Abstract
We describe a method for prediction of genes that encode the RNA component of the signal recognition particle (SRP). A heuristic search for the strongly conserved helix 8 motif of SRP RNA is combined with covariance models that are based on previously known SRP RNA sequences. By screening available genomic sequences we have identified a large number of novel SRP RNA genes and we can account for at least one gene in every genome that has been completely sequenced. Novel bacterial RNAs include that of Thermotoga maritima, which, unlike all other non-gram-positive eubacteria, is predicted to have an Alu domain. We have also found the RNAs of Lactococcus lactis and Staphylococcus to have an unusual UGAC tetraloop in helix 8 instead of the normal GNRA sequence. An investigation of yeast RNAs reveals conserved sequence elements of the Alu domain that aid in the analysis of these RNAs. Analysis of the human genome reveals only two likely genes, both on chromosome 14. Our method for SRP RNA gene prediction is the first convenient tool for this task and should be useful in genome annotation.
INTRODUCTION
Although there are many efficient tools for the identification of protein genes in genome sequences, we lack tools for the analysis of many non-coding RNAs. As a consequence most of the finished genome sequences published today lack annotation information about these RNAs. One difficulty with the identification of non-coding RNAs is that sequence tends to be poorly conserved and, as a consequence, standard tools such as BLAST (1) may be used only in the identification of orthologs in fairly closely related organisms. On the other hand many non-coding RNAs tend to have conserved secondary structure elements that may be used for their identification. Methods that have been used so far include the ‘covariance models’ developed by Eddy and Durbin (2) as well as ‘pattern matching’, which is based on regular expression matching where base-pairing schemes are also taken into account. In the latter category there is PatScan (http://www-unix.mcs.anl.gov/compbio/PatScan/HTML/patscan.html) as well as the ‘rnabob’ tool of Sean Eddy (Washington University School of Medicine, St Louis, MO; http://www.genetics.wustl.edu/eddy/software/#rnabob).
In this work we have studied methods to identify the RNA component of the signal recognition particle (SRP). The SRP plays an important role in membrane targeting (3 and references therein). Some of its structural and functional features have been highly conserved during evolution and SRP has been identified in all three kingdoms of life. In eukaryotic cells, the SRP targets secretory proteins to the endoplasmic reticulum (ER) membrane. Eukaryotic SRP comprises one RNA molecule and six proteins, including SRP54, a GTPase. As the signal sequence of the secretory protein emerges from the ribosome it is recognized by the SRP54 protein. In the resulting ribosome–SRP complex, protein synthesis is arrested and the complex eventually docks to the SRP receptor at the ER membrane. Here, SRP is released, protein synthesis is resumed and translocation of the nascent chain in the ER membrane is initiated. The SRP receptor is composed of two subunits, SRα and SRβ, which are both GTPase proteins.
The SRP pathway in bacteria is similar to that in eukaryotes although the major substrates for the bacterial SRP seem to be integral membrane proteins (4–6). In Escherichia coli the SRP has only two components: a 4.5S RNA and the Ffh protein, homologous to SRP54. The bacterial FtsY protein is homologous to SRα.
The overall design of SRP RNA is somewhat variable, as shown in Figure 1. However, they all share a common motif that will be referred to here as the helix 8 motif. The helix 8 region of the RNA binds to the M domain of the SRP54 protein, a domain that also harbors the signal sequence binding activity. Many studies show that the SRP RNA has an important biological role. For instance, the E.coli 4.5S RNA is necessary for viability (7) and in an in vitro system the SRα can stimulate the GTPase activity of SRP54 only when it is bound to the RNA (8). It has been shown that the E.coli 4.5S RNA plays an important role in the GTPase cycles of Ffh and FtsY (9,10). Furthermore, the three-dimensional structure of the helix 8/SRP54 domain of SRP has recently been elucidated (11) and this structure suggests that the signal sequence binding surface is composed not only of protein but also of RNA elements.
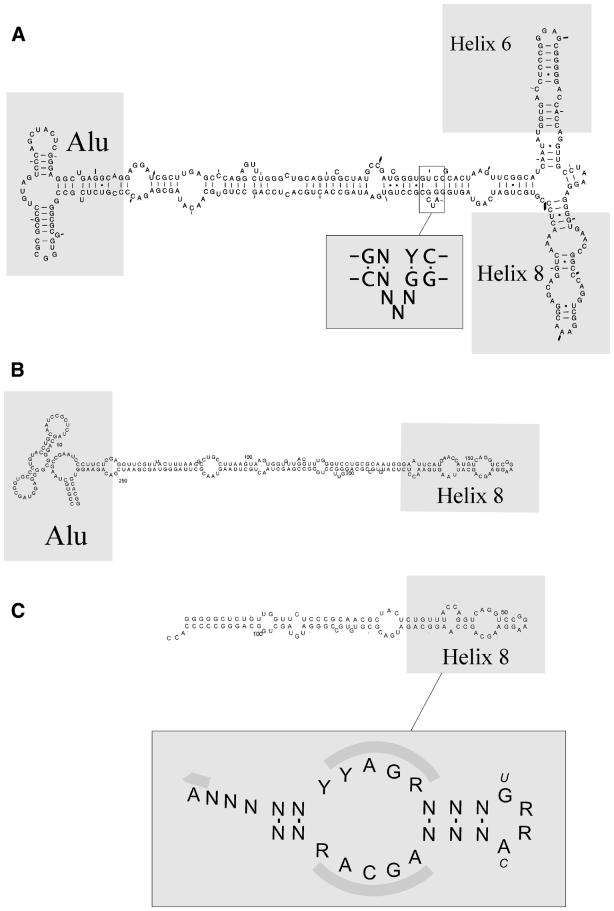
Figure 1.
The three major categories of SRP RNA and conserved elements. (A) Eukaryotic and archaebacterial RNAs that contain helices 6 and 8 as well as an Alu domain. The human SRP RNA is shown together with a highly conserved motif in helix 5 shared between most SRP RNAs that contain an Alu domain, as shown in the present work. (B) Eubacterial RNAs that contain an Alu domain but that lack the helix 6 (Bacillus RNA shown). (C) Eubacterial RNAs without an Alu domain (E.coli RNA shown). The consensus features of the helix 8 region of bacterial SRP RNA are indicated in the lower box. Lactococcus and Staphylococcus have the UGAC tetraloop sequence. The shaded regions in the lower box correspond to bases in contact with the SRP54 protein as described by Batey et al. (11).
The Alu domain is believed to be responsible for a translational arrest activity of SRP and contains the Alu domain of the RNA in complex with the heterodimer SRP9/14. Also this complex has been studied in structural detail (12). The SRP9/14 dimer is bound to a conserved core of the RNA Alu domain, which forms a U-turn that connects two helical stacks. Based on the appearance of the SRP as a rod-like structure, with the SRP54 binding site and Alu domain at opposing ends, it has been speculated that the particle is able to control translation by reaching from the ribosome polypeptide exit site to a site of elongation factor or aminoacyl-tRNA binding. Due to the limited size of the Alu domain it could well enter into a tRNA or elongation factor binding site.
There are three major classes of SRP RNA with respect to its domain organization (Fig. 1). Archaebacterial RNAs are eukaryote-like with an Alu domain as well as a helix 6 domain. A group of gram-positive bacteria, including Bacillus, has an Alu domain but lacks the helix 6 region. The remaining eubacteria-like proteobacteria have a simpler RNA that lacks the Alu domain as well as the helix 6 domain.
The great diversity in terms of SRP RNA structure has hampered the development of convenient methods for their identification in genome sequences. In the context of the Signal Recognition Particle Database (SRPDB) (13) we have previously attempted to identify SRP RNA genes using a combination of pattern searches and sequence similarity searches. However, we have now improved our search strategy considerably and developed an automated method that is based on pattern matching and the COVE programs. Using this method we have systematically searched for SRP RNA genes in public sequence data and have discovered a number of novel SRP RNAs as well as novel features of these RNAs.
MATERIALS AND METHODS
Public sequence data were downloaded from the NCBI and EBI ftp sites. Finished sequence data as well as HTG sections were retrieved. Human expressed sequence tag (EST) sequences were downloaded and used to obtain evidence of expressed SRP RNA genes. Genomes of microorganisms were also obtained from the TIGR CMR database at http://www.tigr.org/tigr-scripts/CMR2/CMRGenomes.spl and sites cited there. Sequences of Candida albicans were downloaded from the Stanford Genome Technology Center website at http://www-sequence.stanford.edu/group/candida (sequencing of C.albicans was accomplished with the support of the NIDR and the Burroughs Wellcome Fund). Neurospora sequences were from the Neurospora Sequencing Project, Whitehead Institute/MIT Center for Genome Research (http://www-genome.wi.mit.edu). The data set used in these studies was neurospora_1.fasta.
Rnabob. Rnabob source code was obtained from http://www.genetics.wustl.edu/eddy/software/#rnabob. Rnabob descriptors were constructed for the different categories: (i) archaebacteria and eubacteria with GRRA loop; (ii) eubacteria with TRRC loop; (iii) plants; (iv) yeasts; and (v) metazoans. They are listed and described in more detail at the website http://bio.lundberg.gu.se/srpscan/. The rnabob source code was slightly modified to produce output with sequences flanking the descriptor pattern and to be able to produce sequence output in fasta format. It was used to scan the sequence databases with the appropriate descriptors for the different classes of SRP RNA genes and to produce outputs in fasta format using the -f option. The -s (skip) option was used to avoid non-significant hits in regions of ‘N’ repeats.
COVE. The COVE programs (version 2.4.2) were obtained from http://www.genetics.wustl.edu/eddy/software/#cove. COVE statistical models were built from the SRP RNA genes alignments available on the SRPDB website at http://bio.lundberg.gu.se/dbs/SRPDB/srprna.html using the -a option of the covet (train) program. The same sequences, unaligned, were used as initial training data for the models. The -m (maximum likelihood) option was used to give more reliable results. Statistical models were built for the following categories: (i) prokaryotes without Alu domain; (ii) prokaryotes with Alu domain; (iii) yeasts; and (iv) non-yeast eukaryotes. Each model was trained on the subset of sequences in the SRPDB belonging to organisms of that particular category. Once novel sequences were found, models were rebuilt on the new set of sequences to obtain a higher accuracy. The covels (local score) program was used to search rnabob fasta outputs for SRP RNA gene candidates. A list was produced with the sequences with the best match to the statistical model used. The -w (window) option was set to a value slightly larger than the expected maximum size for an SRP RNA. Alignments of the sequences were constructed using the covea (align) program and the appropriate statistical model for each category. The -o (outfile) and -s (scorefile) options were used to produce file outputs with alignments and scores of similarity to the model. SRP RNA genes alignments and COVE covariance statistical models are available at the website http://bio.lundberg.gu.se/srpscan/.
Mfold. Mfold version 3.1 was generously provided by Dr M. Zuker (Rensselaer Polytechnic Institute, Troy, NY) (14). It was used to predict the secondary structure of predicted SRP RNAs. To obtain a folding consistent with the secondary structure constraints implicit in the rnabob descriptors or in the COVE models, we used these constraints as input to mfold in many instances.
Automated procedure. Perl scripts were used to produce an automatic procedure where genomic sequences are searched using rnabob with descriptors of the helix8 motif in combination with the COVE programs. As an alternative, rnabob may be used alone, where one makes use of descriptors for helix8 as well as for the Alu domain to obtain candidates for the full RNA sequence.
RESULTS AND DISCUSSION
A protocol for identification of SRP RNA genes
As many other non-coding RNAs, SRP RNA is poorly conserved with respect to sequence and effective methods for RNA identification have to incorporate secondary structure information. The region most highly conserved in SRP RNA is the helix 8 region, which is characterized by a specific base-pairing scheme in combination with primary sequence elements, as indicated in Figure 1. It is interesting to note that there is a strong correlation between the consensus features and the sites of protein interaction as revealed by the structure of an SRP54–RNA complex (Fig. 1C) (11).
We have developed a procedure to identify potential SRP RNA genes in genome sequences where the first step relies on the identification of the helix 8 motif. We search for this motif using a slightly modified version of the RNA motif finder rnabob (http://www.genetics.wustl.edu/eddy/software). The helix 8 motif is retrieved together with appropriate flanking sequences. As a second step, the sequence(s) retrieved in the first step may be further analyzed using rnabob with a more detailed descriptor that also takes into account the Alu domain motif. An example of such a descriptor for plant RNAs is shown in Figure 2. However, a more accurate, although slower, method is to use the probabilistic covariance models implemented in the COVE programs (2) to analyze the sequences retrieved in the first pattern matching step where a model is produced on the basis of previously known SRP RNA sequences. The COVE programs were also highly useful in producing multiple alignments of SRP RNA sequences, to identify conserved motifs in these RNAs and to predict the 5′ and 3′ ends of the RNA, as will be described in more detail below. It should be noted that COVE is computationally demanding and therefore it is not feasible to analyze complete genome sequences without a first heuristic step. It is possible that the pattern-matching approach as a first step will exclude sequences that would obtain a high score from COVE, but we have tried to minimize this problem by using relatively degenerate descriptors in the rnabob search.
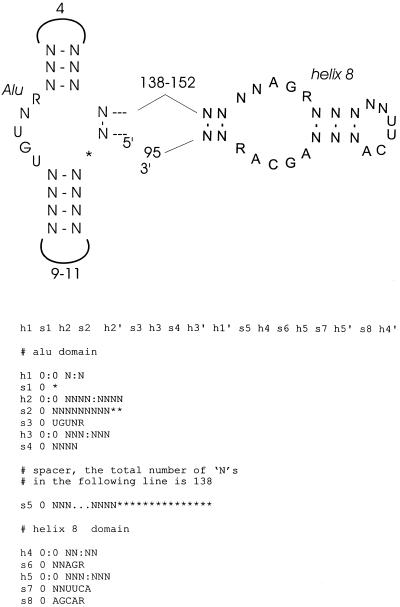
Figure 2.
Characteristics of plant SRP RNAs. Consensus features of helix 8 and Alu domains are shown schematically, as well as descriptor for rnabob based on these features. N represents any nucleotide and an asterisk indicates an optional nucleotide. The numbers refer to allowed number of bases of Alu domain loops and spacers between the Alu and helix 8 domains.
As SRP RNAs are very different in different taxonomic groups, it was not possible to use a single helix 8 descriptor for rnabob or a single COVE model for an efficient identification of all SRP RNAs. For instance, the helix 8 motif is different in plants and certain yeasts that have a 6 nt (GYUUCA) loop instead of the normal tetraloop sequence, and a few bacteria have a UGAC tetraloop (see below). Therefore, for the problem of gene identification we had to consider different classes of SRP RNA such as archaebacteria, eubacteria with and without Alu domain, plants, yeasts and metazoans.
A web server for the prediction of SRP RNA genes is available at http://bio.lundberg.gu.se/srpscan/. These pages also present more detailed information on the work described here. The web service also presents the potential secondary structure of the predicted RNA sequence according to mfold (14). In this procedure the folding is constrained by the base pairing as specified by the rnabob descriptor or by the COVE model used.
Identifying the helix 8 motif in bacterial genome sequences
As mentioned above, the first step of our SRP RNA gene finding procedure is one where we search for the helix 8 motif. As a starting point we designed a pattern based on all previously known SRP RNA sequences as collected from the SRPDB (13). A helix 8 motif NNAGG–xxx–GNRA–yyy– AGCAG (where xxx pairs with yyy) was derived from available SRP RNA genes. We then used rnabob to identify this pattern in genome sequence data. From a majority of bacterial genomes, this motif identified one gene that could be successfully folded into a hairpin structure characteristic of SRP RNAs.
However, in a few genomes, such as in Vibrio cholerae and Lactococcus lactis, no SRP RNA could be identified using this motif. To resolve this problem we made BLAST (1) or FASTA (15) searches using SRP RNA sequence from a closely related organism. Hence, the E.coli sequence was used as a query in a BLAST or FASTA search of the V.cholerae genome. This result revealed one homologous sequence in Vibrio that could be folded into a hairpin structure and that had the consensus features of the helix 8, except that in one of the bulges it had NNAGA instead of the normal NNAGG. We then made a rnabob search in bacterial genomic sequences with a revised pattern, NNAGR–xxx–GNRA–yyy–AGCAG. As expected, this search revealed the SRP RNA gene candidate in Vibrio but also in Deinococcus radiodurans and Buchnera spp., two other strains for which we previously failed to identify an SRP RNA.
The pattern as described still failed to identify an RNA in the strains Staphylococcus aureus and L.lactis. A FASTA search of the Lactococcus genome using the Bacillus subtilis SRP RNA revealed a homolog that had the characteristics of SRP RNA but, instead of the normal GGAA tetraloop, it had the unusual tetraloop sequence UGAC (Fig. 3). This unexpected finding led us to search for a helix 8 pattern where we had substituted the GNRA tetraloop for UGAC. Using rnabob with such a pattern also revealed SRP RNA candidates in S.aureus (Fig. 3) and Staphylococcus equi. Thus, both Lactococcus and Staphylococcus have the helix 8 motif but with the UGAC tetraloop. Even though this is a very unusual tetraloop sequence we believe that we have identified the true SRP RNA gene homologs. This is because the sequence similarity between these RNAs and SRP RNAs of other closely related bacteria is highly significant. Furthermore, there are no other obvious SRP RNA gene candidates in Lactococcus and Staphylococcus. Finally, the SRP RNA gene candidates with the UGAC loop are not part of any known protein-coding regions.
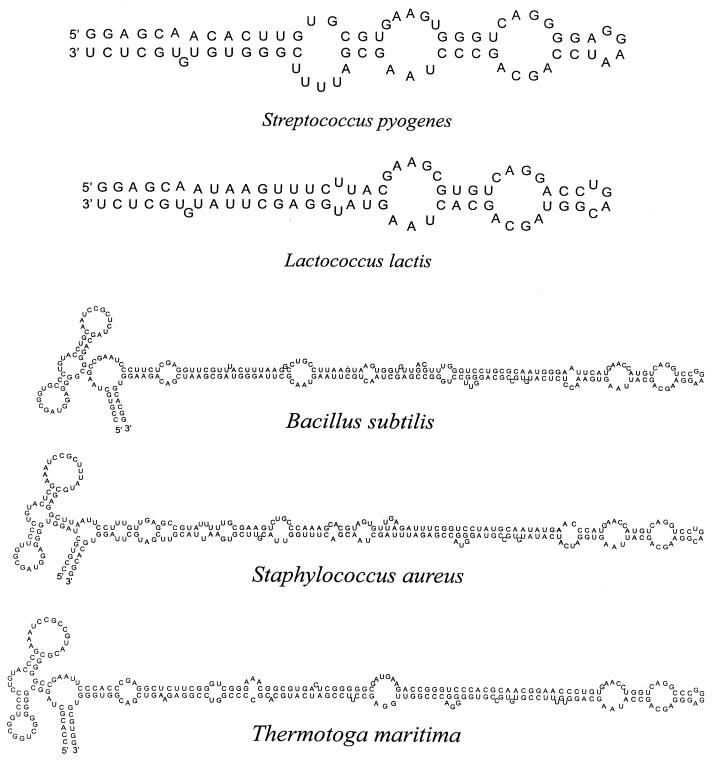
Figure 3.
Comparison of SRP RNAs with GGAA and UGAC tetraloops. Streptococcus pyogenes and Lactococcus are closely related in sequence and secondary structure but differ with respect to their tetraloop sequence. Similarly, the Bacillus and Staphylococcus RNAs, both with an Alu domain, are closely related but differ in their tetraloop sequence. Also shown is the predicted RNA of T.maritima, which was unexpectedly found to have an Alu domain like the Bacillus group of RNAs.
It is interesting to note that the UGAC tetraloop is found in two species that both have close relatives with the normal loop sequence. Thus, Staphylococcus is most closely related to Bacillus, and Lactococcus is most closely related to Streptococcus. A phylogenetic tree with these sequences is shown in Figure 4. It would therefore seem as if the change from GGAA to UGAC loop sequence occurred at least twice during evolution. This suggests that the UGAC loop confers some biological advantage in these gram-positive bacteria. Recently, evidence has been presented that the tetraloop sequence of E.coli SRP RNA (4.5S RNA) is important for the interaction of FtsY, the receptor for bacterial SRP (16). In the context of this evidence it will be interesting to test the function of the UGAC tetraloop experimentally as well as to study its structural role in the helix 8 domain.
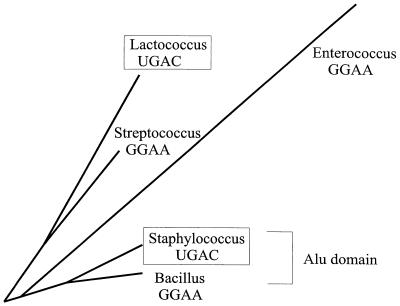
Figure 4.
Relationship of gram-positive SRP RNAs with unusual UGAC tetraloop. Shown in the figure is a part of a phylogenetic tree based on an alignment with COVE of all available SRP RNA sequences from this study. The tree was based on UPGMA and was drawn using Phylodraw (33). As indicated in the figure, the Lactococcus RNA is more closely related to the Streptococcus RNA than to the Staphylococcus RNA, and the Staphylo coccus RNA is more closely related to the Bacillus RNA than to the Lactococcus RNA. Furthermore, the Staphylococcus/Bacillus RNAs have an Alu domain.
For every prokaryotic genome we examined we identified only one possible SRP RNA gene candidate. When we used rnabob alone for prediction there was sometimes more than one gene candidate to one genome, but most often false positives could be identified because they did not fold into the hairpin structure characteristic of SRP RNAs or because they were part of protein-coding regions. At the same time, gene predictions for bacterial genomes with the COVE models were always very specific, i.e. for each genome tested we observed only one gene candidate. When we used a COVE model with the helix 8 domain only to score potential SRP RNA gene candidates obtained from an rnabob search we sometimes observed 16S rRNA genes with a relatively high score. However, these hits are not likely to be significant as the regions in 16S RNA identified by COVE are not the same in the different RNAs.
Bacterial Alu domains
Archaebacteria and certain eubacteria have SRP RNAs with domains that seem to be analogous to the Alu domains of eukaryotic SRP RNAs. Such eubacterial RNAs with Alu domains have been identified in Bacillus, Listeria and Clostridium. We now wanted to know which RNAs that we identified using rnabob and the helix 8 motif descriptor also contain an Alu domain. We used rnabob with a descriptor accounting for consensus features of the Alu domain together with a variable spacer between the helix 8 and the Alu domain. The features of the Alu domain descriptor were based on a careful inspection of available Bacillus, Listeria and Clostridium sequences. The resulting descriptor was used to analyze all the sequences that we identified using the helix 8 motif as described above. In addition to the Bacillus, Listeria and Clostridium sequences that we expected to find, we also identified an Alu domain in the RNA of Thermotoga maritima (Fig. 3). A similar result was obtained when we used the COVE programs with gram-positive Alu domain RNAs in the model. This was a somewhat unexpected finding as Thermotoga is clearly distinct from the Firmicutes group. The Thermotoga RNA does not contain a helix 6 region, and in this respect it belongs to the Bacillus group of RNAs. This finding raises interesting questions as to the relationship between Thermotoga and the Bacillus branch. One possible explanation for the presence of an Alu domain RNA in Thermotoga is a horizontal gene transfer event. However, we find no evidence of such an event, as phylogenetic trees based on SRP RNA sequences are very similar to trees based on 16S rRNA sequences (data not shown).
In a search among the archaebacterial genomes we found as expected an RNA with an Alu domain in all of the species examined, also in the recently sequenced Thermoplasma volcani and Thermoplasma acidophilum. All the sequences we identified also had a helix 6 region according to the structure predicted by mfold. The Aeropyrum pernix RNA differs from the other archaebacterial RNAs in that it seems to have a helix inserted in the Alu domain between the helices 3 and 4 (http://bio.lundberg.gu.se/srpscan).
We noted that the sequence from Sulfolobus solfataricus reported by Kaine (17) is very different from the sequence predicted from the genome sequence of the same bacterium (18). In fact, it is as distant to the genomic sequence as to the gene predicted from the genome of Sulfolobus tokodaii (19). Unless there are very large differences between different isolates of S.solfataricus, it seems likely that there is a mistake as to the identity of the strain.
Prediction of 5′ and 3′ ends
We want our method to predict as accurately as possible the 5′ and 3′ ends of the RNA gene product. For the RNAs with an Alu domain we expect that the ends should be close to the Alu domain. For the eubacterial RNAs without an Alu domain the prediction of ends cannot be guided by such an element, and another difficulty is that there is a variation in size between different species. A few such RNAs have been sequenced, such as those of E.coli (20), a number of mycoplasmas (21–23) and Pseudomonas aeruginosa (24), with a variation in size from 77 to 114 nt. However, analysis by COVE can also make use of the fact that a majority of genes have a T-rich sequence immediately downstream of the 3′ end of the mature sequence. Figure 5 shows an alignment of a number of sequences of RNAs without Alu domain that was produced by COVE and where the starting point was an alignment where the T-rich regions had been manually aligned. However, a similar result was obtained when unaligned sequences were used as input to the program. In spite of the variation in size between these bacterial RNAs, it seems that the COVE model can predict the 5′ and 3′ ends reasonably well. For the RNAs that have been sequenced and for which we have information about the correct ends, these have the predicted site <2–3 bp from the correct one. Of course we cannot exclude the possibility that the ends of a few RNAs are incorrectly predicted because these RNAs have a structure which is different from the expected hairpin structure.
Figure 5.
Alignment of eubacterial RNAs without Alu domain. All eubacterial SRP RNAs without an Alu domain identified in this study were aligned using a COVE model. The figure shows selected sequences from this alignment. The starting point for the alignment was one where T-rich regions at the 3′ end were manually aligned to each other, but a similar result was obtained when unaligned sequences were used as input. The T-rich region is highlighted as well as conserved elements of the helix 8 region (see also Fig. 1).
The T-rich sequence downstream of the gene is highlighted in Figure 5. The hairpin structure of the mature RNA sequence in combination with the T-rich sequence is highly reminiscent of the rho-independent transcription termination signal. It seems likely that this signal is the actual transcription termination signal and that very little, if any, 3′ end processing is needed to obtain the mature 3′ end.
Analysis of eukaryotes
Whereas prokaryotic genomes have only one SRP RNA gene, many eukaryotes contain multiple genes as listed in Table 1. As for the analysis of prokaryotes, it was necessary to define different descriptors and cove models for different taxonomic groups. For COVE analysis, we divided eukaryotes into yeasts and non-yeasts.
Table 1. Eukaryotic SRP RNA genes as predicted from genomic sequences.
| Chromosomal location | Number of genes | GenBank accession numbers | |
|---|---|---|---|
| Neurospora crassa | Unknown | 1 | – |
| Candida albicans | Unknown | 1 | – |
| Arabidopsis thaliana | Chromosome 1 | 2 | AC011807 |
| AC013453 | |||
| Chromosome 2 | 2 | AC005662 | |
| AC005311 | |||
| Chromosome 4 | 3 | AC005275 | |
| AF069442 | |||
| Chromosome 5 | 1 (+ 2 pseudogenes?) | ATF17I14 | |
| AB026661 | |||
| Drosophila | Chromosome 3R | 2 | AC095014 |
| Caenorhabditis elegans | Chromosome 3 | 4 | CEB0285 |
| U00064 | |||
| U23515 | |||
| CEB0284 | |||
| Chromosome 5 | 1 (pseudogene?) | U41993 | |
| Homo sapiens | Chromosome 14 | 2 | CNS01DX7 |
| AC020711 |
The Drosophila genes are identical, in an opposite orientation, and separated by ∼2500 nt. The genes on C.elegans chromosome 3 have 5′ start positions 3172520, 3162782, 3822816 and 4011623, respectively.
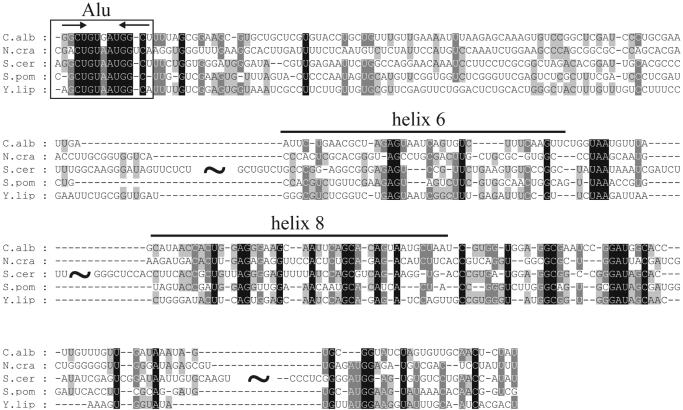
Yeasts. The yeast SRP RNA genes are difficult to identify and analyze as they show a low degree of conservation between different yeast species, and because they are distinct from other eukaryotic RNAs. Previously, SRP RNAs have been identified in Yarrowia (25), Schizosaccharomyces pombe (26) and Saccharomyces cerevisiae (27). The S.cerevisiae sequence is unusual in that the total length of the RNA is 519 nt, whereas most eukaryotic RNAs are ∼300 nt. All the yeast sequences have a helix 8 motif like the metazoan counterparts. However, the Alu domain is very different from the bacterial and metazoan counterparts. It has been suggested that the S.pombe, S.cerevisiae and Yarrowia lipolytica RNAs have a simplified Alu domain that has the consensus features indicated in Figure 6 (28).
Figure 6.
Tentative alignment of yeast RNAs. RNAs shown are from S.cerevisiae, S.pombe and Y.lipolytica as well as RNAs predicted in this study from genome sequence data of N.crassa and C.albicans. The alignment was produced by COVE. COVE had difficulties in correctly aligning helix 8 of the S.cerevisiae RNA to the corresponding part of the other RNAs, using the unaligned sequences as a starting point. Therefore, the starting point for the COVE alignment shown in the figure was one where the helix 8 domain of the S.cerevisiae RNA had been manually aligned with the helix 8 domain of the other RNAs. A conserved motif at the 5′ end is shown with the consensus sequence RCUGURAUGGY, with base pairing as indicated by the arrows. The ‘∼’ symbols indicate insertions in the S.cerevisiae RNA relative to the other RNAs.
We now also analyzed unfinished genome sequence data of Neurospora crassa (http://biology.unm.edu/biology/ngp/home.html) and C.albicans (http://www-sequence.stanford.edu/group/candida/) and were able to use our method to detect SRP RNA candidates in these genomes. This was possible by using in the first screen relatively degenerate helix 8 descriptors for rnabob. In the case of Neurospora it has a 6 nt loop in the helix 8 part like S.cerevisiae.
Interestingly, both C.albicans and N.crassa RNAs have a sequence that is consistent with the simplified Alu motif, suggesting that this motif is highly conserved in yeasts. Figure 6 shows a tentative alignment with the yeast sequences obtained by COVE. It highlights the homology in the Alu domain as well as a predicted structural homology of part of the molecule with the helices 6 and 8. On the basis of this alignment it seems likely that yeast RNAs basically have the same folding as other other SRP RNAs although there are large insertions in the S.cerevisiae RNA with unknown folding. It must be noted that it is difficult to predict the 5′ and 3′ ends of the yeast RNAs and experimental studies will have to be carried out in order to determine these as well as to fully understand the folding of the different yeast RNAs.
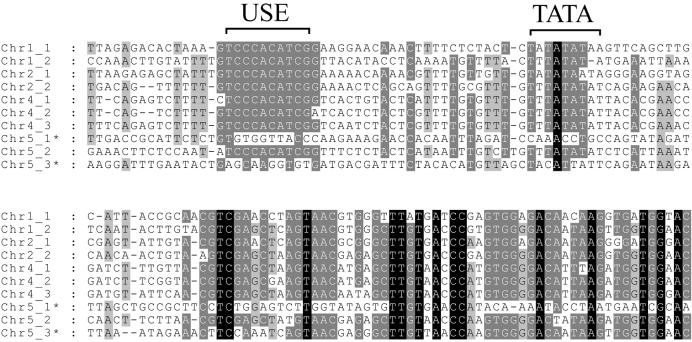
Plants. The plants tend to have fairly conserved SRP RNAs. The predictions of plant RNAs might therefore be considered more reliable than for the other categories of eukaryotes. In Arabidopsis thaliana we have identified eight different gene candidates, as shown in Figure 7. Of these, only two were previously reported (13,29). It will require experimental work to see if all these genes are actually expressed or if some of them are pseudogenes. It has previously been shown that two Arabidopsis SRP RNA genes contain promoter elements, USE and TATA, upstream of the coding region that are identical to the promoters of pol III-specific plant U-snRNA RNA genes (30) and that are important for transcriptional activity. It has been proposed that the plant SRP RNA transcription mainly relies on these elements and that internal promoter elements play a minor role. These upstream elements are indicated in Figure 7 and it is interesting to note that they are found in virtually all the novel genes. We therefore speculate that the majority of these genes are not pseudogenes. However, two of the genes on chromosome 5 do not have these elements (Fig. 7) and may therefore be pseudogenes.
Figure 7.
Predicted genes in Arabidopsis. Multiple alignment of predicted SRP RNA genes in A.thaliana was produced by COVE using unaligned genes as input. Only the 5′ portion of mature RNA together with upstream sequences are shown. Promoter elements that were previously shown to be important for transcription of SRP RNA genes (USE and TATA) in Arabidopsis are indicated (30). Two of the genes on chromosome 4 were previously identified. Asterisks indicate possible pseudogenes.
Metazoans. In Drosophila and Caenorhabditis elegans we found two and five genes, respectively (Table 1). The coding sequences of the Drosophila genes are identical and the genes are located in an opposite orientation, separated by ∼2500 nt. In C.elegans the genes on chromosome 3 were identified before whereas the one on chromosome 5 is a novel finding. However, it might be a pseudogene as the promoter sequence is very different from the genes on chromosome 3. As with the Drosophila genes, the genes on chromosome 3 are in pairs with the genes in opposite orientation (Table 1).
The prediction of SRP RNA genes in the human genome is complicated by the presence of a large number of Alu repeats evolutionarily related to SRP RNA. As a result there is a large background from hits to Alu repeats in the list of high-scoring hits from COVE. The strongest candidate genes are three genes on chromosome 14 (one gene in CNS01DX7 and two genes in AC020711; see Table 1) and one on chromosome 10 (in GenBank accession no. AC091810, not shown in Table 1). The predicted RNAs of these genes can all be folded into the structure characteristic of eukaryotic RNAs. The genes on chromosome 14 have similar sequences immediately upstream of the coding sequence whereas the gene on chromosome 10 is different in this region. We also carried out BLAST searches of human EST database sequences to obtain evidence of expression of these genes. The results show that the major species in EST databases are CNS01DX7 (∼50%) and one of the genes in AC020711 (∼50%). These two variants also correspond to the sequences reported by Ullu et al. (31). The fact that only one gene on the AC020711 segment is expressed also has support from previous experimental studies (32). In conclusion, the likely predominating human SRP RNA genes are the one contained in CNS01DX7 and one of the genes in AC020711 on chromosome 14.
When COVE was used to produce an alignment of all our predicted eukaryotic SRP RNAs, including those of yeasts, the UGU consensus sequence of the predicted Alu domain of the yeast RNAs, as indicated in Figure 6, aligned well with the corresponding sequence in the Alu domains of the metazoans. This gives further support to the structural relationship between the metazoan and yeast Alu domains. The result of this alignment also shows that it is possible to use only one COVE model for the prediction and analysis of all eukaryotic SRP RNAs, with the possible exception of the S.cerevisiae RNA. This model will now be very useful in the identification of SRP RNA genes in genomes that become available in the future.
Alignment of all the eukaryotic SRP RNAs also revealed conserved elements. In addition to the helix 8 and Alu domain motifs already referred to, the most prominent one is the motif shown in Figure 1, which is located in helix 5 near the helices 6 and 8. This region of the RNA is involved in the interaction with the SRP68/72 protein dimer. Possibly, the conserved motif is an important recognition element for these proteins, just as the conserved parts of helix 8 and the Alu domain are recognized by the SRP9/14 dimer and the SRP54 protein, respectively.
Conclusion
Non-coding RNA genes vary in their degree of primary and secondary structure conservation. It is obvious that the lower the degree of conservation, the more difficult is the prediction of such genes from genomic sequence information. Certain RNAs, like rRNAs, are so conserved that they may be effectively detected by primary sequence homology only. Other RNAs, like tRNA and SRP RNA, have to incorporate a search for secondary structure features. With both tRNA and SRP RNA, the degree of conservation in terms of combined primary and secondary structure features is sufficient to develop an accurate prediction method. However, there are more difficult cases, like the yeast SRP RNAs, that quite strongly diverge from a consensus structure. The predictions of yeast RNAs will therefore have to be verified by experimental studies. Experimental work is also required to determine the exact location of the 5′ and 3′ ends of mature RNAs and to distinguish pseudogenes from genes that give rise to functional RNAs.
Nevertheless, the methods described are in general very efficient in identifying SRP RNA gene candidates. In this method we combine a heuristic pattern screen for the conserved helix 8 and Alu domain motifs with the COVE programs. We account for SRP RNA genes in all organisms that have been completely sequenced. We have detected a number of novel RNAs as well as novel features of SRP RNAs such as the unusual tetraloop sequence of the Lactococcus and Staphylococcus RNAs. In particular, our method is very sensitive and specific in the prediction of bacterial RNA genes. For the first time, we now have an automated procedure to detect SRP RNA genes that may be used as a routine tool during genome annotation.
REFERENCES
- 1.Altschul S.F., Madden,T.L., Schaffer,A.A., Zhang, J., Zhang,Z., Miller,W. and Lipman,D.J. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res., 25, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Eddy S.R. and Durbin,R. (1994) RNA sequence analysis using covariance models. Nucleic Acids Res., 22, 2079–2088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Keenan R.J., Freymann,D.M., Stroud,R.M. and Walter,P. (2001) The signal recognition particle. Annu. Rev. Biochem., 70, 755–775. [DOI] [PubMed] [Google Scholar]
- 4.Ulbrandt N.D., Newitt,J.A. and Bernstein,H.D. (1997) The E. coli signal recognition particle is required for the insertion of a subset of inner membrane proteins. Cell, 88, 187–196. [DOI] [PubMed] [Google Scholar]
- 5.Newitt J.A., Ulbrandt,N.D. and Bernstein,H.D. (1999) The structure of multiple polypeptide domains determines the signal recognition particle targeting requirement of Escherichia coli inner membrane proteins. J. Bacteriol., 181, 4561–4567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lee H.C. and Bernstein,H.D. (2001) The targeting pathway of Escherichia coli presecretory and integral membrane proteins is specified by the hydrophobicity of the targeting signal. Proc. Natl Acad. Sci. USA, 98, 3471–3476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brown S. and Fournier,M.J. (1984) The 4.5 S RNA gene of Escherichia coli is essential for cell growth. J. Mol. Biol., 178, 533–550. [DOI] [PubMed] [Google Scholar]
- 8.Miller J.D., Wilhelm,H., Gierasch,L., Gilmore,R. and Walter,P. (1993) GTP binding and hydrolysis by the signal recognition particle during initiation of protein translocation. Nature, 366, 351–354. [DOI] [PubMed] [Google Scholar]
- 9.Peluso P., Herschlag,D., Nock,S., Freymann,D.M., Johnson,A.E. and Walter,P. (2000) Role of 4.5S RNA in assembly of the bacterial signal recognition particle with its receptor. Science, 288, 1640–1643. [DOI] [PubMed] [Google Scholar]
- 10.Peluso P., Shan,S.O., Nock,S., Herschlag,D. and Walter,P. (2001) Role of SRP RNA in the GTPase cycles of Ffh and FtsY. Biochemistry, 40, 15224–15233. [DOI] [PubMed] [Google Scholar]
- 11.Batey R.T., Rambo,R.P., Lucast,L., Rha,B. and Doudna,J.A. (2000) Crystal structure of the ribonucleoprotein core of the signal recognition particle. Science, 287, 1232–1239. [DOI] [PubMed] [Google Scholar]
- 12.Weichenrieder O., Wild,K., Strub,K. and Cusack,S. (2000) Structure and assembly of the Alu domain of the mammalian signal recognition particle. Nature, 408, 167–173. [DOI] [PubMed] [Google Scholar]
- 13.Gorodkin J., Knudsen,B., Zwieb,C. and Samuelsson,T. (2001) SRPDB (Signal Recognition Particle Database). Nucleic Acids Res., 29, 169–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zuker M. (1989) On finding all suboptimal foldings of an RNA molecule. Science, 244, 48–52. [DOI] [PubMed] [Google Scholar]
- 15.Pearson W.R. (2000) Flexible sequence similarity searching with the FASTA3 program package. Methods Mol. Biol., 132, 185–219. [DOI] [PubMed] [Google Scholar]
- 16.Jagath J.R., Matassova,N.B., de Leeuw,E., Warnecke,J.M., Lentzen,G., Rodnina,M.V., Luirink,J. and Wintermeyer,W. (2001) Important role of the tetraloop region of 4.5S RNA in SRP binding to its receptor FtsY. RNA, 7, 293–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kaine B.P. (1990) Structure of the archaebacterial 7S RNA molecule. Mol. Gen. Genet., 221, 315–321. [DOI] [PubMed] [Google Scholar]
- 18.She Q., Singh,R.K., Confalonieri,F., Zivanovic,Y., Allard,G., Awayez,M.J., Chan-Weiher,C.C., Clausen,I.G., Curtis,B.A., De Moors,A. et al. (2001) The complete genome of the crenarchaeon Sulfolobus solfataricus P2. Proc. Natl Acad. Sci. USA, 98, 7835–7840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kawarabayasi Y., Hino,Y., Horikawa,H., Jin-no,K., Takahashi,M., Sekine,M., Baba,S., Ankai,A., Kosugi,H., Hosoyama,A. et al. (2001) Complete genome sequence of an aerobic thermoacidophilic crenarchaeon, Sulfolobus tokodaii strain7. DNA Res., 8, 123–140. [DOI] [PubMed] [Google Scholar]
- 20.Hsu L.M., Zagorski,J. and Fournier,M.J. (1984) Cloning and sequence analysis of the Escherichia coli 4.5 S RNA gene. J. Mol. Biol., 178, 509–531. [DOI] [PubMed] [Google Scholar]
- 21.Muto A., Andachi,Y., Yuzawa,H., Yamao,F. and Osawa,S. (1990) The organization and evolution of transfer RNA genes in Mycoplasma capricolum. Nucleic Acids Res., 18, 5037–5043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Samuelsson T. and Guindy,Y. (1990) Nucleotide sequence of a Mycoplasma mycoides RNA which is homologous to E. coli 4.5S RNA. Nucleic Acids Res., 18, 4938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Simoneau P. and Hu,P.C. (1992) The gene for a 4.5S RNA homolog from Mycoplasma pneumoniae: genetic selection, sequence, and transcription analysis. J. Bacteriol., 174, 627–629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Toschka H.Y., Struck,J.C. and Erdmann,V.A. (1989) The 4.5S RNA gene from Pseudomonas aeruginosa. Nucleic Acids Res., 17, 31–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.He F., Yaver,D., Beckerich,J.M., Ogrydziak,D. and Gaillardin,C. (1990) The yeast Yarrowia lipolytica has two, functional, signal recognition particle 7S RNA genes. Curr. Genet., 17, 289–292. [DOI] [PubMed] [Google Scholar]
- 26.Brennwald P., Liao,X., Holm,K., Porter,G. and Wise,J.A. (1988) Identification of an essential Schizosaccharomyces pombe RNA homologous to the 7SL component of signal recognition particle. Mol. Cell. Biol., 8, 1580–1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Felici F., Cesareni,G. and Hughes,J.M. (1989) The most abundant small cytoplasmic RNA of Saccharomyces cerevisiae has an important function required for normal cell growth. Mol. Cell. Biol., 9, 3260–3268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Strub K., Fornallaz,M. and Bui,N. (1999) The Alu domain homolog of the yeast signal recognition particle consists of an Srp14p homodimer and a yeast-specific RNA structure. RNA, 5, 1333–1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Marques J.P., Gualberto,J.M. and Palme,K. (1993) Sequence of the Arabidopsis thaliana 7SL RNA gene. Nucleic Acids Res., 21, 3581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Heard D.J., Filipowicz,W., Marques,J.P., Palme,K. and Gualberto,J.M. (1995) An upstream U-snRNA gene-like promoter is required for transcription of the Arabidopsis thaliana 7SL RNA gene. Nucleic Acids Res., 23, 1970–1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ullu E., Murphy,S. and Melli,M. (1982) Human 7SL RNA consists of a 140 nucleotide middle-repetitive sequence inserted in an alu sequence. Cell, 29, 195–202. [DOI] [PubMed] [Google Scholar]
- 32.Ullu E. and Tschudi,C. (1984) Alu sequences are processed 7SL RNA genes. Nature, 312, 171–172. [DOI] [PubMed] [Google Scholar]
- 33.Choi J.H., Jung,H.Y., Kim,H.S. and Cho,H.G. (2000) PhyloDraw: a phylogenetic tree drawing system. Bioinformatics, 16, 1056–1058. [DOI] [PubMed] [Google Scholar]