Abstract
The crystal structure of the recombinant hepatitis C virus (HCV) RNA-dependent RNA polymerase (RdRp) revealed extensive interactions between the fingers and the thumb subdomains, resulting in a closed conformation with an established template channel that should specifically accept single-stranded templates. We made circularized RNA templates and found that they were efficiently used by the HCV RdRp to synthesize product RNAs that are significantly longer than the template, suggesting that RdRp could exist in an open conformation prior to template binding. RNA synthesis using circular RNA templates had properties similar to those previously documented for linear RNA, including a need for higher GTP concentration for initiation, usage of GTP analogs, sensitivity to salt, and involvement of active-site residues for product formation. Some products were resistant to challenge with the template competitor heparin, indicating that the elongation complexes remain bound to template and are competent for RNA synthesis. Other products were not elongated in the presence of heparin, indicating that the elongation complex was terminated. Lastly, recombinant RdRps from two other flaviviruses and from the Pseudomonas phage φ6 also could use circular RNA templates for RNA-dependent RNA synthesis, although the φ6 RdRp could only use circular RNAs made from the 3′-terminal sequence of the φ6 genome.
Keywords: hepatitis C virus, RNA-dependent RNA polymerase, elongation, circular template, ternary complex, heparin
INTRODUCTION
Template-dependent RNA polymerases tend to share a general structure that is analogous to a right hand, in which the palm subdomain contains the catalytic residues while the thumb and the fingers subdomains are involved in interaction with the template (Joyce and Steitz 1995; Hansen et al. 1997). DNA-dependent RNA polymerases (DdRp) have been extensively characterized for the mechanism of RNA synthesis, which can be divided into several biochemically distinct steps: binding of the DdRp to the promoter, formation of a transcriptionally active open complex, synthesis of the first phosphodiester bond, abortive RNA synthesis, promoter clearance, processive elongation, and, finally, termination (McClure 1985). A key step in RNA synthesis for DdRps is template binding, in which the polymerase undergoes a large conformational change involving the thumb subdomain to allow stable binding to the template (von Hippel et al. 1984; Steitz 2004). Binding of the substrate NTP causes the fingers subdomain of the polymerase to rotate from an open to a closed conformation (Steitz 1999).
Viral RNA-dependent RNA polymerases are less well-characterized, but where characterizations exist they follow steps similar to those used by DNA-dependent RNA polymerases (Sun et al. 1996; Sun and Kao 1997a,b; Adkins et al. 1998). Several RdRps could also recognize specific RNA promoters for accurate initiation of RNA synthesis (Siegel et al. 1997; Choi et al. 2004b; Li and Stollar 2004). Several RdRps have now been crystallized, including those from HCV, poliovirus, bacteriophage φ6, foot and mouth disease virus, human rhinovirus, bovine viral diarrhea virus (BVDV), and rabbit hemorrhagic disease virus (Hansen et al. 1997; Ago et al. 1999; Bressanelli et al. 1999; Lesberg et al. 1999; Butcher et al. 2001; Ng et al. 2002; Choi et al. 2004a; Ferrer-Orta et al. 2004; Love et al. 2004). The HCV RdRp active site has several novel features, such as a β-loop of 12 residues and the C-terminal tail of the RdRp that lines the active site. The β-loop and the C-terminal tail, along with a GTP bound at the NTPi site, will suppress RNA synthesis from a primed template (Ranjith-Kumar et al. 2003a). Another notable feature of the HCV RdRp is that, unlike the relatively open conformation of DNA polymerase I (Ollis et al. 1985a; Li et al. 1998), the T7 RNA polymerase (Ollis et al. 1985b), and the retroviral reverse transcriptase (Sarafianos et al. 2002), the extensive interactions between the fingers and the thumb subdomains allows the HCV RdRp to form a closed catalytic pocket in the absence of template and NTP. The closed catalytic pocket and the template channel should favor the use of templates with single-stranded 3′-termini, a requirement observed in vitro (Kao et al. 2000; Vo et al. 2003). The encircled active site is not unique to the HCV RdRp since the RdRp from the Pseudomonas phage φ6 also contains an encircled active site with a well-defined template channel (Butcher et al. 2001). Furthermore, several de novo initiating viral RdRps, such as those from phage Qβ and several plant RNA viruses, require single-stranded 3′-ends for de novo initiation and have mechanisms to protect the 3′-termini (Rao et al. 1989; Rao and Hall 1993; Nagy et al. 1997; Chapman and Kao 1999; Dreher 1999; Schuppli et al. 2000; Pogany et al. 2003).
The structure of the HCV RdRp raises the question of whether the RdRp can only accept linear templates that thread through the template channel and/or undergo conformational changes associated with the initiation of RNA synthesis. A prediction of a stably enclosed active site is that a circular RNA template would not be able to direct RNA-dependent RNA synthesis, since the thumb and the fingers domains would have to open to allow a circular template to be bound. In this work, we demonstrate that circular templates are used for RNA synthesis by the HCV and the φ6 RdRps, functionally demonstrating that these RdRps undergo transitions between closed and open states.
RESULTS
Circular RNAs are templates for the HCV RdRp
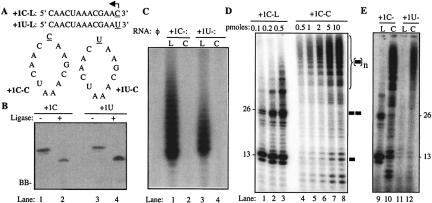
We attempted to circularize several RNAs before finding that a 13-nt RNA sequence named +1C could be efficiently ligated to form a covalently closed circle (Fig. 1A ▶). In fact, +1C was readily and nearly completely ligated into the circular form after a 1-h incubation with T4 RNA ligase. RNA +1C contains four cytidylates. Based on previous characterization of short linear templates (Kao et al. 2000), the cytidylate at the 3′-terminus should be efficiently used as the T1 nucleotide. We also made a second 13-nt sequence named +1U that should decrease de novo initiation from a linear template drastically since the next available cytidylate from the 3′-end would be at the +5 position (Fig. 1A ▶).
FIGURE 1.
Linear and circular 13-nt templates for RNA-dependent RNA synthesis. (A) Sequences of the linear and circular templates are shown with the arrow indicating the direction of RNA synthesis. RNAs +1C-L and +1U-L differ only at the 3′-most nucleotide (underlined). (B) Circularization of the RNAs. Both +1C and +1U were circularized with T4 RNA ligase to result in a conformation that migrated at a lower position in a 7.5 M urea–20% polyacrylamide gel. Each lane contains between 30 and 50 pmol of RNA. The RNA bands were visualized by staining with toluidine blue. Treatment with the T4 ligase is indicated by +. The faint band at the bottom of the gel image is bromophenol blue (BB). (C) A demonstration that the ligated RNAs exist in a covalently closed form. The purified RNAs were adjusted to 0.1 μM and then incubated in poly(A) polymerase buffer, 0.1 units of poly(A) polymerase (Ambion Inc.), and [α-32P]ATP. The reaction products were then precipitated with ethanol, centrifuged, and electrophoresed in a 7.5 M urea–15% polyacrylamide gel. (D) An image from a denaturing polyacrylamide gel containing products made by Δ21 with increasing concentrations of linear or circular template. Flaviviral RdRps can synthesize one product molecule from two or more linear molecules of the template RNA. These template-switch products are therefore multiples of the length of the template (Kim and Kao 2001). The names and amounts of the RNAs are listed above the appropriate lanes of the gel image. The lengths of the product RNAs from the linear template, in nucleotides, are shown to the left of the gel image. The bar to the right of the gel signifies a monomer template-sized product, two bars correspond to dimer, and the bar within parentheses with subscript “n” corresponds to multimeric forms. (E) Initiation of RNA synthesis from linear and circular templates. The 3′-terminal cytidylate is the primary initiation nucleotide for RNA synthesis by the HCV Δ21 from linear, but not circular, templates. All four reactions were performed with 0.1 pmol of linear and 10 pmol of circular template RNA and 0.04 μM Δ21.
Several properties were observed in the ligated versions of +1C and +1U that led us to conclude that covalently closed-circle RNAs were made. First, both migrated at a lower position in a denaturing gel than the linear molecule (Fig. 1B ▶). Second, the ligated RNAs were resistant to the treatment by calf intestinal phosphatase, while their unligated counterparts were readily dephosphorylated (data not shown). Third, the ligated RNAs were unable to serve as templates for the addition of 3′-terminal adenylates by poly(A) polymerase (Fig. 1C ▶). To facilitate identification of the templates used in this work, we refer to the linear and circular forms of these RNAs with an “-L” and a “-C”, respectively. For example, +1C-L refers to the linear form of +1C.
The version of the HCV RdRp (nonstructural protein 5B, NS5B) lacking the C-terminal 21 hydrophobic residues, named Δ21, can generate four classes of products with short linear RNAs: (1) de novo initiation products that are the same length of the template or one that terminated 1 nt short of the 5′-end of the template, (2) templates elongated by the addition of one or a few terminal nucleotides, (3) primer-extended products that arose from the duplexes formed by two RNAs, and (4) template-switch products generated when the HCV RdRp ternary complex uses two or more noncovalently linked templates to synthesize one product (Ranjith-Kumar et al. 2001, 2002a, b, 2003b). When tested for RNA synthesis by the HCV RdRp, +1C-L directed the synthesis of several products, the most abundant being 13 nt in length, which likely initiated using the 3′-terminal cytidylate (Fig. 1D ▶, lanes 1–3). These products were much reduced with +1U-L (Fig. 1E ▶, lane 11), as would be expected since it lacks the 3′-terminal cytidylate preferred for de novo initiation. Since the process of de novo initiation is required for the formation of the template-switch products (Kim and Kao 2001), +1U-L is also deficient for the formation of the higher-molecular-weight RNAs (Fig. 1E ▶, cf. lanes 9 and 11). With +1C-C, however, we observed several products, most of which formed a ladder that reached the top of the 15% gel (Fig. 1D ▶, lanes 4–8), suggesting that the HCV RdRp could use a circular template to generate products akin to rolling circle replication. These higher-molecular-weight products increased with template concentration and were distinct from the template-switch products seen with +1C-L (Fig. 1D ▶, cf. lanes 1–3 and 4–8). Notably, the +1U-C template also generated higher-molecular-weight products. One of the other three cytidylates within +1U-C likely served as the initiation cytidylate, but not at the same efficiency as the two consecutive cytidylates within +1C-C (Fig. 1E ▶). With +1C-C as template, there were two prominent products of 9 and 10 nt, which appeared to be due to the premature termination of RNA synthesis (Fig. 1D ▶, lanes 4–8). Formation of radiolabeled products required the presence of all four NTPs (data not shown), indicating that the smear of products is due to template-dependent RNA synthesis.
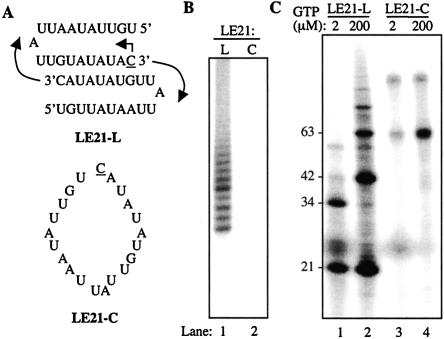
To analyze whether Δ21 can use larger circular RNA, we circularized RNA LE21, which was used to characterize the initiation properties of Δ21 (Fig. 2A ▶; Ranjith-Kumar et al. 2002b, 2003a). Circularization of LE21 was less efficient than with +1C, but purification of the uncircularized and circularized RNAs from a denaturing gel resulted in the recovery of LE21-L (linear) and LE21-C (circular). As with +1C-C, LE21-C was unable to serve as a template for poly(A) polymerase (Fig. 2B ▶). Analysis of the products from LE21-C that will be presented later confirms that it is a covalently closed circle.
FIGURE 2.
Δ21 can use a 21-nt circular RNA template. (A) The sequence of linear LE21, LE21-L along with the possible dimer formation between two molecules of RNA is shown. The arrows indicate the direction of RNA synthesis. The initiation nucleotide is underlined. (B) A demonstration that the ligated RNAs exist in a covalently closed form. The purified RNAs were treated with poly(A) polymerase and [α-32P]ATP as described in Figure 1 ▶. (C) Initiation of RNA synthesis from linear and circular templates. The reactions were performed with 0.2 pmol of linear and 5 pmol of circular template RNA and 0.04 μM Δ21. The two different concentrations of GTP, 2 μM and 200 μM, used are given on the top, and the sizes of RNA products are given at the left of the gel.
LE21-L can generate 21-nt de novo initiated RNA product along with 42 and 63, and its multimers by end-to-end template switch (Fig. 2C ▶, lanes 1,2). Also, two molecules of LE21 can anneal together and direct the synthesis of 34-nt primer extension products (Fig. 2A,C ▶). In contrast, LE21-C generated a product close to 60 nt and one of ~120 nt that are not observed in reactions with LE21-L (Fig. 2C ▶, lanes 3,4). Interestingly, very few 21-nt products were seen with LE21-C, a situation different from that of synthesis with +1C-C (cf. Fig. 2C ▶ and Fig. 1D ▶).
RNA synthesis by a catalytic mutant and full-length NS5B
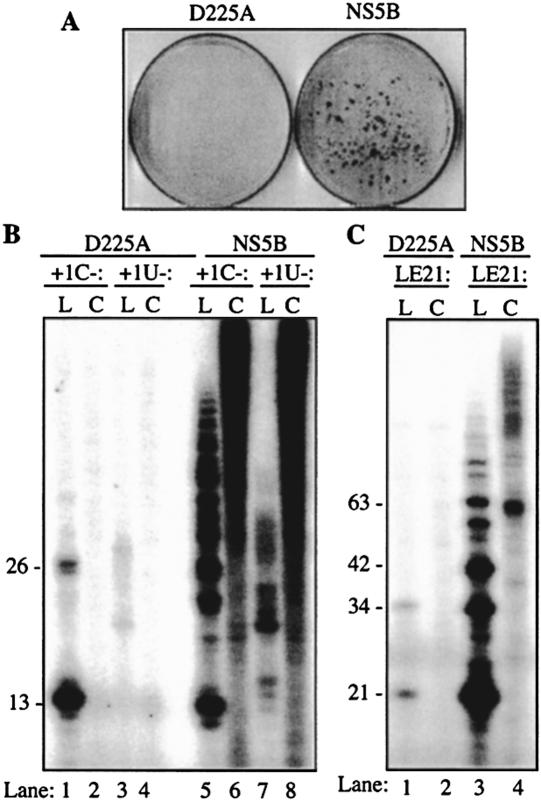
Asp225 is one of the residues in the HCV RdRp that binds the NTPi by recognizing the ribose 2′-hydroxyl of the NTP (Bressanelli et al. 1999, 2002; Lesberg et al. 1999). An HCV subgenomic replicon with a change of NS5B Asp225 to an alanine, D225A, was lethal to HCV replication in the sub-genomic replicon assay, demonstrating its importance for HCV replication (Fig. 3A ▶). In vitro, D225A was severely defective for RNA synthesis when the buffer contained Mg2+ as the sole divalent metal. However, the presence of 1 mM Mn2+ partially rescued initiation by D225A, but it remained defective for generating longer template-switch RNA synthesis products (Ranjith-Kumar et al. 2004). With template +1C-L, D225A was capable of synthesizing a 13-nt product in a buffer containing 1 mM Mn2+ (Fig. 3B ▶, lane 1). However, the dimeric product was significantly reduced relative to the reaction with Δ21, as expected. Furthermore, RNA synthesis from +1U-L did not result in a 13-nt product, demonstrating that initiation took place primarily from the +1C position (Fig. 3B ▶, lanes 1,3). D225A was unable to significantly direct RNA synthesis from +1C-C or +1U-C (Fig. 3B ▶, lanes 2,4). D225A generated several-fold lower amounts of products with LE21-L and was unable to use LE21-C RNA as a template (Fig. 3C ▶, lanes 1,2). These results indicate that the active site of the HCV RdRp is required for RNA synthesis from circular templates.
FIGURE 3.
RNA synthesis from linear and circular templates by full-length and an active-site mutant, D225A, of the HCV RdRp. (A) Huh7 cells transformed by the wild-type subgenomic replicon (designated as NS5B) or one with the mutation D225A. Transfections were performed as described in Materials and Methods and selected for cells resistant to G418 treatment. Cell colonies were visualized 3 wk after transfection by Coomassie blue staining. The results show that D225A is a critical mutation for HCV RNA replication in cells. (B) RNA synthesis by D225A (expressed in the context of Δ21) and by the full-length NS5B proteins from 13-nt +1C-L, +1C-C, +1U-L, and +1U-C templates and (C) 21-nt LE21-L and LE21-C templates. The templates used in each reaction are shown above the gel image.
We also tested the full-length NS5B protein, henceforth referred to as NS5B, since the presence of the intact C-terminal tail had been reported to increase RNA binding (Vo et al. 2004). NS5B produced the de novo initiated products and recombination products from both +1C-L and +1U-L, with +1C-L being the preferred template (Fig. 3B ▶, cf. lanes 5 and 7). NS5B could also efficiently use the linear form of LE21 as a template (Fig. 3C ▶, lane 3). However, with the circularized versions of the templates, a large smear of products that migrated up to the top of the gel was generated (Fig. 3B ▶, lanes 6,8, and C, lane 4). Our results demonstrate that the HCV RdRp could use circular templates of different lengths and sequences. However, there are two notable differences between the products of NS5B and Δ21 from circular templates +1C-C and +1U-C: (1) NS5B generated larger amounts of higher-molecular-weight products with +1U-C than did Δ21; and (2) the 9- and 10-nt products and the discrete ladder of bands characteristic of products from Δ21 were significantly less abundant in the reactions with NS5B (Fig. 3B ▶). Also, NS5B generated a ladder of higher-molecular-weight products with LE21-C unlike Δ21 (cf. Figs. 3B ▶ and 2C ▶). These features could be attributed to the higher affinity for RNA by NS5B in comparison to Δ21 (Vo et al. 2004).
Initiation of RNA synthesis by the HCV RdRp on circular templates
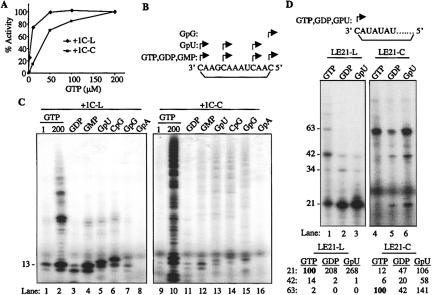
GTP is the preferred NTPi for efficient de novo RNA synthesis and is required at a higher concentration than other nucleotides in order for RNA synthesis to take place in vitro (Ranjith-Kumar et al. 2002b, 2004). Higher GTP concentration was found to increase RNA synthesis from both LE21-L and LE21-C (Fig. 2C ▶). We also examined the effects of increasing GTP concentration on RNA synthesis from +1C-L and +1C-C and found that GTP stimulated RNA synthesis from both RNAs (Fig. 4A ▶). Notably, the GTP concentration needed for a half-maximal level of 13-nt RNA synthesis from +1C-L was lower (4 μM) than for +1C-C (20 μM).
FIGURE 4.
Initiation of RNA synthesis by Δ21 using linear and circular RNAs. (A) Requirement for GTP for RNA synthesis by Δ21. The RdRp reactions using +1C-L and +1C-C were separated on a gel and the 13-nt RNA product for +1C-L and ~65-nt product for +1C-C were quantified and plotted against different concentrations of GTP. The amount of product formed with 200 μM GTP was considered as 100%. (B) Schematic showing the RNA synthesis initiation sites on +1C RNA. Arrows correspond to the possible initiation sites for the nucleotides analogs given on the left. The RNA sequence is given in the 3′-to-5′ direction, and the line underneath shows the bond formed during circularization of the RNA. (C) Analysis with GTP analogs. A representative gel picture showing the RNA synthesis products obtained by using different GTP analogs with templates +1C-L (left) and +1C-C (right). The analogs used are given on the top, and the position of 13-nt product is shown at the left of the gel. All reactions were supplemented with 1 μM GTP, which is needed for elongative RNA synthesis. (D) Analysis of LE21-L and LE21-C using GTP analogs. Since LE21 has only one C (at the 3′-end of linear RNA), the analogs used can initiate RNA synthesis only from one position. The analogs used are given above and the sizes of RNA products to the left of the gel. The bottom of the gel shows the quantitative analysis of the 21-, 42-, and 63-nt products obtained with respective analogs. For LE21-L, the amount of 21-nt product generated with GTP was considered as 100%, and for LE21-C, 100% is the amount of ~60-nt product generated with GTP. All the values are after normalizing for the number of radiolabeled CMPs incorporated.
GTP analogs have been informative in characterizing de novo initiation by the HCV RdRp (Zhong et al. 2000; Ranjith-Kumar et al. 2002a). We used analogs including dinucleotide mimics of the NTPi to characterize RNA synthesis from the cRNAs. Since GTP analogs can be used for initiation but not elongation, all reactions were supplemented with 1 μM GTP, a level that cannot efficiently drive initiation (Ranjith-Kumar 2002b; Fig. 4A ▶). Based on complementarity with the sequence in +1C-C and +1C-L, all analogs except dinucleotide GpA should be capable of initiating RNA synthesis at one to three positions along the template (Fig. 4B ▶).
Indeed, we determined that synthesis from +1C-L and +1C-C can use all of the analogs except GpA (Fig. 4C ▶). Interestingly, there are several products initiated with GpU, CpG, and GpG from template +1C-C that are of distinct sizes when compared to the ones produced with GTP or with each other, suggesting that the products initiated from different positions in the template (Fig. 4C ▶, cf. lanes 11–15). In contrast, RNA synthesis using template +1C-L primarily initiated at the 3′-most cytidylate. The shifted migration with GpU is likely due to the lack of 5′-phosphates. The product initiated with GpG was previously determined to have the 3′-guanylate base-paired with the T1 nucleotide (the 5′-G of GpG was not base-paired), thus resulting in a 14-nt product (Ranjith-Kumar et al. 2002a).
We also analyzed the products initiated from LE21-L and LE21-C using GTP analogs (Fig. 4D ▶). GDP and GpU could initiate RNA synthesis and generate a 21-nt product at a two- to threefold-higher level than with GTP using LE21-L (Fig. 4D ▶, lanes 1–3). Since there is only one cytidine in the template LE21 (at the +1 position) (Fig. 2A ▶), products initiated with GTP analogs should terminate after one round of RNA synthesis and generate a 21-nt product. We observed that the 21-nt products were increased with LE21-C when GDP and GpU were the initiation nucleotides (Fig. 4D ▶, lanes 5,6), further confirming that LE21-C is functioning as a circular template and can direct three or more rounds of RNA synthesis by Δ21. Unexpectedly, reactions containing GDP and GpU directed synthesis of the ~60-nt products at 40% and 140%, respectively, of the amounts observed in reactions with GTP. Some amount of 42-nt products was also observed. These products could be attributed to NTP misincorporation or contamination of nucleotide stocks with trace amounts of GTP. The generation of these products needs to be examined in a future work.
Elongation of RNA synthesis by the HCV RdRp on circular templates
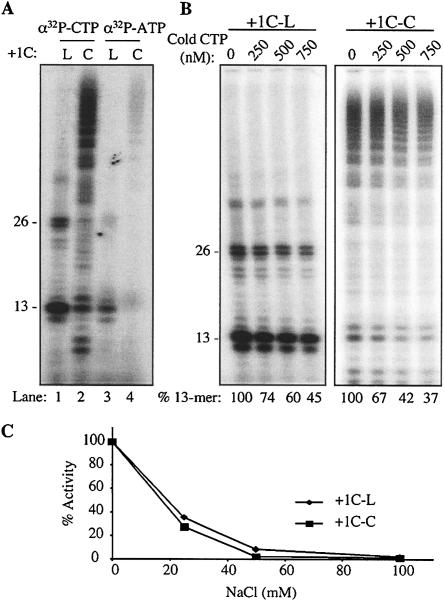
With both LE21-C and +1C-C, we observed a ladder of bands suggesting that the RdRp ternary complex had terminated at specific sites. These products are especially prominent with +1C-C. One hypothesis for this ladder is that some substrate, such as radiolabeled 32P-CTP (present at 250 nM), was limiting in our reaction. To examine the role of substrate requirements in RNA synthesis from +1C-C, we first analyzed the products made using 32P-CTP or 32P-ATP as the radiolabel (Fig. 5A ▶). All of the other nucleotides were present at 100 μM, except for GTP, which was present at 200 μM. Based on previous characterizations of NTP usage, pyrimidine triphosphates are needed at lower concentrations than purine triphosphates (Ferrari et al. 1999; Kao et al. 1999; Oh et al. 1999; Ranjith-Kumar et al. 2002b); hence, we expected that labeling with 32P-CTP would be more efficient than 32P-ATP. Indeed, reactions with both labeled NTPs generated a ladder of products using +1C-L or +1C-C as templates, indicating that low concentration of one specific nucleotide was not the primary reason for the ladder of bands (Fig. 5A ▶). Furthermore, we increased the abundance of unlabeled CTP in the reaction from two- to fourfold that of [α-32P]CTP and found that while the specificity of radiolabeling decreased, as expected for synthesis from both the linear and circular templates, the relative abundance of the bands in the ladder was unchanged (Fig. 5B ▶).
FIGURE 5.
NTP requirements for RNA synthesis by Δ21 from linear and circular templates. (A) Labeling of the products from linear and circular templates with [α-32P]CTP and [α-32P]ATP. The conditions used in each reaction are indicated above the gel image. The final concentration of the radiolabel was 250 nM. GTP is present at 0.2 mM in all of the reactions, and NTPs that are not used as radiolabel are present at 0.1 mM. (B) Titration of nonradiolabled CTP into reactions radiolabeled with [α-32P]CTP (250 nM final concentration). The templates used for RNA synthesis by Δ21 and the concentrations of the nonradiolabeled CTP added to the reactions are shown above the gel image. The decrease in the signals is due to decreased specificity of labeling in the presence of nonradiolabeled CTP. The amount of 13-nt product was normalized to the reaction not amended with cold CTP. (C) Effect of increasing salt on RNA synthesis from +1C-L and +1C-C. RNA synthesis was performed with increasing concentrations of NaCl (up to 100 mM), and the 13-nt product obtained was quantified and plotted against NaCl concentration.
Another explanation for the ladder of bands is that the stability of the HCV RdRp ternary complex may be affected by the conformation of the template. To examine whether there is a difference in the stability of the RNA synthesis complex, we challenged the reactions with increasing concentrations of NaCl (Fig. 5C ▶). RNA syntheses from both the +1C-L and +1C-C templates were equally sensitive to higher salt concentrations. Furthermore, salt equally affected both initiation and elongation of RNA synthesis, suggesting that RNA syntheses from linear and circular templates share similar properties.
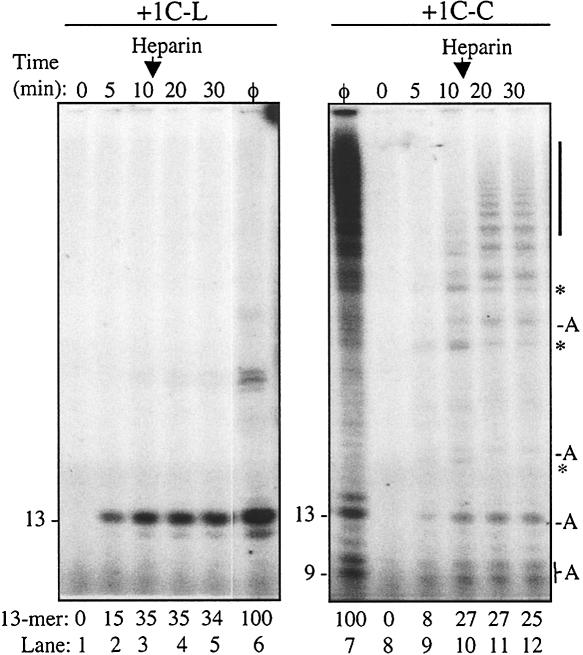
The higher-molecular-weight bands produced by the HCV RdRp on +1C-C likely represent a mixture of products generated by the RdRp ternary complex that terminated at different positions. To determine whether this is the case, we allowed RNA synthesis to begin on the linear or circular template for 10 min, then challenged the reaction with heparin sulfate at 250 ng/μL (Fig. 6 ▶). Heparin is a nucleic acid mimic that binds RdRps that are not stably associated with template, and prevents productive RNA synthesis. Along with the heparin, we also adjusted the CTP from the initial concentration of 250 nM to 50 μM to prevent it from becoming limiting during elongative RNA synthesis (Fig. 6 ▶). As a control for the multiple rounds of RNA synthesis from linear and circular templates, a reaction was incubated for 1 h without heparin (Fig. 6 ▶, lanes 6,7). The amount of product formed from +1C-L did not increase further after the addition of heparin (Fig. 6 ▶, lanes 3–5), as would be expected for ternary complexes that would terminate upon reaching the 5′-end of the template. For +1C-C, however, heparin did not prevent RdRp from making higher-molecular-weight products (Fig. 6 ▶, lanes 8–12; see bar on right side of gel image). Some of the products present in the 10-min reaction could at least partially resume elongative synthesis (labeled with asterisks), while others, especially those of 9, 10, and 13 nt, were either released by the RdRp or were otherwise incapable of continuing RNA synthesis (labeled “A”). Based on these results, we conclude that RNA synthesis by the HCV RdRp on the circular template will contain functionally distinguishable ternary complexes, some of which can continue synthesis after one length of the template is used.
FIGURE 6.
Effect of heparin on RNA synthesis by Δ21 from linear and circular templates. RNA synthesis over time was examined with templates +1C-L and +1C-C. Heparin (250 ng/μL) was added to the reaction after 10 min. (φ) This lane was allowed to synthesize products for 60 min in the absence of heparin. (Right) (A) The RdRp products that were not able to resume RNA synthesis in the presence of heparin; (*) the RdRp products that were able. The vertical bar identifies the higher-molecular-weightproducts made by the HCV RdRp in the presence of heparin. Quantifications of the 13-nt product made at each time point were normalized to the amount made in the sample labeled φ.
A common theme for related RdRps
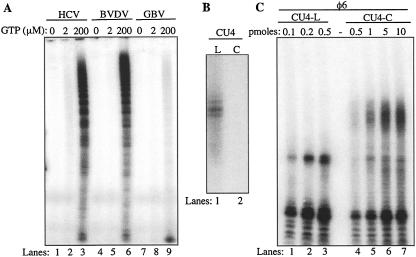
We wanted to determine whether related RdRps are capable of directing RNA synthesis from circular templates. The highly purified RdRp from BVDV was able to synthesize products from the circular RNA in a manner similar to that of the HCV RdRp (Fig. 7A ▶, lanes 4–6). Furthermore, synthesis required GTP concentrations >2 μM, consistent with de novo initiation being responsible for the initiation of product synthesis. The RdRp from the GB virus-B (GBV-B) did not prefer the use of either +1C-L or +1C-C as a template (Fig. 7A ▶; data not shown). However, it was capable of generating higher-molecular-weight products, as shown by the smear reaching toward the top of the gel (Fig. 7A ▶, lanes 7–9).
FIGURE 7.
Use of linear and circular RNA templates by several recombinant RdRps. (A) RNA synthesis from +1C-C by purified recombinant RdRps from HCV, BVDV, and GBV. Each reaction was performed with different GTP concentrations to show that RNA synthesis required higher levels of GTP. Radiolabeled [α-32P]CTP, ATP, and UTP were present at 250 nM, 100 μM, and 100 μM, respectively. (B) Circularization of an RNA (CU4) from bacteriophage φ6. CU4 is a 22-nt RNA derived from the 3′-terminal sequence of the φ6 RNA genome (sequence: 5′-GGCCCCUUCGGGGGCUCUCUCU-3′). After ligation with T4 RNA ligase, the linear and circularized RNAs were purified from a denaturing gel and then used for 3′-terminal extension reaction in the presence of [α-32P]ATP with poly(A) polymerase. (C) RNA synthesis by the φ6 RdRp from the linear and circularized versions of CU4. The amount of input template in each reaction is denoted above the gel image.
Next, we examined RNA synthesis from +1C-L and +1C-C by the RdRp from bacteriophage φ6. No product synthesis was observed with either template (data not shown), suggesting that these templates either are not recognized and/or cannot direct initiation by the φ6 RdRp. The φ6 RdRp was reported to prefer templates with more cytosines at the 3′- terminus (Makeyev and Bamford 2000; Laurila et al. 2002). Therefore, we used an RNA sequence from the authentic 3′-sequence of φ6 (CU4: 5′-GGCCCCUUCGGGGGCUCUCU CU-3′). After two rounds of overnight ligations, only ~20% of the linear molecules were converted to apparent circles, as noted by a change in electrophoretic mobility (data not shown). Therefore, we purified the presumed circular forms from the linear ones after preparative denaturing gel electrophoresis. When CU4 was tested for the ability to be extended by the poly(A) polymerase, the linear form, but not the presumed circles, was a substrate for terminal nucleotide addition (Fig. 7B ▶). In terms of RNA synthesis, the CU4-L could efficiently direct RNA synthesis by the φ6 polymerase, while the CU4-C yielded a set of products that included novel ones not found with the linear template (Fig. 7C ▶). Even though the φ6 RdRp structure indicates a closed conformation, these results suggest that the polymerase could open and accept a circular template. It is interesting to note, however, that the products made by φ6 terminated after two to three rounds of synthesis from the circular RNA.
DISCUSSION
Biochemical characterizations of HCV NS5B revealed several requirements for RNA synthesis in vitro (Lohmann et al. 1998; Kao et al. 1999; Ranjith-Kumar et al. 2002a,b). Here we show that HCV NS5B is capable of initiating RNA synthesis from a circular RNA template. The requirements for RNA synthesis from the circular templates are generally similar to those from linear templates, with GTP and a cytidylate as the NTPi and the T + 1 nucleotides. Some notable differences were observed, however, such as a requirement for higher concentration of GTP for initiation from circular templates (Fig. 4A ▶). Since a circular template could not be threaded into the preformed active site, this observation suggests the HCV RdRp can release the interactions between the thumb and the fingers subdomains to accept the circular template. In addition, we demonstrate that the HCV RdRp lacking the C-terminal 21 residues can form at least two elongation complexes that differ in the ability to resume elongation (Fig. 6 ▶).
Several observations demonstrate that the HCV RdRp could initiate from a circular template. First, +1U-L was a poor template for Δ21, likely due to the lack of a cytidylate near the 3′-terminus of the template, but +1U-C could restore RNA synthesis from an internal cytidylate (Fig. 1E ▶). Second, higher concentrations of GTP were required for RNA synthesis, which suggests de novo initiation (Fig. 4A ▶). Third, the products generated from the circular +1C-C and +1U-C are multiples of the lengths of the template molecule (Figs. 1 ▶, 3 ▶–6 ▶ ▶ ▶). RNA synthesis from +1U-C is less robust than from +1C-C. We believe that this could be attributed to +1C-C RNA having two consecutive cytidylates (Fig. 1A ▶). Kao et al. (2000) have previously demonstrated that, in vitro, a template with two accessible cytidylates is a more efficient template. HCV RdRp can also use a 21-nt circular RNA (Fig. 2 ▶). However, with LE21-C, the products generated are not abundant as with shorter circular RNA +1C. Interestingly, with LE21-C, HCV RdRp generates close to a 60-nt product, suggesting that the polymerase can go around the RNA at least three times before it falls off. This difference could possibly be due to some steric constraints that affected the progression of the RdRp on the shorter circular template leading to frequent release of products from the ternary complex. This is further substantiated with the use of dinucleotide GTP analogs, which could initiate RNA synthesis at different positions, hence generating different-sized products (Fig. 4C ▶).
Use of the circular template was observed for four recombinant RdRps (Fig. 7 ▶). The mechanism of initiation of RNA synthesis on a circular RNA might be similar to internal initiation on linear templates as during subgenomic RNA synthesis of viruses like brome mosaic virus and tombus-viruses (Siegel et al. 1998; Panavas et al. 2002). However, inappropriate initiation from internal sites could be detrimental to the success of RNA infection and could trigger anti-viral responses. We note that the use of the circular template does not abrogate specificity requirements, as seen by the use of the dinucleotides and by the requirement for a cytidylate as the initiation nucleotide. Also, φ6 RdRp preferred a template with sequence corresponding to its own genomic RNA and was not capable of using +1C-C or LE21-C (Fig. 7C ▶; data not shown). This requirement should discourage initiation from some internal site. Additional regulation of the use of inappropriate internal initiation sites could be by interaction with other replicase subunits and/or by the structure of the RNA template.
Our functional results can have implications for the structure of RdRps. All known polymerases have similar structures with fingers, thumb, and palm subdomains. However, the enclosed active sites of the HCV and φ6 RdRps were suggested to contribute to the specific recognition of single-stranded RNAs (Butcher et al. 2001; Hong et al. 2001; Bressanelli et al. 2002). The enclosed state likely represents only one conformation for the two RdRps. Recently, crystal structure analysis of HCV RdRp from genotype 2a revealed two conformational states, open and closed forms of NS5B (Biswal et al. 2005). Such open and closed forms have been reported for rabbit hemorrhagic disease virus RdRp as well by Ng et al. (2002), who also postulated that a significant movement of the thumb subdomain regulates RdRp activity by converting it from an inactive to an active state. The closed (active) form of 2a RdRp is similar to the known structures of RdRp from genotype 1b. By analogy, 1b RdRp should also exist in an open conformation. We postulate that the open form of the HCV RdRp is the form that recognizes circular templates for initiation. An open form that binds template could help explain the strong preference of the HCV RdRp to extend from primed templates (Behrens et al. 1996; Lohmann et al. 1997; Ishii et al. 1999). The discrimination of the dsRNA could take place after template binding and likely includes features in the RdRp such as the β-loop in the active site and the C-terminal tail and the recognition of the initiating GTP and the NTPi (Hong et al. 2001; Ranjith-Kumar et al. 2002b, 2003a).
Should viral RdRps first recognize the template initiation site in an open conformation and then undergo conformational changes to form stable binary complexes, their initiation would be analogous to that of other template-dependent polymerases. For example, large conformational changes characterize almost every major step in the polymerization process by the T7 RNA polymerase (Steitz 2004). Also, in many DNA and RNA polymerases, a network of conserved residues is involved in the transition between open and closed conformers (Zheng et al. 2005). Kinetic analyses with polymerases have suggested at least five steps for every single nucleotide addition, which includes two possible conformational changes: the first after nucleotide binding and the second after phosphodiester bond formation (Dahlberg and Benkovic 1991; Wong et al. 1991; Arnold and Cameron 2004). Even though crystal structure studies did not support such changes, these observations suggest that local structural changes within the RdRp are still possible in solution and may offer additional steps to regulate viral RNA synthesis. Recently, it was shown that fidelity of poliovirus RdRp can be controlled by a site farther away from the active site, and this type of control is probably a common feature of all polymerases (Arnold et al. 2005). This adds to the evidence that the polymerase can undergo changes in its structure upon ligand binding. These changes should be considered in developing antivirals against the HCV RdRp.
MATERIALS AND METHODS
Reagents
The nucleotides, the heparin, and all chemicals were of the highest purity from Sigma Inc. The nucleotides were dissolved and neutralized with NaOH to a pH between 7 and 7.5, and their concentration was adjusted based on their absorbance at 260 nm. All RNAs used were chemically synthesized by Dharmacon Inc., and purified by denaturing gel electrophoresis, checked for quality by Toluidine blue staining, and quantified by spectrophotometry. The φ6 RdRp was a kind gift from Dennis Bamford. Recombinant flaviviral RdRps were purified as described by Ranjith-Kumar et al. (2003a). NS5B proteins were expressed from pET derivatives in Escherichia coli BL21(DE3). All RdRps were purified through a Talon cobalt affinity column (BD Biosciences) followed by a poly(U)-agarose column (Pharmacia Inc.) in buffers lacking divalent metals. The proteins were adjusted to between 1 and 2 mg/mL after quantification by the Lowry assay with bovine serum albumin as a concentration control.
Making circular RNA
The RNAs were kinased at 37° C for 30 min with T4 polynucleotide kinase in the buffer supplied by the manufacturer and [γ-32P]ATP. The reaction was extracted with a 1:1 mixture of phenol-chloroform and precipitated with six volumes of ethanol. The precipitated RNA was then solubilized in 1 × T4 RNA ligase buffer (Ambion Inc.) and ligated for between 15 min and overnight. Formation of a circular product was detected by altered mobility in a 7.5 M urea–20% polyacrylamide gel and PhosphorImage analysis.
HCV subgenomic replicon assay
The plasmid pET21bNS5B was used for site-directed mutagenesis using the QuikChange Kit according to the manufacturer’s protocol. The entire NS5B coding sequence was analyzed to confirm that the specified mutation was made and that no additional changes had inadvertently taken place. A BglII fragment from the mutated cDNA was then exchanged into the wild-type subgenomic HCV replicon (pFK/I389neo/NS3–3′/5.1). The presence of the mutation in the HCV subgenomic replicon was confirmed by DNA sequencing.
Subgenomic HCV replicon RNAs were transcribed in vitro using the T7 Ampliscribe kit (Epicenter) from the wild-type (WT) and mutant DNA constructs after linearizing with ScaI. The subgenomic HCV replicon RNAs were transfected into Huh7 cells by electroporation with a GenePulser system (BioRad) as described by Krieger et al. (2001). Briefly, 1 μg of the in vitro transcribed RNAs along with total RNA from naive Huh-7 cells to a final amount of 10 μg were electroporated into ~1 × 106 Huh7 cells in 0.4 mL of ice-cold Cytomix (van den Hoff et al. 1992). The electroporation conditions were 960 μF and 270 V. Cells were immediately transferred to 8 mL of complete DMEM medium containing 1.25% DMSO and seeded in a 10-cm dish. At 24 h after transfection, the cell culture medium was replaced with complete DMEM supplemented with 0.5 mg/mL G418. The medium was changed every week. After 3 wk of selection with G418 sulfate, cell colonies in culture dishes were stained with 0.01% Coomassie blue.
RdRp activity assays
The standard RdRp assays consisted of 0.1 pmol of linear and 10 pmol of circular template RNA (unless stated otherwise) and 0.04 μM NS5B in a 20-μL reaction containing 20 mM sodium glutamate (pH 8.2), 4 mM MgCl2, 12.5 mM dithiothreitol, 0.5% (v/v) Triton X-100, 200 μM ATP and UTP, and 250 nM [α-32P]CTP (Amersham, Inc.). Unless otherwise stated, 200 μM GTP was used in the reaction. MnCl2, if used, was added to a final concentration of 1 mM. Due to the buffer used to store the RdRp, the final reaction also contains 10 mM NaCl. RNA synthesis reactions were incubated at 25° C for 60 min and stopped by phenol/chloroform extraction followed by ethanol precipitation in the presence of 5 μg of glycogen and 0.5 M ammonium acetate. Products were usually separated by electrophoresis on denaturing (7.5 M urea) polyacrylamide gels. Gels were wrapped in plastic, and quantification of radiolabeled bands was performed using a PhosphorImager (Molecular Dynamics). Each result shown has been reproduced in at least one, usually two, other independent experiment.
Acknowledgments
We thank the Texas A&M University Cereal Killers, especially Y.C. Kim, for helpful discussions; M. Laurila and D. Bamford for the gift of the φ6 RdRp; L. Guarino for help with the circularization of RNAs; and J. Santos and W. Miller for editing the manuscript. Support by the National Science Foundation grants MCB 0332259 and MCB 461511 is gratefully acknowledged.
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.2163106.
REFERENCES
- Adkins, S., Stawicki, S.S., Faurote, G., Siegel, R.W., and Kao, C.C. 1998. Mechanistic analysis of RNA synthesis by RNA-dependent RNA polymerase from two promoters reveals similarities to DNA-dependent RNA polymerase. RNA 4: 455–470. [PMC free article] [PubMed] [Google Scholar]
- Ago, H., Adachi, T., Yoshida, A., Yamamoto, M., Habuka, N., Yatsunami, K., and Miyano, M. 1999. Crystal structure of the RNA-dependent RNA polymerase of hepatitis C virus. Struct. Fold. Des. 7: 1417–1426. [DOI] [PubMed] [Google Scholar]
- Arnold, J.J. and Cameron, C.E. 2004. Poliovirus RNA-dependent RNA polymerase (3Dpol): Pre-steady-state kinetic analysis of ribonucleotide incorporation in the presence of Mg2+. Biochemistry 43: 5126–5137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold, J.J., Vignuzzi, M., Stone, J.K., Andino, R., and Cameron, C.E. 2005. Remote site control of an active site fidelity checkpoint in a viral RNA-dependent RNA polymerase. J. Biol. Chem. 280: 25706–25716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behrens, S.-E., Tomei, L., and De Francesco, R. 1996. Identification and properties of the RNA-dependent RNA polymerase of hepatitis C virus. EMBO J. 15: 12–22. [PMC free article] [PubMed] [Google Scholar]
- Biswal, B.K., Cherney, M.M., Wang, M., Chan, L., Yannopoulos, C.G., Bilimoria, D., Nicolas, O., Bedard, J., and James, M.N. 2005. Crystal structures of the RNA dependent RNA polymerase genotype 2a of hepatitis C virus reveal two conformations and suggest mechanisms of inhibition by non-nucleoside inhibitors. J. Biol. Chem. 280: 18202–18210. [DOI] [PubMed] [Google Scholar]
- Bressanelli, S., Tomei, L., Roussel, A., Incitti, I., Vitale, R.L., Mathieu, M., and DeFrancesco, R. 1999. Crystal structure of the RNA-dependent RNA polymerase of hepatitis C virus. Proc. Natl. Acad. Sci. 96: 13034–13039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bressanelli, S., Tomei, L., Rey, F.A., and DeFrancesco, R. 2002. Structural analysis of the hepatitis C virus RNA polymerase in complex with ribonucleotides. J. Virol. 76: 3482–3492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butcher, S.J., Grimes, J.M., Makeyev, E.V., Bramford, D.H., and Stuart, D.I. 2001. A mechanism for initiating RNA-dependent RNA polymerization. Nature 410: 235–240. [DOI] [PubMed] [Google Scholar]
- Chapman, M.R. and Kao, C. 1999. A minimal RNA promoter for minus-strand RNA synthesis by the brome mosaic virus polymerase complex. J. Mol. Biol. 286: 709–720. [DOI] [PubMed] [Google Scholar]
- Choi, K.H., Groarke, J.M., Young, D.C., Kuhn, R.J., Smith, J.L., Pevear, D.C., and Rossmann, M.G. 2004a. The structure of the RNA-dependent RNA polymerase from bovine viral diarrhea virus establishes the role of GTP in de novo initiation. Proc. Natl. Acad. Sci. 30: 4425–4430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi, S.K., Hema, M., Gopinath, K., Santos, J., and Kao, C. 2004b. Replicase-binding sites on plus- and minus-strand brome mosaic virus RNAs and their roles in RNA replication in plant cells. J. Virol. 78: 13420–13429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahlberg, M.E. and Benkovic, S.J. 1991. Kinetic mechanism of DNA polymerase I (Klenow fragment): Identification of a second conformational change and evaluation of the internal equilibrium constant. Biochemistry 30: 4835–4843. [DOI] [PubMed] [Google Scholar]
- Dreher, T.W. 1999. Functions of the 3′-untranslated regions of positive strand RNA viral genomes. Annu. Rev. Phytopathol. 37: 151–174. [DOI] [PubMed] [Google Scholar]
- Ferrari, E., Wright-Minogue, J., Fang, J.W.S., Baroudy, B.M., Lau, J.Y.N., and Hong, Z. 1999. Characterization of soluble hepatitis C virus RNA-dependent RNA polymerase expressed in Escherichia coli. J. Virol. 73: 1649–1654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrer-Orta, C., Arias, A., Perez-Luque, R., Escarmis, C., Domingo, E., and Verdaguer, N. 2004. Structure of foot-and-mouth disease virus RNA-dependent RNA polymerase and its complex with a template-primer RNA. J. Biol. Chem. 279: 47212–47221. [DOI] [PubMed] [Google Scholar]
- Hansen, J.L., Long, A.M., and Schultz, S.C. 1997. Structure of the RNA-dependent RNA polymerase of poliovirus. Structure 5: 1109–1122. [DOI] [PubMed] [Google Scholar]
- Hong, Z., Cameron, C.E., Walker, M.P., Castro, C., Yao, N., Lau, J.Y., and Zhong, W. 2001. A novel mechanism to ensure terminal initiation by hepatitis C virus NS5B polymerase. Virology 285: 6–11. [DOI] [PubMed] [Google Scholar]
- Ishii, K., Tanaka, Y., Yap, C.C., Aizaki, H., Matsuura, Y., and Miyamura, T. 1999. Expression of hepatitis C virus NS5B protein: Characterization of its RNA polymerase activity and RNA binding. Hepatology 29: 1227–1235. [DOI] [PubMed] [Google Scholar]
- Joyce, C.M. and Steitz, T.A. 1995. Polymerase structures and function: Variations on a theme? J. Bacteriol. 177: 6321–6329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kao, C.C., Del Vecchio, A.M., and Zhong, W. 1999. De novo initiation of RNA synthesis by a recombinant flaviviridae RNA-dependent RNA polymerase. Virology 253: 1–7. [DOI] [PubMed] [Google Scholar]
- Kao, C.C., Yang, X., Kline, A., Wang, Q.M., Barket, D., and Heinz, B.A. 2000. Template requirements for RNA synthesis by a recombinant hepatitis C virus RNA-dependent RNA polymerase. J. Virol. 74: 11121–11128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, M.-J. and Kao, C.C. 2001. Factors regulating template switch in vitro by viral RNA-dependent RNA polymerases: Implications for RNA–RNA recombination. Proc. Natl. Acad. Sci. 98: 4972–4977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krieger, N., Lohmann, V., and Bartenschlager, R. 2001. Enhancement of hepatitis C virus RNA replication by cell culture-adaptive mutations. J. Virol. 75: 4614–4624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurila, M.R., Makeyev, E.V., and Bamford, D.H. 2002. Bacteriophage φ 6 RNA-dependent RNA polymerase: Molecular details of initiating nucleic acid synthesis without primer. J. Biol. Chem. 277: 17117–17124. [DOI] [PubMed] [Google Scholar]
- Lesberg, C.A., Cable, M.B., Ferrari, E., Hong, Z., Mannarino, A.F., and Weber, P.C. 1999. Crystal structure of the RNA-dependent RNA polymerase from hepatitis C virus reveals a fully encircled active site. Nat. Struct. Biol. 6: 937–943. [DOI] [PubMed] [Google Scholar]
- Li, M.L. and Stollar, V. 2004. Identification of the amino acid sequence in Sindbis virus nsP4 that binds to the promoter for the synthesis of the subgenomic RNA. Proc. Natl. Acad. Sci. 101: 9429–9434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Y., Korolev, S., and Waksman, G. 1998. Crystal structures of open and closed forms of binary and ternary complexes of the large fragment of Thermus aquaticus DNA polymerase I: Structural basis for nucleotide incorporation. EMBO J. 17: 7514–7525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohmann, V., Korner, F., Herian, U., and Bartenschlager, R. 1997. Biochemical properties of hepatitis C virus NS5B RNA-dependent RNA polymerase and identification of amino acid sequence motifs essential for enzymatic activity. J. Virol. 71: 8416–8428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohmann, V., Roos, A., Korner, F., Koch, J.O., and Bartenschlager, R. 1998. Biochemical and kinetic analyses of NS5B RNA-dependent RNA polymerase of the hepatitis C virus. Virology 249: 108–118. [DOI] [PubMed] [Google Scholar]
- Love, R.A., Maegley, K.A., Yu, X., Ferre, R.A., Lingardo, L.K., Diehl, W., Parge, H.E., Dragovich, P.S., and Fuhrman, S.A. 2004. The crystal structure of the RNA-dependent RNA polymerase from human rhinovirus: A dual function target for common cold antiviral therapy. Structure 12: 1533–1544. [DOI] [PubMed] [Google Scholar]
- Makeyev, E.V. and Bamford, D.H. 2000. Replicase activity of purified recombinant protein P2 of double-stranded RNA bacteriophage φ6. EMBO J. 19: 124–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClure, W.R. 1985. Mechanism and control of transcription initiation in prokaryotes. Annu. Rev. Biochem. 54: 171–204. [DOI] [PubMed] [Google Scholar]
- Nagy, P.D., Carpenter, C.D., and Simon, A.E. 1997. A novel 3′-end repair mechanism in an RNA virus. Proc. Natl. Acad. Sci. 94: 1113–1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng, K.K., Cherney, M.M., Vazquez, A.L., Machin, A., Alonso, J.M., Parra, F., and James, M.N. 2002. Crystal structures of active and inactive conformations of a caliciviral RNA-dependent RNA polymerase. J. Biol. Chem. 277: 1381–1387. [DOI] [PubMed] [Google Scholar]
- Oh, J.W., Ito, T., and Lai, M.M. 1999. A recombinant hepatitis C virus RNA-dependent RNA polymerase capable of copying the full-length viral RNA. J. Virol. 73: 7694–7702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ollis, D.L., Brick, P., Hamlin, R., Xuong, N.G., and Steitz, T.A. 1985a. Structure of large fragment of Escherichia coli DNA polymerase I complexed with dTMP. Nature 313: 762–766. [DOI] [PubMed] [Google Scholar]
- Ollis, D.L., Kline, C., and Steitz, T.A. 1985b. Domain of E. coli DNA polymerase I showing sequence homology to T7 DNA polymerase. Nature 313: 818–819. [DOI] [PubMed] [Google Scholar]
- Pogany, J., Fabian, M.R., White, K.A., and Nagy, P.D. 2003. A replication enhancer element in a plus-strand RNA virus. EMBO J. 22: 5602–5611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranjith-Kumar, C.T., Gajewski, J., Gutshall, L., Maley, D., Sarisky, R., and Kao, C. 2001. Viral RNA-dependent RNA polymerase has terminal transferase activity: Implications for viral RNA synthesis J. Virol. 75: 8615–8623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranjith-Kumar, C.T., Gutshall, L., Kim, M.-J., Sarisky, R.T., and Kao, C.C. 2002a. Requirements for de novo initiation of RNA synthesis by recombinant flaviviral RNA-dependent RNA polymerases. J. Virol. 76: 12526–12536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranjith-Kumar, C.T., Kim, Y.C., Gutshall, L., Silverman, C., Khandekar, S., Sarisky, R.T., and Kao, C.C. 2002b. Mechanism of de novo initiation by the hepatitis C virus RNA-dependent RNA polymerase: Role of divalent metals. J. Virol. 76: 12513–12525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranjith-Kumar, C.T., Gutshall, L., Sarisky, R.T., and Kao, C.C. 2003a. Multiple interactions within the hepatitis C virus RNA polymerase repress primer-dependent RNA synthesis. J. Mol. Biol. 330: 675–685. [DOI] [PubMed] [Google Scholar]
- Ranjith-Kumar, C.T., Santos, J.L., Gutshall, L.L., Johnston, V.K., Lin-Goerke, J., Kim, M.-J., Porter, D.J., Maley, D., Greenwood, C., Earnshaw, D.L., et al. 2003b. Enzymatic activities of the GB virus-B RNA-dependent RNA polymerase. Virology 312: 270–280. [DOI] [PubMed] [Google Scholar]
- Ranjith-Kumar, C.T., Sarisky, R.T., Gutshall, L., Thomson, M., and Kao, C.C. 2004. De novo initiation pocket mutations have multiple effects on hepatitis C virus RNA-dependent RNA polymerase activities. J. Virol. 78: 12207–12217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao, A.L.N. and Hall, T.C. 1993. Recombination and polymerase error facilitate restoration of infectivity in brome mosaic virus. J. Virol. 67: 969–979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao, A.L.N., Dreher, T.W., Marsh, L.E., and Hall, T.C. 1989. Telo-meric function of the transfer RNA-like structure of brome mosaic virus RNA. Proc. Natl. Acad. Sci. 86: 5335–5339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarafianos, S.G., Clark Jr., A.D., Das, K., Tuske, S., Birktoft, J.J., Ilankumaran, P., Ramesha, A.R., Sayer, J.M., Jerina, D.M., Boyer, P.L., et al. 2002. Structures of HIV-1 reverse transcriptase with pre-and post-translocation AZTMP-terminated DNA. EMBO J. 21: 6614–6624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuppli, D., Georgijevic, J., and Weber, H. 2000. Synergism of mutations in bacteriophage Qβ affecting host factor dependence of Qβ replicase. J. Mol. Biol. 295: 149–154. [DOI] [PubMed] [Google Scholar]
- Siegel, R.W., Adkins, S., and Kao, C.C. 1997. Sequence-specific recognition of a subgenomic promoter by a viral RNA polymerase. Proc. Natl. Acad. Sci. 94: 11238–11243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel, R.W., Bellon, L., Beigelman, L., and Kao, C.C. 1998. Moieties in an RNA promoter specifically recognized by a viral RNA-dependent RNA polymerase. Proc. Natl. Acad. Sci. 95: 11613–11618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steitz, T.A. 1999. DNA polymerases: Structural diversity and common mechanisms. J. Biol. Chem. 274: 17395–17398. [DOI] [PubMed] [Google Scholar]
- ———. 2004. The structural basis of the transition from initiation to elongation phases of transcription, as well as translocation and strand separation, by T7 RNA polymerase. Curr. Opin. Struct. Biol. 14: 4–9. [DOI] [PubMed] [Google Scholar]
- Sun, J.H. and Kao, C.C. 1997a. RNA synthesis by the brome mosaic virus RNA-dependent RNA polymerase: Transition from initiation to elongation. Virology 233: 63–73. [DOI] [PubMed] [Google Scholar]
- ———. 1997b. Characterization of RNA products associated with or aborted by a viral RNA-dependent RNA polymerase. Virology 236: 348–353. [DOI] [PubMed] [Google Scholar]
- Sun, J.H., Adkins, S., Faurote, G., and Kao, C.C. 1996. Initiation of (−)-strand RNA synthesis catalyzed by the BMV RNA-dependent RNA polymerase: Synthesis of oligonucleotides. Virology 226: 1–12. [DOI] [PubMed] [Google Scholar]
- van den Hoff, M.J., Moorman, A.F., and Lamers, W.H. 1992. Electroporation in ‘intracellular’ buffer increases cell survival. Nucleic Acids Res. 20: 2902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vo, N.V., Oh, J.W., and Lai, M.M. 2003. Identification of RNA ligands that bind hepatitis C virus polymerase selectively and inhibit its RNA synthesis from the natural viral RNA templates. Virology 307: 301–316. [DOI] [PubMed] [Google Scholar]
- Vo, N.V., Tuler, J.R., and Lai, M.M. 2004. Enzymatic characterization of the full-length and C-terminally truncated hepatitis C virus RNA polymerases: Function of the last 21 amino acids of the C terminus in template binding and RNA synthesis. Biochemistry 43: 10579–10591. [DOI] [PubMed] [Google Scholar]
- von Hippel, P.H., Bear, D.G., Morgan, W.D., and McSwiggen, J.A. 1984. Protein–nucleic acid interactions in transcription: A molecular analysis. Annu. Rev. Biochem. 53: 389–446. [DOI] [PubMed] [Google Scholar]
- Wong, I., Patel, S.S., and Johnson, K.A. 1991. An induced-fit kinetic mechanism for DNA replication fidelity: Direct measurement by single-turnover kinetics. Biochemistry 30: 526–537. [DOI] [PubMed] [Google Scholar]
- Zheng, W., Brooks, B.R., Doniach, S., and Thirumalai, D. 2005. Network of dynamically important residues in the open/closed transition in polymerases is strongly conserved. Structure 13: 565–577. [DOI] [PubMed] [Google Scholar]
- Zhong, W., Ferrari, E., Lesburg, C.A., Maag, D., Ghosh, S.K., Cameron, C.E., Lau, J.Y., and Hong, Z. 2000. Template/primer requirements and single nucleotide incorporation by hepatitis C virus nonstructural protein 5B polymerase. J. Virol. 74: 9134–9143. [DOI] [PMC free article] [PubMed] [Google Scholar]