Abstract
Localizing mRNAs within the cytoplasm gives cells the ability to spatially restrict protein production, a powerful means to regulate gene expression. Localized mRNA is often visible in microscopically observable particles or granules, and the association of mRNA localization with these structures is an indication that particles or granules may be essential to the localization process. Understanding how such structures form will therefore be important for understanding the function of localization RNPs (L-RNPs). We previously identified a novel component of an L-RNP from the Vg1 mRNA from Xenopus oocytes called 40LoVe. 40LoVe interaction with the Vg1-localization element (Vg1LE) was previously shown to be dependent on the VM1 and E2 sequence motifs within the Vg1LE that cross-link to hnRNP I and Vg1RBP/Vera, respectively. We report interaction of these motif-binding proteins with 40LoVe and identify a 40LoVe-Xenopus hnRNP D/AUF1 interaction. We further demonstrate that titration of VM1 and E2 motif binding activity in vivo surprisingly suggests that the motif binding proteins have differing roles during Vg1LE-dependent mRNA localization.
Keywords: mRNA localization, RNP complex, hnRNP D, RNA binding protein, oogenesis, Xenopus
INTRODUCTION
The Vg1 mRNA encodes a member of the TGF-β family that is involved in mesoderm induction during early embryogenesis in Xenopus laevis (Weeks and Melton 1987). Vg1 mRNA is found highly enriched at the vegetal pole of stage II–VI oocytes (Weeks and Melton 1987; Kessler and Melton 1995). The localization of Vg1 mRNA is thought to be necessary for accurate production of Vg1 protein during embryogenesis. Vegetal mRNA localization in Xenopus oogenesis occurs through two pathways, early/METRO and late, distinguished primarily by the timing of localization (King et al. 1999, 2005; Rand and Yisraeli 2001; Kloc et al. 2002). Early and late mRNAs localize to the same region of the cytoplasm, but the timing of localization indicates that the machinery is not identical. Nonetheless, evidence suggests common machinery involving ER, and/or cytoskeletal associations underlie localization by the two pathways (Kloc and Etkin 1998; Betley et al. 2002; Chang et al. 2004; Claussen et al. 2004; Choo et al. 2005).
Deletion analysis revealed a 340-nucleotide (nt) minimal element within the 3′ UTR (Vg1-localization element, Vg1LE) that was sufficient to drive localization of a nonlocalizing reporter mRNA to the vegetal pole in a manner identical to that of the endogenous Vg1 mRNA (Mowry and Melton 1992). Mutagenesis studies found several primary sequence features that appear to be necessary for the function of this element. Two different motifs that can be found multiple times throughout the Vg1LE, and that of the VLE of the VegT mRNA, form the core of the L-RNP of Vg1 mRNA. Mutating the VM1 motifs or deleting the five E2 motifs abolish localization of the Vg1LE, whereas a large number of mutations and deletions throughout the rest of the 340 nt had no effect on localization (Deshler et al. 1997; Gautreau et al. 1997; Havin et al. 1998; Cote et al. 1999; Kwon et al. 2002). Interestingly, the complete primary sequence of Vg1LE is not necessary for localization, since the first 135 nt of the Vg1LE will suffice to drive localization when duplicated, therefore no factors that bind exclusively to the 3′ 205 nt of the Vg1LE are required for the localization process. The 5′ 135 nt contains two VM1 motifs and one E2 motif, consistent with the hypothesis that these motifs are necessary for vegetal localization.
Several trans-acting factors that bind to the Vg1LE have been identified, although of these only three have been demonstrated to be dependent on the VM1 and E2 motifs for binding. Xenopus hnRNP I (VgRBP60) and Vg1RBP/Vera have been identified as the VM1 and E2 interacting proteins, respectively, and evidence suggests that these two proteins may directly interact (Deshler et al. 1998; Havin et al. 1998; Cote et al. 1999; Kwon et al. 2002; Kress et al. 2004; Lewis et al. 2004). An hnRNP-D family protein called 40LoVe was identified by affinity chromatography using the Vg1 and VegT LE, and its binding to the Vg1LE was affected by either VM1 or E2 motif mutations (Czaplinski et al. 2005).
Other proteins have been identified as binding to the Vg1LE, however no VM1 or E2 motif dependence has been examined. VgRBP71 and Prrp were identified as Vg1LE binding proteins by phage display, and both demonstrate binding to vegetally localizing RNA, as well as some other RNAs, and interact with each other in a yeast-two hybrid assay (Zhao et al. 2001; Kroll et al. 2002; Claussen and Pieler 2004). One high affinity binding site for VgRBP71 has been identified near the VM1 motif in the 3′ 205 nt of the Vg1LE, however this proposed VgRBP71 site is absent in the duplicated 1–135-nt element that promotes vegetal localization (Kolev and Huber 2003). Potential binding sites for Prrp have not been directly determined, but SELEX using the highly conserved mouse homolog of Prrp, DAZAP, has revealed likely consensus sites for Prrp binding in Vg1 mRNA (Hori et al. 2005). None of them lie within the first 135 nt of Vg1LE, and all of the indicated sites within the 3′ portion of the VLE could be mutated or deleted without apparent effect on the ability of the Vg1LE to localize (Gautreau et al. 1997; Havin et al. 1998). These data do not rule out that VgRBP71 and Prrp are components of the localizing RNP (L-RNP) but suggest their direct binding to proposed sites of interaction may not be related to the localization process, although it could be involved in other activities of the Vg1LE (Kolev and Huber 2003).
In addition to the directly identified VLE binding proteins, there are several other factors whose activities are likely required for localization to occur. Two Staufen protein homologs in Xenopus are expressed co-incident with the onset of mRNA localization (Allison et al. 2004), and expression of the conserved central core of the XStau1 protein had a dominant negative effect on mRNA localization (Yoon and Mowry 2004), although neither XStau1 nor XStau2 demonstrated specificity for Vg1LE in affinity chromatography (Czaplinski et al. 2005). Also, Kinesin II has been proposed to be involved in localization, since inhibiting Kinesin II activity in oocytes blocked vegetal localization (Betley et al. 2004). Moreover, data suggest that localized mRNA is anchored at the vegetal pole by a mechanism that involves the cytokeratin filament network (Pondel and King 1988; Elinson et al. 1993; Alarcon and Elinson 2001; Bubunenko and King 2001), and some surprising recent data suggest that vegetally localized mRNAs may actually play a structural role in the network (Kloc et al. 2005).
We wished to analyze the role of 40LoVe in the formation of the L-RNP, since, among the motif-dependent binding factors, it demonstrated specificity for the Vg1LE over a non-localizing control RNA in Vg1LE affinity chromatography, and antibodies against 40LoVe inhibited Vg1LE-dependent localization in oocytes (Czaplinski et al. 2005). Here we examine the distribution of 40LoVe protein among tissues and within cells, and report that different isoforms of 40LoVe have differential nucleo/cytoplasmic distributions. Then, consistent with the role of the VM1 and E2 motifs in 40LoVe association with the Vg1LE, we found that 40LoVe’s dependence on these motifs for VLE interaction is at least in part facilitated through protein–protein interaction with the repeat binding proteins. Finally, by competing the motif binding proteins in vivo, we found that the motif binding proteins may have surprisingly different roles during the localization of the Vg1LE.
RESULTS
40LoVe is distributed between the nucleoplasm and the cytoplasm throughout oogenesis
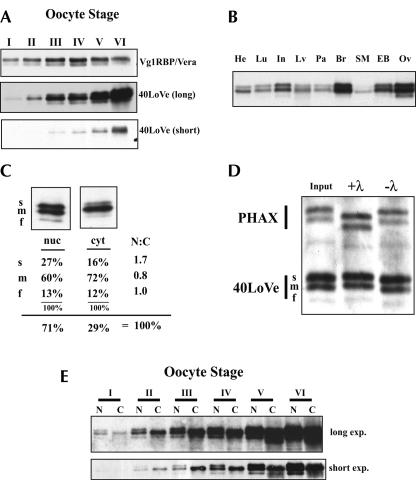
We previously isolated 40LoVe as a novel specific component of the vegetal localization RNP complex that forms on Vg1 and VegT RNA (Czaplinski et al. 2005). To extend our studies on the role of 40LoVe in mRNA localization, we analyzed the distribution of 40LoVe throughout oogenesis and found that the amount of 40LoVe protein increases continuously throughout oogenesis, with clear increases in 40LoVe signal with each stage (Fig. 1A ▶, lower panels). This constant increase is in contrast to other VLE binding proteins, such as Vg1RBP, which shows only a modest increase early in oogenesis and then remains relatively constant (Zhang et al. 1999). As oogenesis progresses the volume of an oocyte increases dramatically (Dumont 1972). Therefore, the relatively constant amount of protein per oocyte for proteins such as Vg1RBP/Vera translates into a decreasing concentration as oogenesis progresses. Conversely, a constant increase in protein per oocyte for proteins such as 40LoVe reflects a relatively constant concentration throughout oogenesis. We do not know the concentration of these factors during oogenesis, but the data allow the conclusion that the relative concentration of these factors, and most likely many others, is changing as oogenesis progresses.
FIGURE 1.
40LoVe distribution in oocytes and tissues. (A) Total protein from one oocyte, stage I–VI as indicated (Dumont 1972), was separated in a 12% SDS-PAGE gel and Western blots were probed for both Vg1RBP/Vera and 40LoVe as indicated. A short (lower panel) and long (middle panel) exposure of identical blots is shown to reflect the large increase in 40LoVe as compared with Vg1RBP/Vera (upper panel). (B) Western blot of 25 μg total protein extracted from the indicated tissues probed with 40LoVe antiserum. Tissues are: heart (He), lung (Lu), small intestine (In), liver (Lv), pancreas (Pa), brain (Br), skeletal muscle (SM), eyeball (EB), and ovary (Ov). (C) One oocyte equivalent of protein from a pool of 10 manually dissected stage III/IV oocyte nuclei (nuc) or cytoplasms (cyt) was separated on an SDS-PAGE gel, probed with 40LoVe antibodies and developed with a fluorescent secondary antibody. The blot was scanned on a fluor-imager to quantify the distribution of the three 40LoVe isoforms indicated fast (f), medium (m), or slow (s). Multiple samples were quantified and the summary of the distribution of the three isoforms is provided below the representative blots. (D) Western blot of isolated oocyte nuclear extract (Input) treated with 200 Units λ-PPase (+λ) or under phosphatase conditions without PPase (−λ). The Western blot was probed with 40LoVe and PHAX antisera sequentially to identify the proteins. (E) One oocyte equivalent from a pool of manually dissected nuclei (N) or cytoplasms (C) of staged oocytes (as indicated) was separated on an SDS-PAGE gel, Western blotted, and probed for 40LoVe, and a short (lower panel) and long (upper panel) exposure of the same gel is shown for comparison.
40LoVe protein expression is not limited to the ovary. Western blotting readily detected 40LoVe in all adult tissues analyzed (Fig. 1B ▶). Ovary and brain contained the highest amounts of 40LoVe signal, while skeletal muscle contained the lowest amount (Fig. 1B ▶). Three isoforms of 40LoVe are observed in all tissues with ratios that are similar to those in the ovary and do not display tissue specificity among the tissues analyzed here.
Nucleo-cytoplasmic shuttling has been demonstrated for several members of the hnRNP D family (Mili et al. 2001; Kawamura et al. 2002; Sarkar et al. 2003; Suzuki et al. 2005), and potential shuttling sequences are conserved in 40LoVe. Since immunofluorescence studies revealed the presence of both nuclear and cytoplasmic 40LoVe (Czaplinski et al. 2005), we determined the nucleo-cytoplasmic distribution of 40LoVe by manual dissection of stage IV oocytes and found approximately two-thirds of the total (combining all three isoforms) nuclear and one-third cytoplasmic (Fig. 1C ▶). Interestingly, the three isoforms of 40LoVe are differently distributed between the compartments. Fluorimetric quantification of the isoforms revealed that the slow isoform demonstrates a higher nuclear/cytoplasmic ratio (1.7) than the middle (0.8) or fast (1.0) isoform (Fig 1C ▶). To determine whether the three 40LoVe isoforms differed in their phosphorylation state, we treated oocyte nuclear extract with λ protein phosphatase. No alteration in the distribution of the three isoforms was observed, while PHAX (Ohno et al. 2000), a positive control for phosphatase activity, increased in mobility after treatment (Fig. 1D ▶). Therefore, the isoforms do not represent different phosphorylation states. By manual dissection, we also found no alteration in the distribution of these isoforms between the nucleus and cytoplasm from stage I–VI of oogenesis (Fig. 1E ▶). Lack of any detectable α-Tubulin in nuclear fractions confirmed the purity of these fractions (data not shown).
40LoVe associates with Vg1RBP/Vera, hnRNP I, and hnRNP D
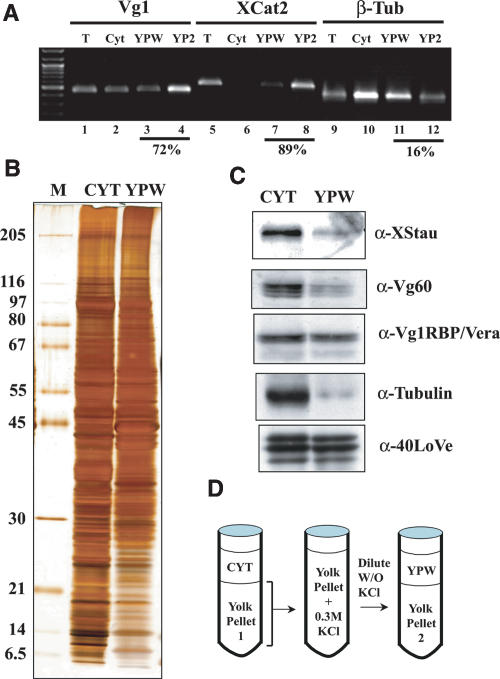
To begin to elaborate the role of 40LoVe in vegetal mRNA localization, we sought to identify 40LoVe cofactors using immunoprecipitation. Vegetally localized RNAs can be found enriched in insoluble fractions during oocyte fractionation (Pondel and King 1988; Bubunenko and King 2001), suggesting that this fraction contains important vegetal localization factors. We therefore reasoned that releasing material from the large insoluble yolk pellet of oocyte fractionation might release these localization factors. We observed that most of the material in the large yolk pellet of an S26 extract was solubilized in 300 mM KCl and that yolk re-precipitates subsequently to dilution of the salts. After a cycle of solubilization and re-precipitation of the yolk pellet (illustrated in Fig. 2D ▶), some pelleted RNA is released into the soluble fraction (Yolk Pellet Wash fraction, YPW). Consistent with previous studies, much of these RNAs remain associated with the second yolk pellet (Fig. 2A ▶). Numerous differences were observed among the proteins present in the total cytoplasmic and YPW fractions demonstrating that this fraction is distinct from soluble cytoplasm (Fig. 2B ▶). Microtubules de-polymerize during preparation of these extracts in the cold, therefore we assayed depletion of soluble cytoplasmic proteins from the yolk wash using anti-α-Tubulin antibodies. Western blotting demonstrated significant depletion of α-Tubulin (Fig. 2C ▶), indicating that the presence of a protein in the YPW fraction is largely due to release from the yolk pellet after the KCl treatment. Probing for hnRNP I and XStau proteins revealed that these proteins are depleted in YPW relative to cytoplasm. However, they are not as depleted as α-Tubulin indicating some release from the pellet. Vg1RBP/Vera and 40LoVe each demonstrates similar levels of protein between the soluble cytoplasm and the YPW fraction, demonstrating that, among putative localization factors, these proteins are highly enriched in the yolk pellet along with vegetal localizing RNAs.
FIGURE 2.
Analysis of the yolk pellet associated localization machinery. (A) RNA from a pool of 40 isolated stage III–IV oocytes was extracted from fractions prepared as illustrated in panel D and RT-PCR from 5 μg of RNA prepared from the fractions indicated was performed using primers specific to Vg1, Xcat-2, or β-Tubulin mRNA. The result demonstrates that the two vegetally localizing RNAs are enriched in the yolk pellet to a much greater extent than tubulin mRNA, and that 0.3 M KCl is sufficient to partially release them from the pellet. Quantification of the fraction of RNA present in the yolk pellet (calculated by adding YPW and YP2 and dividing by the sum of cytoplasm, YPW, and YP2) is indicated below these lanes. (B,) 30 μg of Cytosolic protein (Cyt) or Yolk Pellet Wash protein (YPW) was analyzed by 10%–15% gradient SDS-PAGE. Proteins in these lanes are from a single preparation, sequentially extracted from the same complete ovary. The conditions for extraction were identical to those in panel A. (C) An equivalent gel to that in panel B was Western blotted and probed sequentially with antibodies against several factors implicated in vegetal mRNA localization (indicated to the right of each blot) to assay their release upon washing the yolk pellet. (D) A diagram of the fractionation scheme.
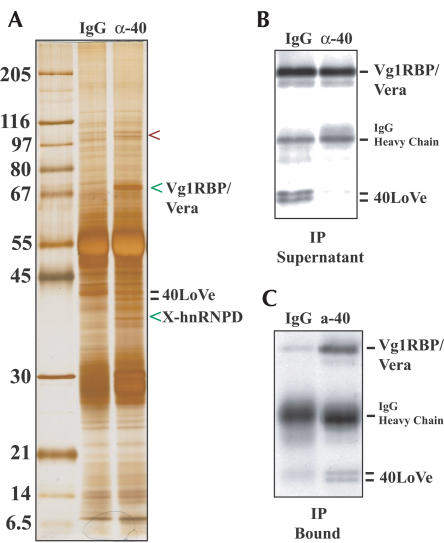
We searched for 40LoVe interaction partners by immunoprecipitation from the YPW fraction. Using affinity purified 40LoVe antibodies we observed three copurifying species (Fig. 3A ▶). The 68-kDa band appears as a doublet and yielded five peptides that all correspond to Vg1RBP/Vera. Western blotting confirmed the presence of Vg1RBP/Vera in the immunoprecipitated fraction (Fig. 3C ▶). We note that, although we deplete approximately 80% of 40LoVe, we only slightly deplete Vg1RBP/Vera (Fig. 3B ▶). An approximately 35-kDa band (Fig. 3A ▶) yielded three peptides that correspond to the X-hnRNP D/AUF1 protein (Czaplinski et al. 2005). The 100-kDa band could not be identified by mass spectrometry.
FIGURE 3.
40LoVe coimmunoprecipitates three proteins from the Yolk Pellet Wash fraction. (A) Eluate from a large-scale immunoprecipitation was analyzed on a 10%–13% gradient SDS-PAGE gel. The position of 40LoVe in the gel is indicated. The location of three proteins specifically present in the 40LoVe IP is indicated by arrowheads. Sequencing of the Vg1RBP/Vera band yielded five peptides, all corresponding to Vg1RBP/Vera. The 35-kDa band yielded three peptides, all matching the Xenopus-hnRNP D/AUF1 protein (Accession AY640106). The 100-kDa band could not be identified. (B) The supernatant of the immunoprecipitation was analyzed by Western blotting for the presence of both Vg1RBP/Vera and 40LoVe (indicated to the right). (C) An aliquot of the immune-precipitates was analyzed to confirm the presence of 40LoVe and Vg1RBP/Vera in the precipitate. The weak 40-kDa signal in the IgG lane is due to a degradation product of control IgG present in this antibody preparation.
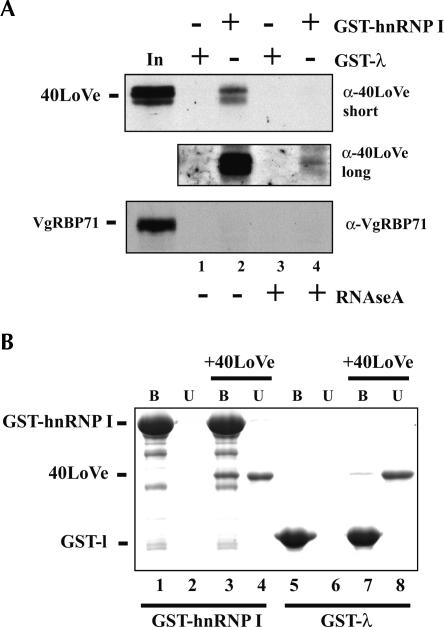
Since Vg1RBP/Vera, a previously identified VLE binding protein, was identified as a 40LoVe interacting partner, we wondered whether hnRNP I might also interact with 40LoVe. IgG heavy chain prevented us from being able to assay for the presence of hnRNP I in these immunoprecipitates owing to the similar molecular weight, so we tested interaction directly between 40LoVe and hnRNP I using GST-hnRNP I affinity purification. GST fused to the Phage-Lambda Box B RNA binding peptide (GST-λ) was used as a control for specificity of interaction. Initially GST fusions were bound to glutathione resin that was incubated in Xenopus total ovary extract. Subsequently, the glutathione resin was washed, eluted, and analyzed for the presence of 40LoVe by SDS-PAGE, followed by Western blotting. 40LoVe was specifically associated with GST-hnRNP I and not GST-λ (Fig. 4A ▶, cf. lanes 1 and 2). Binding in extracts is strongly influenced by RNA, since addition of RNAse A in the washes reduced, but did not eliminate, interaction (Fig. 4A ▶, cf. lanes 2 and 4). To control for the specificity of interaction with other RNA binding proteins, we probed with antibodies to VgRBP71, a four KH-domain containing protein. No interaction between hnRNP I and VgRBP71 was evident (Fig. 4A ▶). Identical results were observed when the YPW fraction was used in place of ovary extract (data not shown).
FIGURE 4.
40LoVe interacts with GST-hnRNP I. (A) Glutathione resin bound to either GST-λ or GST-hnRNP I was incubated in Xenopus ovary extract (Czaplinski et al. 2005) and bound proteins analyzed by Western blotting against 40LoVe or VgRBP71 as indicated. Beads were washed with or without RNAse A as indicated. A short and long exposure of the same 40LoVe blot is shown to reflect the quantitative differences in bound 40LoVe. (B) Purified recombinant GST-λ, GST-hnRNP I, and 6His-40LoVe were combined as indicated, and then total bound (B) and one half of unbound fractions (U) were analyzed by SDS-PAGE gel followed by Coomassie blue staining.
The GST-hnRNP I chromatography established that 40LoVe can associate with hnRNP I in an extract, and we tested if the two proteins can interact directly by using purified recombinant proteins. 6His-40LoVe was mixed with either GST-hnRNP I or GST-λ, and then proteins bound to glutathione resin were analyzed by SDS-PAGE gel. 40LoVe could clearly be detected bound to GST-hnRNP I to a much greater extent than to GST-λ, supporting direct hnRNP I-40LoVe interaction (Fig. 4B ▶, cf. lanes 3 and 7).
Differential roles for E2 and VM1 motif binding factors in vegetal localization
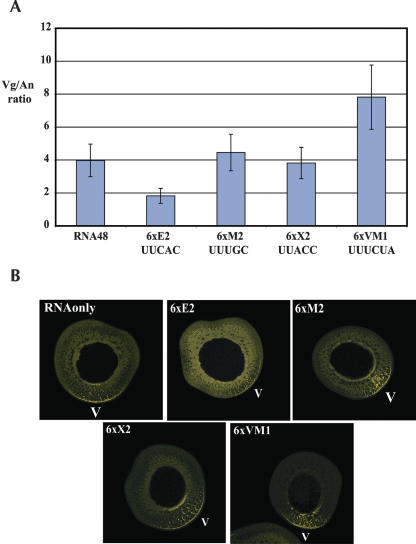
Mutation of the multiple E2 or VM1 motifs in the Vg1LE impairs vegetal localization, suggesting that factors bound to these sequences are required for localization. However, multiple tandem copies of either of these motifs alone cannot promote localization (Lewis et al. 2004), so it is formally possible that mutation of these sites in the Vg1LE alters a different feature that is required for localization and the repeat binding proteins do not play a role in localization. To examine the requirements for motif binding proteins in Vg1LE localization in vivo, we co-injected oocytes with fluorescently labeled Vg1LE containing mRNA (Vg1LE-Xβ-Globin fusion mRNA) and a large molar excess of motif containing RNA. In these experiments we co-injected short RNAs containing six copies of either the E2 motif, two different mutated E2 motifs (M2 or X2) that do not cross-link to Vg1RBP/Vera, or VM1 motifs (Fig. 5 ▶). After 48 h, the amount of localization was quantified and plotted (Fig.5A ▶). Co-injection of E2 repeat RNA strongly inhibited localization, decreasing the Vg/An ratio from 4.0 to 1.9. Surprisingly, co-injection of VM1 repeat containing RNA improved mRNA localization, increasing the Vg/An ratio from 4.0 to 7.9. Co-injection of the M2 or X2 repeats had no effect on the localization process, since the Vg/An ratio in these oocytes remained very close to 4. This result confirms that in vivo E2 repeat element binding proteins are required for localization. Additionally, it shows that VM1 binding proteins functionally interact with the Vg1LE, but since the effect of excess VM1 motif RNA is better localization, VM1 motif binding proteins in the L-RNP may be inhibitory to vegetal localization.
FIGURE 5.
E2 and VM1 repeat binding proteins have opposing effects on vegetal localization. (A) 7 nl of Alexa-546 labelled Xβ-Globin-Vg1LE RNA (conc. 260 nM) was injected to eight Stage III oocytes (RNA48) or co-injected with small RNAs (conc. 45 μM) containing six copies of E2 (UUCACA), M2 (UUUGCA), X2 (UUACCA), or VM1 (UUUCUA) repeats. After 48 h oocytes were fixed, localization was quantified (Czaplinski et al. 2005) and plotted. Error bars represent the 25% variation in ratio that is typically seen in these experiments. (B) Representative oocytes used for quantification are shown; they were co-injected with the indicated repeat containing RNA. The vegetal pole is indicated in all pictures (V).
DISCUSSION
We isolated 40LoVe previously as a novel component of a vegetally localizing RNP (Czaplinski et al. 2005) and characterized its tissue and subcellular distribution to gain further insight into its role in mRNA localization. 40LoVe is present at varying levels in all adult tissues in Xenopus and, therefore, not unique to oocytes (Fig. 1B ▶). The isoforms of 40LoVe that are readily detected by Western blot are not tissue specific but show a different subcellular distribution between nucleus and cytoplasm (Fig. 1C ▶) and may, therefore, demonstrate a differential ability to shuttle between these compartments. Alternative splicing, that specifically removes a glycine–tyrosine rich region, occurs in mammalian 40LoVe homologs (Smidt et al. 1995; Dean et al. 2002). However, the size difference between these isoforms is not consistent with this as an explanation for the different isoforms and we have found no evidence in EST databases that alternative splicing of this region occurs in Xenopus. We do not know the difference between the 40LoVe isoforms, but untagged recombinant 40LoVe (protein accession AAH43814) expressed in Escherichia coli comigrates with the uppermost isoform, suggesting that this isoform likely represents the full-length unmodified protein (data not shown). We demonstrated that the differential mobility of the isoforms is not due to phosphorylation. It is possible that other modifications such as methylation, which are expected to occur on the RGG rich region (Liu and Dreyfuss 1995), or allelic variation for which evidence exists in the EST database, may alter the migration of 40LoVe in an SDS-PAGE gel.
By looking for 40LoVe interacting proteins from oocyte extracts, we identified Vg1RBP/Vera and hnRNP I, both previously implicated in Vg1 mRNA localization. In addition, we isolated the Xenopus hnRNP D homolog. To our knowledge, hnRNP D has not been implicated in mRNA localization, although mammalian homologs of this protein are well known to be involved in regulation of mRNA stability (Wilson and Brewer 1999; Mitchell and Tollervey 2000). Interaction between AUF1 and a mammalian homolog of Vg1RBP/Vera (IMP2) has been observed, consistent with the interaction we observed by coimmunoprecipitation (Moraes et al. 2003). This is the first report describing the isolation of the Xenopus homolog of this protein, and the potential interaction of 40LoVe with this protein as well as a possible role in L-RNPs are interesting areas for future investigation. We currently do not know the precise configuration of protein–protein interactions among these coimmunoprecipitating factors or whether these interactions are RNAse sensitive.
The copurification of Vg1RBP/Vera and hnRNP I with 40LoVe extends our previous observation that interaction of 40LoVe with the Vg1LE involves the E2 and VM1 motifs (Czaplinski et al. 2005), and supports the hypothesis that the requirement of these motifs for 40LoVe binding at least in part reflects an interaction with the cognate motif binding proteins. Direct binding of hnRNP I and Vg1RBP/Vera to the different motifs is extensively supported by many UV cross-linking studies (Deshler et al. 1998; Cote et al. 1999; Kwon et al. 2002; Lewis et al. 2004). We have not found evidence that 40LoVe binds specifically to either VM1 or E2 motifs (data not shown), so the protein–protein interactions reported provide a plausible explanation for the dependence on the motifs for 40LoVe binding. It is important to note that simple protein–protein interactions cannot be sufficient for 40LoVe recruitment, since Vg1RBP/Vera and hnRNP I both interacted well with non-localizing RNA, while 40LoVe did not (Czaplinski et al. 2005). Some unidentified characteristic (a sequence or structure) of the Vg1LE RNA is likely required for stable 40LoVe binding, and by immunoprecipitation subsequent to UV cross-linking we have demonstrated cross-linking of 40LoVe to the Vg1LE (Czaplinski et al. 2005). It has been proposed that the unique configuration of E2 and VM1 elements in Vg1 and VegT LEs accounts for late localization (Bubunenko et al. 2002). Therefore, an explanation for 40LoVe’s E2 and VM1 motif dependence is that 40LoVe can be recruited to RNAs by interactions with Vg1RBP/Vera and hnRNP I uniquely in situations where binding sites for the two proteins are appropriately organized. This model differs from the first possibility only in that it is the arrangement of the E2 and VM1 sites, and therefore the cognate binding proteins, that is the unique characteristic of the RNA.
Our data allow us to propose a model for the initial steps of L-RNP formation. Since depletion of 40LoVe had no effect on the cross-linking of hnRNP I or Vg1RBP/Vera (Czaplinski et al. 2005), but 40LoVe depends on these factors to bind the Vg1LE, we can hypothesize that interaction of the two motif binding proteins with the Vg1LE is the initial step in L-RNP formation. 40LoVe can then join the L-RNP, likely by both protein–protein interaction as well as direct RNA binding. In addition to our reported interactions, interaction between hnRNP I and Vg1RBP/Vera has been characterized (Kress et al. 2004). All three of these components are associated with the oocyte nucleus (Kress et al. 2004; Czaplinski et al. 2005); therefore it is interesting that the formation of this complex for cytoplasmic localization initially occurs in that compartment. However, proper localization occurs following cytoplasmic RNA injection, therefore nuclear complex formation cannot be a requirement for cytoplasmic localization, but it is interesting to speculate that forming such a complex in the nucleus serves some mechanistic purpose.
The observation that titration of either E2 or VM1 motif binding proteins by co-injection of excess motif RNA affected the localization of wild-type VLE is evidence that functional interaction of localization factors with these motifs occurs in vivo. To date, this point has only been examined by mutagenesis, and therefore secondary effects such as structural rearrangements remained a possible explanation for the effects of mutation. Our results support the hypothesis that interaction of RNA binding proteins at these sites is involved in the formation of the L-RNP in vivo. We anticipate that the previously identified E2 and VM1 binding motif proteins are titrated out by excess repeat containing RNA, but it is possible that the titrating RNAs may also target different factors, such as the Xenopus Staufen homologs which are part of the Vg1LE-RNP (Allison et al. 2004; Yoon and Mowry 2004).
The surprising result was that titration of VM1-binding activity in vivo stimulated localization (Fig. 5 ▶). Although this was unexpected because mutation of VM1 motifs blocks localization of Vg1LE (Havin et al. 1998; Cote et al. 1999), it is important to emphasize that this result does not contradict the results of mutagenesis, owing to the very different experimental design. The fact that titration of VM1 binding proteins effects localization is evidence that a VM1 binding factor is a part of the in vivo L-RNP, but the co-injected VLE-RNA localizes when VM1 factor is titrated, therefore such a factor is probably not required for the transport process. We point out that it is not inconsistent for a specific component of an L-RNP to be dispensable for the transport process, and that better localization in its absence is indicative of an inhibitory role in Vg1 localization. We have not directly demonstrated that hnRNP I binding to the Vg1LE is in fact titrated out by injection of the VM1 motifs and, therefore, must carefully consider the conclusions that can be made with regard to hnRNP I. However, we have not found any evidence for other specific VM1 binding proteins by affinity chromatography (data not shown), which is consistent with other studies showing that hnRNP I is the only evident VM1 binding protein (Cote et al. 1999; Lewis et al. 2004). Although we cannot eliminate the possibility that some other unidentified factor is in fact titrated out by the VM1 motif, evaluation of all available evidence suggests that hnRNP I is the most likely target of the excess VM1 motif RNA.
The existence of an activity inhibitory to Vg1LE localization is very consistent with the late mRNA localization pathway. In early oogenesis Vg1 mRNA is uniformly cytoplasmic — at a time when early/METRO pathway localizing mRNAs are localized — and then begins localizing at stage II–III. Several studies indicate mechanistic overlap between these two pathways (Kloc and Etkin 1998; Betley et al. 2002; Chang et al. 2004; Claussen et al. 2004; Choo et al. 2005), therefore an inhibitory activity can function to effect the timing of localization by such a common pathway. Mutating both VM1 motifs blocks localization, therefore VLE recognition by the localization machinery must still require the sequence of the Vg1LE surrounding and including the VM1 motifs, but not the VM1 binding proteins. Given this situation, we consider it possible that an unidentified transport factor recognizes sequences that overlap the VM1 motifs. Overlapping recognition sites explain how the VM1 sites can be required and VM1 binding proteins can be inhibitory, because in this model VM1 binding proteins can sterically hinder the access of localization factors. This possibility simply explains how co-injected excess VM1 motifs can stimulate localization, as long as the unidentified localization factor cannot be titrated out by co-injection of the VM1 sequence. All published Vg1LE mutations are consistent with this explanation. Such a cryptic localization factor need not be a novel protein, although that is possible. Other Vg1LE binding proteins that have not yet been recognized to bind directly to this region (even 40LoVe itself) or RNA–RNA interactions (secondary structure or transcript multimerization) that incorporate this region of the Vg1LE can fulfill the role for such a factor.
Our model for late L-RNP formation predicts that early in oogenesis, activity inhibitory to elements for late localization predominates and, therefore, no localization is observed. Either an increase in positive effectors or a decrease in negative effectors at stage II–III can result in a change in the L-RNP that allows localization to occur. The fact that 40LoVe increases constantly during oogenesis as compared with Vg1RBP/Vera (Fig. 1 ▶) is evidence that ratios of at least these two Vg1LE binding proteins are changing throughout oogenesis. It is also possible that protein modifications positively or negatively effecting localization may occur, thus the ratio of factors as well as their relative activities could change leading to localization. Since several factors in addition to those studied here are involved in vegetal localization, we cannot say at this time which changes could be the most critical. It is interesting to note that hnRNP I phosphorylated at Serine 16 accumulates specifically in the cytoplasm of oocytes as well as other cells, suggesting some link between this modification and cytoplasmic function of hnRNP I (Xie et al. 2003). A combination of these possibilities may also be involved in localizing late mRNAs.
40LoVe is an interesting example of an RNA binding protein whose specificity in the case of the Vg1LE does not appear to derive solely from RNA sequence, but also from protein–protein interaction. L-RNP formation cannot be explained by a single RNA–protein or protein–protein interaction, and mRNA localization is, therefore, the result of multiple subtle interactions among factors that may both positively and negatively affect the ability of the RNP to localize. This example may be generally applicable to understand the formation and function of the large granules characteristic of localizing mRNA, as well as entire UTR regulatory domains that potentially exist in many RNAs.
MATERIALS AND METHODS
Oocyte maintenance and dissection
Ovary from an adult Xenopus laevis was treated with Liberase Blendzyme 3 (Roche) at 100 μg/mL in OR2 at room temperature until most of the oocytes were freed from the surrounding tissue (2.5–3 h). Oocytes were then maintained and injected at 19°C in OR2. Nuclei and cytoplasms were dissected manually in J-Buffer (70 mM NH4Cl, 7 mM MgCL2, 0.1 mM EDTA, 10 mM HEPES pH 7.9) and then immediately transferred to SDS-PAGE loading buffer. Analysis of nuclear and cytoplasmic fractions was from five pooled oocyte nuclei or cytoplasms, and one oocyte equivalent per lane was used. For Phosphatase treatment, several oocyte nuclei isolated in J-Buffer were transferred to 1× Lambda Protein Phosphatase buffer (from NEB), homogenized by pipetting and an aliquot was treated as per manufacturer’s suggestion.
YPW extract preparation
For large-scale YPW, total oocytes from an adult female were obtained by Liberase 3 treatment (see oocyte injection) and washed and maintained overnight at 19°C in Barth’s-Ca-Ab (Barth’s with 0.34 mM Ca(NO3)2 0.4 mM CaCl2, 10 μg/mL each ampicillin and streptomycin). Oocytes were washed several times in cold Barth’s buffer (10 mM HEPES pH 7.6, 87.8 mM NaCl, 1 mM KCl, 2.38 mM NaHCO3, 0.8 mM MgSO4) and packed to remove excess buffer. All further steps were at 4°C. Oocytes were lysed by centrifugation for 20 min at 26,000 ×g. This supernatant is cytosolic extract in Figure 2 ▶, and 1 mM DTT, 5 μg/mL Cytochalasin B, 0.1mM EGTA and 2× protease inhibitor cocktail was added before freezing and storage. The light lipid layer above the yolk pellet was rinsed away after cytosol removal and the thick pellet fraction was resuspended in several small portions with Barth’s buffer containing 0.1 mM EGTA and 1 mM DTT until the entire pellet was resuspended in three volumes of buffer. This was passed several times through a dounce homogenizer to rupture any remaining oocytes. This resuspended pellet was centrifuged 10 min at 12,000 ×g and the supernatant removed and discarded. The pellet was again resuspended in Barth’s, then Barth1200 (Barth with 1.2M KCl) was added such that the final concentration of K plus Na was 300 mM (23.5% the volume of the suspension) and this was dounced several times (becomes clearer and less viscous). This was then diluted with two volumes of BarthNS (Barth lacking KCl and NaCl) with 0.1 mM EGTA and 1 mM DTT (a greenish yellow precipitate forms) and centrifuged 10 min at 12,000 ×g. The supernatant was dialyzed for 2 h in 20 volumes Barth’s with 0.1 mM EGTA, 0.1 mM DTT, and 10% glycerol with changes every 40 min. This was centrifuged for 15 min at 17,000 ×g frozen in aliquots in liquid nitrogen and stored at −70°C.
RT-PCR
RT-PCR for Vg1 (fwd: GACAATGTGGTGCTGAGAC, rev: CAGC TAACAGTCAAGGC), Xcat-2 (fwd: GCTTTGACTCATGGAGCGA, rev: GCCGAGTGAGACATCAGTG), and β-Tubulin (fwd: ATGA GGGAAATCGTGCACTTG, rev: GAGCAGTGTACCCATGCCAGA) mRNA was performed from 5 μg of RNA. RNA was isolated from either whole stage IV oocytes (T), soluble extract of 40 stage IV oocytes homogenized in Barth’s with 5% Sucrose (cyt), the YPW fraction from these oocytes or the final insoluble pellet. Cytosol, YPW, and YP2 in Figure 2 ▶ derive from the same oocytes.
Immunoprecipitation of 40LoVe
Immunoprecipitation of 40LoVe was performed at 4°C, 12 μg of affinity purified anti-40LoVe antibody or non-immune rabbit IgG was added to 720 μg of YPW extract and incubated for 30 min end-over-end. 30 μL of 1:1 protein A Sepharose in BIP (Barth’s, 0.1 mM EGTA, 0.1 mM DTT, 0.05% NP40, 1 mM PMSF, 0.1 mg/mL tRNA, 10% glycerol (v/v)) was added and incubation continued for 1 h. Resin was washed three times in 600 μL of BIP and eluted in 5 × 20 μL aliquots with 0.1% SDS, 5 mM Tris pH 6.8 and combined fractions analyzed by SDS-PAGE.
Recombinant protein
GST-λ production was as described (Czaplinski et al. 2005). 6His-40LoVe (Czaplinski et al. 2005) was expressed in E. coli and purified by Ni-NTA chromatography, followed by Heparin Sepharose chromatography. Binding to Ni-NTA was performed in 50 mM Tris pH 8.0, 500 mM NaCl, 1mM PMSF 0.1% Triton X-100, 15 mM imidazole, 5% glycerol. The column was washed by alternating four times each 500 mM NaCl or 0 mM NaCl in 10 mM Tris pH 8.0, 0.01% Triton X-100, 15 mM imidazole, 5% glycerol. Elution was performed by increasing imidazole to 250 mM in 0 mM NaCl. 0.1 mM DTT was added and then applied to Heparin Sepharose. After extensive washing, elution was performed with a linear gradient of 0–700 mM NaCl in 10 mM Tris 8.0, 0.01% Triton X-100, 0.1 mM DTT. Eluted 40LoVe was dialyzed against 50 mM Tris pH 8.0, 100 mM NaCl, 20% glycerol, 0.1 mM DTT, then stored frozen at −80°C.
GST-hnRNP I was purified by Glutathione Sepharose chromatography, followed by Heparin Sepharose. Human GST-hnRNP I was expressed in E. coli and bound to Glutathione Sepharose in the same conditions as 40LoVe to Ni-NTA, omitting imidazole, adding 2 mM DTT. Wash was the same as 40LoVe, omitting imidazole, with 2 mM DTT. Elution was performed in 20 mM Tris pH 8.0, 250 mM NaCl, 0.01% Triton X-100, 40 mM Glutathione, 5 mM DTT, 5% glycerol. Eluate was diluted with 10 mM Tris pH 8.0 until the NaCl concentration reached 100 mM, then applied to Heparin Sepharose and washed extensively in 10 mM Tris pH 8.0, 0.1 mM DTT. Elution was performed with a 0–1 M NaCl gradient in 10 mM Tris pH 8.0, 0.1 mM DTT. Eluted GST-hnRNP I was dialyzed against 50 mM Tris pH 8.0, 100 mM NaCl, 20% glycerol, 0.1 mM DTT, then stored frozen at −80°C.
GST-pulldown assay
For GST-pulldown from oocyte extract, 250 pmol of GST-hnRNP I or GST-λ was bound to 10 μL Glutathione Sepharose in GST-Pulldown Buffer (GB — 20 mM HEPES 7.9, 100 mM NaCl, 5% glycerol, 1 mM DTT, 0.01% Triton X-100 and 0.01 mg/mL Heparin) for 30 min. Unbound material was removed and replaced with 500 μg total ovary extract (Czaplinski et al. 2005). After 1 h incubating end-over-end in the cold room, beads were washed one time in ice cold GB, followed by 10 min end-over-end in ice cold GB with 0.1 mg/mL RNAse A where indicated. Beads were collected and eluted with SDS-PAGE buffer.
Five micromolar GST-λ or GST-hnRNP I was incubated with or without 5 μM 40LoVe in a 50 μL binding reaction including 10 μL (packed vol) of Glutathione Sepharose in GB. After 45 min rotating in cold room, supernatant from the binding reaction was removed; then beads were washed two times for 5 min in GB and eluted with SDS-PAGE Buffer.
VgRBP71 antibody was raised by expressing a 6His-VgRBP71 alternative splice variant found in the Xenopus Tropicalis EST database (BQ389414, BQ521745, CF218759) lacking the 3rd and 4th KH domains, although other full-length clones containing the 3rd and 4th KH domains could be found. Antibody raised against this fragment recognized full-length VgRBP71 in Xenopus laevis oocyte extracts (data not shown).
RNA localization assay
The RNA localization assay and quantification were as described previously (Czaplinski et al. 2005), except that fluorescent RNA was combined with competitor, to yield the final concentrations in the injection mixtures indicated in the Figure 5 ▶ legend, without lyophilization.
Acknowledgments
K.C. was supported by a National Research Service Award from the NIH and a grant from the Deutsche Forschungsgemeinschaft.
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.2820106.
REFERENCES
- Alarcon, V.B. and Elinson, R.P. 2001. RNA anchoring in the vegetal cortex of the Xenopus oocyte. J. Cell Sci. 114: 1731–1741. [DOI] [PubMed] [Google Scholar]
- Allison, R., Czaplinski, K., Git, A., Adegbenro, E., Stennard, F., Houliston, E., and Standart, N. 2004. Two distinct Staufen isoforms in Xenopus are vegetally localized during oogenesis. RNA 10: 1751–1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betley, J.N., Frith, M.C., Graber, J.H., Choo, S., and Deshler, J.O. 2002. A ubiquitous and conserved signal for RNA localization in chordates. Curr. Biol. 12: 1756–1761. [DOI] [PubMed] [Google Scholar]
- Betley, J.N., Heinrich, B., Vernos, I., Sardet, C., Prodon, F., and Deshler, J.O. 2004. Kinesin II mediates Vg1 mRNA transport in Xenopus oocytes. Curr. Biol. 14: 219–224. [DOI] [PubMed] [Google Scholar]
- Bubunenko, M. and King, M.L. 2001. Biochemical characterization of a cellular structure retaining vegetally localized RNAs in Xenopus late stage oocytes. J. Cell. Biochem. 80: 560–570. [DOI] [PubMed] [Google Scholar]
- Bubunenko, M., Kress, T.L., Vempati, U.D., Mowry, K.L., and King, M.L. 2002. A consensus RNA signal that directs germ layer determinants to the vegetal cortex of Xenopus oocytes. Dev. Biol. 248: 82–92. [DOI] [PubMed] [Google Scholar]
- Chang, P., Torres, J., Lewis, R.A., Mowry, K.L., Houliston, E., and King, M.L. 2004. Localization of RNAs to the mitochondrial cloud in Xenopus oocytes through entrapment and association with endoplasmic reticulum. Mol. Biol. Cell 15: 4669–4681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choo, S., Heinrich, B., Betley, J.N., Chen, Z., and Deshler, J.O. 2005. Evidence for common machinery utilized by the early and late RNA localization pathways in Xenopus oocytes. Dev. Biol. 278: 103–117. [DOI] [PubMed] [Google Scholar]
- Claussen, M. and Pieler, T. 2004. Xvelo1 uses a novel 75-nucleotide signal sequence that drives vegetal localization along the late pathway in Xenopus oocytes. Dev. Biol. 266: 270–284. [DOI] [PubMed] [Google Scholar]
- Claussen, M., Horvay, K., and Pieler, T. 2004. Evidence for overlapping, but not identical, protein machineries operating in vegetal RNA localization along early and late pathways in Xenopus oocytes. Development 131: 4263–4273. [DOI] [PubMed] [Google Scholar]
- Cote, C.A., Gautreau, D., Denegre, J.M., Kress, T.L., Terry, N.A., and Mowry, K.L. 1999. A Xenopus protein related to hnRNP I has a role in cytoplasmic RNA localization. Mol. Cell 4: 431–437. [DOI] [PubMed] [Google Scholar]
- Czaplinski, K., Kocher, T., Schelder, M., Segref, A., Wilm, M., and Mattaj, I.W. 2005. Identification of 40LoVe, a Xenopus hnRNP D family protein involved in localizing a TGF-beta-Related mRNA during oogenesis. Dev. Cell 8: 505–515. [DOI] [PubMed] [Google Scholar]
- Dean, J.L., Sully, G., Wait, R., Rawlinson, L., Clark, A.R., and Saklatvala, J. 2002. Identification of a novel AU-rich-element-binding protein which is related to AUF1. Biochem. J. 366: 709–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshler, J.O., Highett, M.I., and Schnapp, B.J. 1997. Localization of Xenopus Vg1 mRNA by Vera protein and the endoplasmic reticulum. Science 276: 1128–1131. [DOI] [PubMed] [Google Scholar]
- Deshler, J.O., Highett, M.I., Abramson, T., and Schnapp, B.J. 1998. A highly conserved RNA-binding protein for cytoplasmic mRNA localization in vertebrates. Curr. Biol. 8: 489–496. [DOI] [PubMed] [Google Scholar]
- Dumont, J.N. 1972. Oogenesis in Xenopus laevis (Daudin). I. Stages of oocyte development in laboratory maintained animals. J. Morphol. 136: 153–179. [DOI] [PubMed] [Google Scholar]
- Elinson, R.P., King, M.L., and Forristall, C. 1993. Isolated vegetal cortex from Xenopus oocytes selectively retains localized mRNAs. Dev. Biol. 160: 554–562. [DOI] [PubMed] [Google Scholar]
- Gautreau, D., Cote, C.A., and Mowry, K.L. 1997. Two copies of a subelement from the Vg1 RNA localization sequence are sufficient to direct vegetal localization in Xenopus oocytes. Development 124: 5013–5020. [DOI] [PubMed] [Google Scholar]
- Havin, L., Git, A., Elisha, Z., Oberman, F., Yaniv, K., Schwartz, S.P., Standart, N., and Yisraeli, J.K. 1998. RNA-binding protein conserved in both microtubule- and microfilament-based RNA localization. Genes & Dev. 12: 1593–1598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hori, T., Taguchi, Y., Uesugi, S., and Kurihara, Y. 2005. The RNA ligands for mouse proline-rich RNA-binding protein (mouse Prrp) contain two consensus sequences in separate loop structure. Nucleic Acids Res. 33: 190–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawamura, H., Tomozoe, Y., Akagi, T., Kamei, D., Ochiai, M., and Yamada, M. 2002. Identification of the nucleocytoplasmic shuttling sequence of heterogeneous nuclear ribonucleoprotein D-like protein JKTBP and its interaction with mRNA. J. Biol. Chem. 277: 2732–2739. [DOI] [PubMed] [Google Scholar]
- Kessler, D.S. and Melton, D.A. 1995. Induction of dorsal mesoderm by soluble, mature Vg1 protein. Development 121: 2155–2164. [DOI] [PubMed] [Google Scholar]
- King, M.L., Zhou, Y., and Bubunenko, M. 1999. Polarizing genetic information in the egg: RNA localization in the frog oocyte. Bioessays 21: 546–557. [DOI] [PubMed] [Google Scholar]
- King, M.L., Messitt, T.J., and Mowry, K.L. 2005. Putting RNAs in the right place at the right time: RNA localization in the frog oocyte. Biol. Cell 97: 19–33. [DOI] [PubMed] [Google Scholar]
- Kloc, M. and Etkin, L.D. 1998. Apparent continuity between the messenger transport organizer and late RNA localization pathways during oogenesis in Xenopus. Mech. Dev. 73: 95–106. [DOI] [PubMed] [Google Scholar]
- Kloc, M., Zearfoss, N.R., and Etkin, L.D. 2002. Mechanisms of subcellular mRNA localization. Cell 108: 533–544. [DOI] [PubMed] [Google Scholar]
- Kloc, M., Wilk, K., Vargas, D., Shirato, Y., Bilinski, S., and Etkin, L.D. 2005. Potential structural role of non-coding and coding RNAs in the organization of the cytoskeleton at the vegetal cortex of Xenopus oocytes. Development 132: 3445–3457. [DOI] [PubMed] [Google Scholar]
- Kolev, N.G. and Huber, P.W. 2003. VgRBP71 stimulates cleavage at a polyadenylation signal in Vg1 mRNA, resulting in the removal of a cis-acting element that represses translation. Mol. Cell 11: 745–755. [DOI] [PubMed] [Google Scholar]
- Kress, T.L., Yoon, Y.J., and Mowry, K.L. 2004. Nuclear RNP complex assembly initiates cytoplasmic RNA localization. J. Cell Biol. 165: 203–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroll, T.T., Zhao, W.M., Jiang, C., and Huber, P.W. 2002. A homolog of FBP2/KSRP binds to localized mRNAs in Xenopus oocytes. Development 129: 5609–5619. [DOI] [PubMed] [Google Scholar]
- Kwon, S., Abramson, T., Munro, T.P., John, C.M., Kohrmann, M., and Schnapp, B.J. 2002. UUCAC- and vera-dependent localization of VegT RNA in Xenopus oocytes. Curr. Biol. 12: 558–564. [DOI] [PubMed] [Google Scholar]
- Lewis, R.A., Kress, T.L., Cote, C.A., Gautreau, D., Rokop, M.E., and Mowry, K.L. 2004. Conserved and clustered RNA recognition sequences are a critical feature of signals directing RNA localization in Xenopus oocytes. Mech. Dev. 121: 101–109. [DOI] [PubMed] [Google Scholar]
- Liu, Q. and Dreyfuss, G. 1995. In vivo and in vitro arginine methylation of RNA-binding proteins. Mol. Cell. Biol. 15: 2800–2808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mili, S., Shu, H.J., Zhao, Y., and Pinol-Roma, S. 2001. Distinct RNP complexes of shuttling hnRNP proteins with pre-mRNA and mRNA: Candidate intermediates in formation and export of mRNA. Mol. Cell. Biol. 21: 7307–7319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell, P. and Tollervey, D. 2000. mRNA stability in eukaryotes. Curr. Opin. Genet. Dev. 10: 193–198. [DOI] [PubMed] [Google Scholar]
- Moraes, K.C., Quaresma, A.J., Maehnss, K., and Kobarg, J. 2003. Identification and characterization of proteins that selectively interact with isoforms of the mRNA binding protein AUF1 (hnRNP D). Biol. Chem. 384: 25–37. [DOI] [PubMed] [Google Scholar]
- Mowry, K.L. and Melton, D.A. 1992. Vegetal messenger RNA localization directed by a 340-nt RNA sequence element in Xenopus oocytes. Science 255: 991–994. [DOI] [PubMed] [Google Scholar]
- Ohno, M., Segref, A., Bachi, A., Wilm, M., and Mattaj, I.W. 2000. PHAX, a mediator of U snRNA nuclear export whose activity is regulated by phosphorylation. Cell 101: 187–198. [DOI] [PubMed] [Google Scholar]
- Pondel, M.D. and King, M.L. 1988. Localized maternal mRNA related to transforming growth factor beta mRNA is concentrated in a cytokeratin-enriched fraction from Xenopus oocytes. Proc. Natl. Acad. Sci. 85: 7612–7616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rand, K. and Yisraeli, J. 2001. RNA localization in Xenopus oocytes. Results Probl. Cell Differ. 34: 157–173. [DOI] [PubMed] [Google Scholar]
- Sarkar, B., Lu, J.Y., and Schneider, R.J. 2003. Nuclear import and export functions in the different isoforms of the AUF1/heterogeneous nuclear ribonucleoprotein protein family. J. Biol. Chem. 278: 20700–20707. [DOI] [PubMed] [Google Scholar]
- Smidt, M.P., Russchen, B., Snippe, L., Wijnholds, J., and Ab, G. 1995. Cloning and characterisation of a nuclear, site specific ssDNA binding protein. Nucleic Acids Res. 23: 2389–2395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki, M., Iijima, M., Nishimura, A., Tomozoe, Y., Kamei, D., and Yamada, M. 2005. Two separate regions essential for nuclear import of the hnRNP D nucleocytoplasmic shuttling sequence. FEBS J. 272: 3975–3987. [DOI] [PubMed] [Google Scholar]
- Weeks, D.L. and Melton, D.A. 1987. A maternal mRNA localized to the vegetal hemisphere in Xenopus eggs codes for a growth factor related to TGF-beta. Cell 51: 861–867. [DOI] [PubMed] [Google Scholar]
- Wilson, G.M. and Brewer, G. 1999. The search for trans-acting factors controlling messenger RNA decay. Prog. Nucleic Acid Res. Mol. Biol. 62: 257–291. [DOI] [PubMed] [Google Scholar]
- Xie, J., Lee, J.A., Kress, T.L., Mowry, K.L., and Black, D.L. 2003. Protein kinase A phosphorylation modulates transport of the polypyrimidine tract-binding protein. Proc. Natl. Acad. Sci. 100: 8776–8781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon, Y.J. and Mowry, K.L. 2004. Xenopus Staufen is a component of a ribonucleoprotein complex containing Vg1 RNA and kinesin. Development 131: 3035–3045. [DOI] [PubMed] [Google Scholar]
- Zhang, Q., Yaniv, K., Oberman, F., Wolke, U., Git, A., Fromer, M., Taylor, W.L., Meyer, D., Standart, N., Raz, E., et al. 1999. Vg1 RBP intracellular distribution and evolutionarily conserved expression at multiple stages during development. Mech. Dev. 88: 101–106. [DOI] [PubMed] [Google Scholar]
- Zhao, W.M., Jiang, C., Kroll, T.T., and Huber, P.W. 2001. A proline-rich protein binds to the localization element of Xenopus Vg1 mRNA and to ligands involved in actin polymerization. EMBO J. 20: 2315–2325. [DOI] [PMC free article] [PubMed] [Google Scholar]