the satC 3′ end
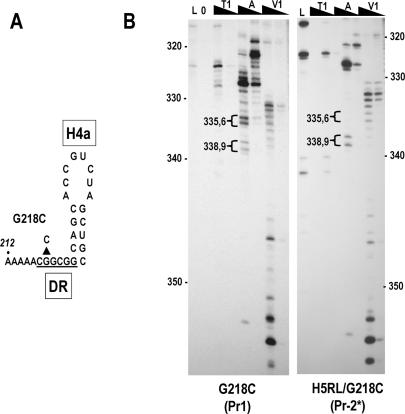
The unchanged RNase reactivity of the 3′ terminal 16 bases of wild-type satC, H5RL, and G304C was unexpected, since the residues in the H5 LSL (G297–299) that pair with the 3′ terminal cytidylates for normal transcription initiation in vitro are strongly cleaved by RNase T1 in G304C, and contain two alterations in H5RL. One possible explanation is that the 3′ end is paired with the H5 LSL in wild-type satC, but becomes paired with an alternative sequence when H5 is disrupted in H5RL and G304C. A possible location of sequence that could base-pair with the 3′ end is the DR, since this interaction is predicted in some mFold-generated structures. Alteration of the DR at position 218 (G218C) was previously shown to reduce satC accumulation by 3.6-fold in protoplasts and reduce transcription in vitro to near undetectable levels (Zhang et al. 2004b; see Fig. 7A ▶, below). The DR is also highly sequence-specific as established by in vivo functional selection (Sun et al. 2005), indicating that it plays an important role in satC (−)-strand synthesis both in vivo and in vitro.
To determine if the DR is an alternative binding site for the 3′ end in H5RL, solution structure analysis was conducted on transcripts containing G218C alone or in combination with the H5RL mutations (Fig. 6B ▶). The G218C Pr contained the hallmarks of the Pr-1 form, including nonspecific cleavages and enhanced RNase A reactivity at U335/C336 compared with U338/C339. In the combined H5RL/G218C mutant, the Pr structure was identical to that of H5RL (cf. the H5RL Pr-2* structure in Fig. 5B ▶). For both G218C and H5RL/G218C, the terminal 16 nt had identical RNase V1 reactivities as wild-type satC and H5RL, indicating that the DR is not an alternative binding site for the 3′ end.
FIGURE 6.
DR mutation G218C has no effect on the structure of Pr. (A) Location of the G218C alteration in the satC DR. (B) Cleavage patterns of G218C and combined G218C/H5RL. G218C gives the Pr-1 pattern, while H5RL/G218C gives the Pr-2* pattern. Note that neither mutant causes any change to the cleavages of the 3′ terminal 16 nt. See legend to Figure 3 ▶ for abbreviations.
Possible role of the DR region in transcription initiation in vitro and accumulation in vivo
The G218C mutation had no discernable effect on the structure of the Pr (Fig. 6B ▶) or H5 regions (J. Zhang, G. Zhang, R. Guo, B.A. Shapiro, and A.E. Simon, in prep.); thus, the loss of activity of G218C transcripts appears to be due to an ancillary inhibitory function. We previously reported that deleting the 3′ terminal three cytidylates obviated the inhibitory effect of G218C in vitro, with CΔ3C/G218G transcripts generating the high levels of aberrant synthesis associated with CΔ3C alone (Zhang et al. 2004b). The loss of requirement for the DR in the CΔ3C/G218C construct may be connected to a role for the DR in promoting the conformational rearrangement necessary for template activation of wild-type satC, but not for CΔ3C, which already contains rearrangements in the H5 and Pr regions. If this explanation is valid, then the H5RL mutations, which also cause the Pr-2 to Pr-2* structural rearrangement and promote aberrant transcription by the TCV RdRp in vitro, should also preclude G218C-mediated transcription inhibition. This was tested by subjecting wild-type satC, H5RL, G218C, and H5RL/G218C transcripts to in vitro transcription using TCV RdRp (Fig. 7A ▶). As previously shown, G218C transcripts were poor templates for in vitro transcription, while H5RL transcripts generated high levels of full-length and shorter than full-length products. Combining the G218C and H5RL mutations eliminated the inhibitory effect of G218C, supporting a possible requirement for the DR in the satC conformational change.
If the DR region helps to promote the conformational changes needed to activate satC transcripts, then stabilizing the active structure of satC should reduce the negative effects of G218C. To stabilize the satC active structure, a satC mutant (C279A) was used that pairs the lowest positions in the H5 LSL, and thus should contain a more stable H5 stem in the inferred H5 structure. This mutant was previously reported to enhance satC accumulation by 30% (Zhang et al. 2004a), although repeated assays for this report gave an average enhancement of 10%. SatC containing C279A, with and without alteration G218C, was assayed for accumulation in protoplasts (Fig. 7B ▶). At 40 h postinoculation, satC with G218C accumulated to 28% of wild type, similar to the previously reported level (29%). Inclusion of the C279A mutation reduced the negative effect of the DR mutation twofold, with the satRNA now accumulating to 57% of the C279A level. These results support a role for the DR region in the conformational changes leading to satC transcription initiation.
DISCUSSION
The past few years have led to increasing recognition that regulation of both gene expression and RNA function can be controlled by switches in RNA structural arrangements (Sudarsan et al. 2003; Mandal and Breaker 2004; Nudler and Mirovov 2004). These riboswitches fold into metastable structures that require an event, such as allosteric binding to a small metabolite, to rearrange into an alternative structure with reduced free energy that promotes or inhibits transcription or translation (Micura and Hobartner 2003; Mandal and Breaker 2004; Nudler and Mirovov 2004). It would be surprising if RNA viruses do not also make extensive use of RNA switches given the need to precisely regulate numerous events in their life cycle, including switching between translation and replication, switching between (−)- and (+)-strand synthesis, and switching off transcription of newly synthesized (+)-strands.
Wild-type satC transcripts appear to adopt an initial, pre-active configuration that does not contain either the canonical carmoviral H5 or Pr, but rather folds into a form stabilized by extensive tertiary contacts throughout the 3′ region. Analysis of RNase T1 cleavages in the absence or presence of increasing concentrations of Mg2+ indicated that 16/42 guanylates in the 3′ terminal 140 nt had reduced RNase T1 susceptibility in the presence of Mg2+ with no obvious cleavage differences upstream of this region (Fig. 2A ▶). Since Mg2+ promotes and stabilizes RNA tertiary structure (Woodson 2005), the 3′ portion of satC appears to be substantially stabilized by tertiary elements. No differences in digestion patterns were discernable between untreated or heated/slow-cooled RNA transcripts, suggesting that the initial population of satC transcripts was homogeneous and did not contain detectable amounts of kinetically trapped intermediates (Emerick and Woodson 1993; Pan et al. 1997).
This conclusion was supported by TGGE analysis of satC transcripts. TGGE as a method to study RNA structure employs qualitative and quantitative observations to interpret changes in RNA electrophoretic mobility resulting from conformational changes impacting the hydrodynamic shape. TGGE was applied to satC transcripts to determine the RNA’s potential to form one or more different structures. SatC transcripts appeared to fold into a single structure in 0.2× Tris-borate buffer supplemented with EDTA or magnesium acetate. This buffer system was chosen since electrophoretic mobility changes are generally larger in lower ionic strength (Riesner and Steger 2005). Even though there is no absolute criteria about how small a difference between two similar RNA conformations can be resolved by TGGE, it is unlikely that two structures would have the same hydrodynamic properties over the entire range of the temperature gradients analyzed, from the native state across one or more denaturation steps. One main transition at ~24°C was observed in 0.2× TBE, which is equivalent in ionic strength to ~3 mM NaCl (G. Steger, pers. comm.). The Tm value of RNA structural transitions increases with the ionic strength, and Tm increases per log of [Na+] between 5°C and >20°C have been reported for different RNAs (Steger et al. 1980; Riesner and Steger 1990). From this relationship we conclude that the observed Tm of ~24°C for satC in 0.2× TBE translates into stability of the pre-transition structure >30°C–35°C for physiological Na+ concentrations.
This argument is strengthened by the increase in stability of the satC structure to >40°C in the presence of 10 μM Mg2+. In other examples in which tertiary structures were studied using TGGE, magnesium concentrations in the hundreds of micromolar to millimolar range were necessary to stabilize the functional three-dimensional conformation (Brion et al. 1999; Guo and Cech 2002). For satC RNA transcripts, the 16°C shift in the main transition by as little as 10 μM Mg2+ is indicative of a strong potential to adopt tertiary structural folding and the utilization of specific magnesium binding sites.
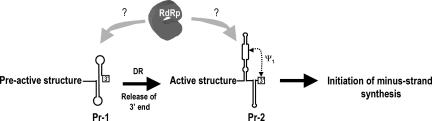
We propose that this initial structure assumed by satC transcripts in vitro represents a pre-active conformation of the RNA, and that a conformational switch to the active structure is important for initiation of minus-strand synthesis (Fig. 8 ▶). Since the 3′ regions of both C+18b and CΔ3C transcripts assume a similar configuration in vitro that differs from the structure of wild-type satC, we believe that release of the 3′ end likely promotes the structural switch. The identical susceptibility of the 3′ terminal 16 nt to RNases in transcripts of all constructs tested to date (with the exception of C+18b and CΔ3C), including those with mutations in the H5 LSL, suggests that the 3′ end does not pair with H5 in the initial pre-active conformation (Figs. 3A ▶, 5B ▶). It is important to note, however, that all wild-type and mutant transcripts contained an additional 3′ cytidylate due to labeling the 3′ end with [32P]-pCp, and, therefore, it is possible that the natural 3′ cytidylates are highly reactive with RNase V1 due to the presence of the additional cytidylate. Nevertheless, ψ1 is necessary for generation of normal (reduced) levels of minus-strands in vitro (Zhang et al. 2004b). One possible explanation is if ψ1 is a constituent of the active structure that also contains phylogenetically conserved H5 and Pr-2. Transcripts with mutations in H5 (H5RL and G304C), or deletion of the 3′ CCC (Δ3C) or 5′ GG (Δ2G), contain the Pr-2-like structure Pr-2* and are substantially more active for transcription than wild-type satC (Zhang et al. 2004b). We therefore propose that the pre-active structure adopted by satC transcripts in vitro requires reorganization to an active conformation containing the phylogenetically conserved H5, Pr-2, and Ψ1, prior to initiation of (−)-strand synthesis (Fig. 8 ▶).
FIGURE 8.
Conformational switch model for initiation of (−)-strand satC synthesis. See text for details. It is currently unknown at what stage the template is recognized by the RdRp, or if the RdRp is involved in mediating the switch. Putative structure of Pr-1 is shown.
An important question is whether the pre-active satC structure exists in vivo. We have recently discovered a second pseudoknot, termed Ψ2, that forms between the H4b loop sequence UGGA and sequence just 3′ of H5 (UCCG) (J. Zhang, G. Zhang, R. Guo, B.A. Shapiro, and A.E. Simon, in prep.). Disruption of Ψ2 reduced satC accumulation in vivo, which was partially restored by compensatory mutations. Results from in vitro RdRp transcription assays and solution structure probing suggest that Ψ2 is present in, and stabilizes, the pre-active satC structure (J. Zhang, G. Zhang, R. Guo, B.A. Shapiro, and A.E. Simon, in prep.). The apparent association of Ψ2 with the pre-active structure is consistent with our current finding that the alternative “active” conformation assumed by C+18b and CΔ3C transcripts has new RNase A cleavage sites in the Ψ2 UCCG element, and thus the active conformation may not contain Ψ2 (Fig. 4A ▶).
The DR was originally identified as an important sequence-specific element for satC accumulation using in vivo functional selection (Sun et al. 2005). Fitness of satC in plants and high-level accumulation in protoplasts was associated with recovery of the wild-type DR sequence 217CGGCGG or two related sequences. The proposal was made that the DR was important for initiation of minus-strand synthesis through an alternative base-pairing interaction with the 3′ terminus, as all three sequences recovered by in vivo functional selection could maintain this putative interaction that was also predicted by mFold (Sun et al. 2005). Our current results do not support such an association, since the DR G218C alteration, in the presence or absence of H5 LSL mutations, had no effect on RNase susceptibility of the 3′ terminal 16 bases (Fig. 6B ▶).
While the DR does not apparently interact directly with the 3′ end, three recent observations suggest an association between the DR and H5 (or the nearby Ψ2) that has important implications for the conformational switch. (1) The majority of progeny accumulating in plants inoculated with satC containing a replacement H5 from the related carmovirus Cardamine chlorotic fleck virus (CCFV) had the G218C alteration (J. Zhang, G. Zhang, R. Guo, B.A. Shapiro, and A.E. Simon, in prep.). While this alteration enhanced accumulation of satC with H5 of CCFV (J. Zhang, G. Zhang, R. Guo, B.A. Shapiro, and A.E. Simon, in prep.), it reduced accumulation of wild-type satC in protoplasts and transcription of complementary strands in vitro (Zhang et al. 2004b). G218C had no effect on transcription of H5RL (this report) or CΔ3C (Zhang et al. 2004b), both of which are proposed to already contain the active conformation of Pr. (2) Recent solution structure analyses indicated that transcripts containing mutations in Ψ2 base-paired sequences had strong new RNase T1 cleavages in the DR, suggesting a physical connection between the DR and Ψ2 (J. Zhang, G. Zhang, R. Guo, B.A. Shapiro, and A.E. Simon, in prep.). (3) Transcripts with G218C exhibited structural rearrangements in the DR/H4a region and new RNase V1 cleavages in the UCCG Ψ2 sequence at the base of H5 (J. Zhang, G. Zhang, R. Guo, B.A. Shapiro, and A.E. Simon, in prep.). Altogether, these results suggest that DR mutation G218C inhibits conversion of the pre-active to the active structure by altering the satC pre-active conformation including Ψ2. This model is supported by our current finding that stabilizing the active structure of satC by inclusion of the C279A H5 mutation along with G218C reduced the negative effects of G218C by twofold in vivo (Fig. 7B ▶).
We have not yet conducted a systematic investigation of the structural configuration of the TCV genomic RNA 3′ end region that is the parental sequence for the 3′ region of satC. However, the 10 positional differences between satC and TCV, some of which allow the TCV Pr hairpin to assume a highly stable 10 base-pair stem, suggest that TCV does not adopt the satC pre-active structure. Chimeric constructs containing exchanges of the 3′ 100 bases replicate poorly in vivo due primarily to differences in the Pr region (J. Zhang, G. Zhang, and A.E. Simon, unpubl.). Since TCV is translated, it is possible that the TCV pre-active structure requires 5′ sequences not found in satC, to assume a form that is inactive for replication while active for translation.
MATERIALS AND METHODS
Construction of satC mutants
Construction of CΔ3C, CΔ2G, G304C, H5RL, G218C, and C279A were previously described (Zhang et al. 2004a,b). Mutant G218C/C279A was constructed by replacing the SpeI/SmaI fragment of G218C with the corresponding fragment of C279A. G218C/H5RL was generated in a similar fashion. C+18b, a satC mutant with 18 additional bases derived from plasmid sequence at its 3′ end, was generated by in vitro transcription of EcoRI-digested pT7C(+), which contains the full-length cDNA of satC (Song and Simon 1994).
In vitro preparation of RNA
In vitro transcription was carried out for 2 h at 37°C in a 60 μL reaction mixture containing 40 mM Tris-HCl (pH 8.0); 10 mM dithiothreitol; 8 mM MgCl2; 25 mM NaCl; 2 mM spermidine; 1 mM each of ATP, CTP, GTP, and UTP; 8 μg of DNA template ,and 40 U of T7 RNA polymerase (Promega). For RNA structure probing, the DNA template was removed by further incubation for 20 min in the presence of 8 U of RNase-free DNase (Promega). RNA transcripts were then phenol–chloroform extracted and ammonium acetate–isopropanol precipitated, and purified through a 6% sequencing gel.
Temperature-gradient gel electrophoresis
RNA pretreatments prior to TGGE analysis were performed essentially as previously described (Baumstark and Riesner 1995). Briefly, RNA in vitro transcripts were purified from the T7 transcription reaction using RNeasy (Qiagen) and eluted from the columns in 80 μL of 0.2 × TE buffer (1 × TE: 10 mM Tris-HCl, pH 8.0, 1 mM EDTA), then stored at −20°C. Low-salt snap-cooling at low ionic strength was performed in TE buffer by denaturing the RNA for 10 min at 90°C, then rapidly transferring the tube to ice immediately before loading the sample on a pre-cooled temperature gradient gel. For renaturation of RNA at high ionic strength, RNA was first snap-cooled in TE; the buffer was adjusted to 500 mM NaCl, 4 M urea, 1 mM Na-cacodylate, 0.1 mM EDTA; and then the RNA was denatured for 10 min at 85°C, followed by slow cooling to room temperature. Prior to TGGE analysis, the sample was dialyzed at 4°C against electrophoresis buffer on swim filters to remove salt (Millipore, #VSWP02500).
TGGE was performed on a commercially available instrument (Biometra TGGE Maxi System). Gels were 20 × 19 × 0.1 cm on film support (GelBond-PAG, GE/Amersham) and contained 5% (w/v) acrylamide, 0.17% bisacrylamide, 0.05% (v/v) TEMED, 0.05% (w/v) ammonium peroxidisulfate for initiating the polymerization, and either 0.2 × TBE (1 × TBE: 89 mM Tris, 89 mM boric acid, 10 mM EDTA) or 0.2 × TB with 10 μM magnesium acetate. Electrophoresis was performed in three steps: (1) samples were applied to the 16 × 0.4-cm sample slot of the horizontally mounted, precooled gel, and RNA was allowed to migrate several millimeters into the matrix at a uniform temperature of 10°C and 400 V for 10 min; (2) a constant temperature gradient was established in the gel (either from 15°C–45°C or 20°C–60°C as indicated); and (3) electrophoresis was resumed with the applied temperature gradient for 1.5 h at 400 V. RNA was detected by silver staining (Schumacher et al. 1986).
RNA solution structure probing with 3′ end-labeled transcripts
RNA structure probing of satC and its mutants was performed by using protocol and reagents obtained from Ambion. Briefly, gel-purified transcripts were labeled at the 3′ end in a final volume of 20 μL containing 6 μg of transcripts, 50 μCi of [32P]-pCp (Amersham), 20 U of T4 RNA ligase, 50 mM Tris-HCl (pH 7.8), 10 mM MgCl2, 10 mM dithiothreitol, and 1mM ATP. After overnight incubation at 4°C, reactions were terminated by phenol–chloroform extraction. Labeled transcripts were separated in a 6% sequencing gel and eluted by soaking overnight with constant shaking in a buffer containing 25 mM Tris-HCl (pH 7.5), 400 mM NaCl, and 0.1% SDS. Labeled transcripts were added to a mixture containing RNA structure buffer (10 mM Tris-HCl pH 7.0, 100 mM KCl, 10 mM Mg2+), yeast tRNA, and either water or RNase T1 (0.01 or 0.001 U/μL), RNase A (0.01 or 0.001 μg/mL), or RNase V1 (0.001 or 0.0001 U/μL), and incubated for 15 min at 22°C. Samples were precipitated, resuspended in 10 μL of loading buffer, and subjected to electrophoresis through 10% or 20% sequencing gels. Alkaline hydrolysis ladders were obtained by treatment of 3′ end-labeled transcripts with alkaline hydrolysis buffer (Ambion) for 5 min at 95°C. To obtain RNase T1 ladders, 3′ end-labeled transcripts were heated for 5 min at 95°C in buffer supplied by the manufacturer (Ambion), then cooled on ice, followed by RNase T1 (0.1 or 0.01 U/μL) digestion for 15 min at 22°C.
To investigate how prior treatment of RNA template and Mg2+ concentration affect RNA structure probing, 3′ end-labeled transcripts were either not treated or heated at 60°C in 10 mM Tris-HCl, 100 mM KCl, and different concentrations of Mg2+ (0, 0.5, 1.0, 5, and 10 mM) for 5 min and slow cooled to 35°C, then placed on ice. Transcripts were then digested with RNase T1 (0.01 U/μL) or RNase V1 (0.001 U/μL) at 22°C and analyzed as described above.
In vitro RdRp assay
In vitro RdRp assays were carried out using recombinant TCV p88. The p88-expressing plasmid, which was a kind gift from P.D. Nagy (University of Kentucky), was transformed into E. coli Rosetta (DE3) pLacI competent cells (Novagen), and expression and purification of the recombinant protein was carried out as described (Rajendran et al. 2002). In vitro RdRp assays were performed in the presence of 10 mM Mg2+, the same concentration as that used for RNA structure probing. Briefly, 1 μg of purified RNA template was added to a 25-μL reaction mixture containing 50 mM Tris-HCl (pH 8.2); 100 mM potassium glutamate; 10 mM MgCl2; 10 mM dithiothreitol; 1 mM each of ATP, CTP, GTP; 0.01 mM UTP; 10 μCi of [α-32P] UTP (Amersham); and 2 μg of recombinant p88. After a 90-min incubation at 20°C, 1 μg of tRNA was added, and the mixture was subjected to phenol–chloroform extraction and ammonium acetate–isopropanol precipitation. Radiolabeled products were analyzed by denaturing 8M urea–5% polyacrylamide gel electrophoresis followed by autoradiography.
Protoplast inoculations, RNA extraction, and Northern blots
Protoplasts (5 × 106) prepared from callus cultures of Arabidoposis thaliana (ecotype Col-0) were inoculated with 20 μg of TCV genomic RNA transcripts and 2 μg of satC transcripts as previously described (Kong et al. 1997). Equal amounts of total RNA extracted from protoplasts at 40 h post-inoculation were subjected to electrophoresis through a 1.5% non-denaturing agarose gel. After blotting, (+)-strand TCV and satC RNAs were probed with a [α-32P]-dATP labeled oligonucleotide complimentary to positions 3950–3970 of TCV genomic RNA and 249–269 of satC (Zhang and Simon 2003).
Acknowledgments
Funding was provided by grants from the U.S. Public Health Service (GM61515-01) and the National Science Foundation (MCB-0086952) to A.E.S.
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.2166706.
REFERENCES
- Barton, D.J., O’Donnell, B.J., and Flanegan, J.B. 2001. 5′ cloveleaf in poliovirus RNA is a cis-acting replication element required for negative-strand synthesis. EMBO J. 20: 1439–1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumstark, T. and Riesner, D. 1995. Only one of four possible secondary structures of the central conserved region of potato spindle tuber viroid is a substrate for processing in a potato nuclear extract. Nucleic Acids Res. 23: 4246–4254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumstark, T., Schroeder, A.R.W., and Riesner, D. 1997. Viroid processing: Switch from cleavage to ligation is driven by a change from a tetraloop to a loop E conformation. EMBO J. 16: 599–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brion, P., Michel, F., Schroeder, R., and Westhof, E. 1999. Analysis of the cooperative thermal unfolding of the td intron of bacteriophage T4. Nucleic Acids Res. 27: 2494–2502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buck, K.W. 1996. Comparison of the replication of positive-stranded RNA viruses of plants and animals. Adv. Virus Res. 47: 159–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpenter, C.D. and Simon, A.E. 1998. Analysis of sequences and putative structures required for viral satellite RNA accumulation by in vivo genetic selection. Nucleic Acids Res. 26: 2426–2432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrington, J.C., Heaton, L.A., Zuidema, D., Hillman, B.I., and Morris, T.J. 1989. The genome structure of turnip crinkle virus. Virology 170: 219–226 [DOI] [PubMed] [Google Scholar]
- Chapman, M.R. and Kao, C.C. 1999. A minimal RNA promoter for minus-strand RNA synthesis by the brome mosaic virus polymerase complex. J. Mol. Biol. 286: 709–720. [DOI] [PubMed] [Google Scholar]
- Draper, D. 1996. Stategies for RNA folding. Trends Biochem. Sci. 21: 145–149. [PubMed] [Google Scholar]
- Dreher, T.W. 1999. Functions of the 3′-untranslated regions of positive strand RNA viral genomes. Annu. Rev. Phytopathol. 37: 151–174. [DOI] [PubMed] [Google Scholar]
- Duggal, R., Lahser, F.C., and Hall, T.C. 1994. Cis-acting sequences in the replication of plant viruses with plus-sense RNA genomes. Annu. Rev. Phytopathol. 32: 287–309. [Google Scholar]
- Eckerle, L.D. and Ball, L.A. 2002. Replication of the RNA segments of a bipartite viral genome is coordinated by a transactivating subgenomic RNA. Virology 296: 165–176. [DOI] [PubMed] [Google Scholar]
- Emerick, V.L. and Woodson, S.A. 1993. Self-splicing of the Tetrahymena pre-rRNA is decreased by misfolding during transcription. Biochemistry 32: 14062–14067. [DOI] [PubMed] [Google Scholar]
- Eun, H.-M. 1996. Enzymology primer for recombinant DNA technology. Academic Press, San Diego, CA.
- Frolov, I., Hardy, R., and Rice, C.M. 2001. Cis-acting RNA elements at the 5′ end of Sindbis virus genome RNA regulate minus- and plus-strand RNA synthesis. RNA 7: 1638–1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goebel, S.J., Hsue, B., Dombrowski, T.F., and Masters, P.S. 2004. Characterization of the RNA components of a putative molecular switch in the 3′ untranslated region of the murine coronavirus genome. J. Virol. 78: 669–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan, H. and Simon, A.E. 2000. Polymerization of non-template bases prior to transcription initiation by an RNA-dependent RNA polymerase: A novel activity involved in 3′-end repair of viral RNAs. Proc. Natl. Acad. Sci. 97: 12451–12456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan, H., Carpenter, C.D., and Simon, A.E. 2000. Requirement of a 5′-proximal linear sequence on minus strands for plus-strand synthesis of a satellite RNA associated with TCV. Virology 68: 355–363. [DOI] [PubMed] [Google Scholar]
- Guo, F. and Cech, T.R. 2002. Evolution of Tetrahymena ribozyme mutants with increased structural stability. Nat. Struct. Biol. 9: 855–861. [DOI] [PubMed] [Google Scholar]
- Hacker, D.L., Petty, I.T.D., Wei, N., and Morris, T.J. 1992. Turnip crinkle virus genes required for RNA replication and virus movement. Virology 186: 1–8. [DOI] [PubMed] [Google Scholar]
- Herold, J. and Andino, R. 2001. Poliovirus RNA replication requires genome circularization through a protein–protein bridge. Mol. Cell 7: 581–591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kao, C.C., Singh, P., and Ecker, D.J. 2001. De novo initiation of viral RNA-dependent RNA synthesis. Virology 287: 251–260. [DOI] [PubMed] [Google Scholar]
- Khromykh, A.A., Meka, H., Guyatt, K.J., and Westaway, E.G. 2001. Essential role of cyclization sequences in flavivirus RNA replication. J. Virol. 75: 6719–6728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, Y.-N. and Makino, S. 1995. Characterization of a murine coronavirus defective interfering RNA internal cis-acting replication signal. J. Virol. 69: 4963–4971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klovins, J., Berzins, V., and van Duin, J. 1998. A long-range interaction in Qβ RNA that bridges the thousand nucleotides between the M-site and the 3′ end is required for replication. RNA 4: 948–957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koev, G., Liu, S., Beckett, R., and Miller, W.A. 2002. The 3′-terminal structure required for replication of barley yellow dwarf virus RNA contains an embedded 3′ end. Virology 292: 114–126. [DOI] [PubMed] [Google Scholar]
- Kong, Q., Wang, J., and Simon, A.E. 1997. Satellite RNA-mediated resistance to turnip crinkle virus in Arabidopsis involves a reduction in virus movement. Plant Cell 9: 2051–2063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandal, M. and Breaker, R.R. 2004. Gene regulation by riboswitches. Nature Rev. Mol. Cell. Biol. 5: 451–463. [DOI] [PubMed] [Google Scholar]
- McCormack, J. and Simon, A.E. 2004. Biased hypermutagensis associated with mutations in an untranslated hairpin of an RNA virus. J. Virol. 78: 7813–7817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Micura, R. and Hobartner, C. 2003. On secondary structure rearrangements and equilibria of small RNAs. ChemBioChem 4: 984–990. [DOI] [PubMed] [Google Scholar]
- Murray, K.E. and Barton, D.J. 2003. Poliovirus CRE-dependent VPg uridylylation is required for positive-strand RNA synthesis but not for negative-strand RNA synthesis. J. Virol. 77: 4739–4750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy, P.E., Pogany, J., and Simon, A.E. 1999. RNA elements required for RNA recombination function as replication enhancers in vitro and in vivo in a plus strand RNA virus. EMBO J. 18: 5653–5665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nudler, E. and Mirovov, A.S. 2004. The riboswitch control of bacterial metabolism. Trends Biochem. Sci. 29: 11–17. [DOI] [PubMed] [Google Scholar]
- Oh, J.-W., Kong, Q., Song, C., Carpenter, C.D., and Simon, A.E. 1995. Open reading frames of turnip crinkle virus involved in satellite symptom expression and incompatibility with Arabidopsis thaliana ecotype Dijon. Mol. Plant-Microbe Interact. 8: 979–987. [DOI] [PubMed] [Google Scholar]
- Olsthoorn, R.C., Mertens, S., Brederode, F.T., and Bol, J.F. 1999. A conformational switch at the 3′ end of a plant virus RNA regulates viral replication. EMBO J. 18: 4856–4864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan, J., Thirumalai, D., and Woodson, S.A. 1997. Folding of RNA involves parallel pathways. J. Mol. Biol. 273: 7–13. [DOI] [PubMed] [Google Scholar]
- Pogany, J., Fabian, M.R., White, K.A., and Nagy, P.D. 2003. Functions of novel replication enhancer and silencer elements in tombusvirus replication. EMBO J. 22: 5602–5611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajendran, K.S., Pogany, J., and Nagy, P.D. 2002. Comparison of turnip crinkle virus RNA-dependent polymerase preparations expressed in Escherichia coli or derived from infected plants. J. Virol. 76: 1707–1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray, D. and White, K.A. 1999. Enhancer-like properties of an RNA element that modulates Tombusvirus RNA accumulation. Virology 256: 162–171. [DOI] [PubMed] [Google Scholar]
- ———. 2003. An internally located RNA hairpin enhances replication of Tomato bushy stunt virus RNAs. J. Virol. 77: 245–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riesner, D. and Steger, G. 1990. Viroids and viroid-like RNAs. In Landolt-Boernstein New Series (ed. W. Saenger), VII/1D, pp. 194–243. Springer-Verlag GmbH, Berlin.
- ———. 2005. Temperature-gradient gel electrophoresis of RNA. In Handbook of RNA biochemistry (eds. R.K. Hartmann et al.), pp. 398–414. Wiley-VCH Verlag, Weinheim, Germany.
- Riesner, D., Steger, G., Zimmat, R., Owens, R.A., Wagenhofer, M., Hillen, W., Vollbach, S., and Henco, K. 1989. Temperature-gradient gel electrophoresis of nucleic acids: Analysis of conformational transitions, sequence variations, and protein–nucleic acid interactions. Electrophoresis 10: 377–389. [DOI] [PubMed] [Google Scholar]
- Rosenbaum, V. and Riesner, D. 1987. Temperature-gradient gel electrophoresis. Thermodynamic analysis of nucleic acids and proteins in purified form and in cellular extracts. Biophys. Chem. 26: 235–246. [DOI] [PubMed] [Google Scholar]
- Schroeder, A.R.W., Baumstark, T., and Riesner, D. 1998. Chemical mapping of coexisting RNA structures. Nucleic Acids Res. 26: 3449–3450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumacher, J., Meyer, N., Riesner, D., and Weidemann, H.L. 1986. Diagnostic procedure for detection of viroids and viruses with circular RNAs by return-gel electrophoresis. J. Phytopathol. 115: 332–343. [Google Scholar]
- Schuppli, D., Georgijevic, J., and Weber, H. 2000. Synergism of mutations in bacteriophage Qβ RNA affecting host factor dependence of Qβ replicase. J. Mol. Biol. 295: 149–154. [DOI] [PubMed] [Google Scholar]
- Simon, A.E. and Howell, S.H. 1986. The virulent satellite RNA of turnip crinkle virus has a major domain homologous to the 3′-end of the helper virus genome. EMBO J. 5: 3423–3428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sit, T.L., Vaewhongs, A.A., and Lommel, S.A. 1998. RNA-mediated trans-activation of transcription from a viral RNA. Science 281: 829–832. [DOI] [PubMed] [Google Scholar]
- Song, C. and Simon, A.E. 1994. RNA-dependent RNA polymerase from plants infected with turnip crinkle virus can transcribe (+)and (−)-strands of virus-associated RNAs. Proc. Natl. Acad. Sci. 91: 8792–8796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ———. 2005. Requirement of a 3′-terminal stem-loop in in vitro transcription by an RNA-dependent RNA polymerase. J. Mol. Biol. 254: 6–14. [DOI] [PubMed] [Google Scholar]
- Steger, G., Mueller, H., and Riesner, D. 1980. Helix–coil transitions in double-stranded viral RNA. Fine resolution melting and ionic strength dependence. Biochim. Biophys. Acta 606: 274–284. [DOI] [PubMed] [Google Scholar]
- Stupina, V. and Simon, A.E. 1997. Analysis in vivo of turnip crinkle virus satellite RNA C variants with mutations in the 3′ terminal minus strand promoter. Virology 238: 470–477. [DOI] [PubMed] [Google Scholar]
- Sudarsan, N., Barrick, J.E., and Breaker, R.R. 2003. Metabolite-binding RNA domains are present in the genes of eukaryotes. RNA 9: 644–647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, X., Zhang, G., and Simon, A.E. 2005. Short internal sequences involved in RNA replication and virion accumulation in a subviral RNA of Turnip crinkle virus. J. Virol. 79: 512–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tinoco, I. and Bustamante, C. 1999. How RNA folds. J. Mol. Biol. 293: 271–281. [DOI] [PubMed] [Google Scholar]
- Turner, R.L. and Buck, K.W. 1999. Mutational analysis of cis-acting sequences in the 3′- and 5′-untranslated regions of RNA2 of Red clover necrotic mosaic virus. Virology 253: 115–224. [DOI] [PubMed] [Google Scholar]
- van Dijk, A.A., Kakeyev, E.V., and Bamford, D.H. 2004. Initiation of viral RNA-dependent RNA polymerization. J. Gen. Virol. 85: 1077–1093. [DOI] [PubMed] [Google Scholar]
- Woodson, S.A. 2005. Metal ions and RNA folding: A highly charged topic with a dynamic future. Curr. Opin. Chem. Biol. 9: 104–109. [DOI] [PubMed] [Google Scholar]
- You, S., Falgout, B., Markoff, L., and Padmanabhan, R. 2001. In vitro RNA synthesis from exogenous Dengue viral RNA templates requires long-range interactions between 5′- and 3′-terminal regions that influence RNA structure. J. Biol. Chem. 276: 15581–15591. [DOI] [PubMed] [Google Scholar]
- Zhang, G. and Simon, A.E. 2003. A multifunctional Turnip crinkle virus replication enhancer revealed by in vivo functional SELEX. J. Mol. Biol. 326: 35–48. [DOI] [PubMed] [Google Scholar]
- Zhang, J. and Simon, A.E. 2005. Importance of sequence and structural elements within a viral replication repressor. Virology 333: 301–315. [DOI] [PubMed] [Google Scholar]
- Zhang, J., Stuntz, R., and Simon, A.E. 2004a. Analysis of a viral replication repressor: Sequence requirements for a large symmetrical internal loop. Virology 326: 90–102. [DOI] [PubMed] [Google Scholar]
- Zhang, G., Zhang, J., and Simon, A.E. 2004b. Repression and derepression of minus-strand synthesis in a plus-strand RNA virus replicon. J. Virol. 78: 7619–7633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuker, M. 2003. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 31: 3406–3415. [DOI] [PMC free article] [PubMed] [Google Scholar]