Abstract
We describe a general and efficient two-step strategy for lariat RNA synthesis. In the first step, a deoxyribozyme synthesizes 2′,5′-branched RNA. In the second step, T4 RNA ligase closes the loop that completes the lariat. The loop-closure reaction can form either a natural or unnatural lariat isomer, depending on which of the two 3′-termini of the branched RNA reacts with the lone 5′-end. We demonstrate two approaches to control formation of either lariat isomer. In conjunction with other routes for lariat RNA synthesis, the two-step strategy described here will facilitate biochemical studies that require lariat RNAs of varying nucleotide sequence.
Keywords: lariat RNA, branched RNA, deoxyribozyme, T4 RNA ligase, debranching enzyme
INTRODUCTION
Branched nucleic acid strands play an integral role in biological RNA splicing. During the first step of intron splicing by the spliceosome (Burge et al. 1999) and also by self-splicing RNAs such as group II introns (Saldanha et al. 1993; Michel and Ferat 1995; Jacquier 1996; Fedorova et al. 2002), a conserved branch-site adenosine within the intron uses its 2′-hydroxyl group to attack the 5′-splice site phosphodiester linkage. The 5′-exon is expelled, forming a lariat intermediate that has the 3′-exon attached (lariat/3′-exon intermediate). During the second step of splicing, the 5′-exon attacks the 3′-splice site, releasing the ligated exons along with the lariat intron. This lariat is degraded via initial branch-site hydrolysis catalyzed by de-branching enzyme, e.g., Dbr1p in yeast (Nam et al. 1994; Ooi et al. 2001). Lariat RNAs are common to all of these biochemical transformations, and studies of splicing pathways will be facilitated by synthetic access to lariat RNAs of wide sequence composition.
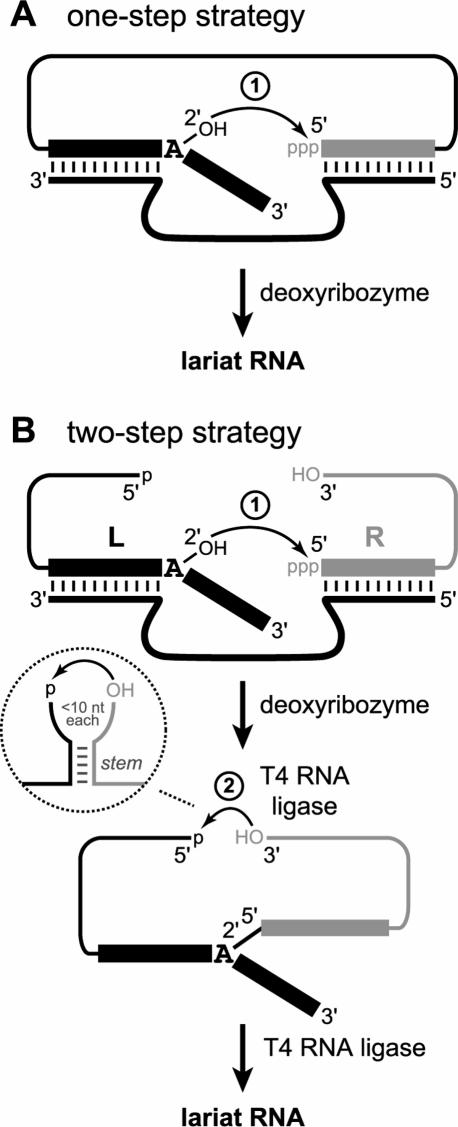
Synthesis of lariat RNAs using chemical or biochemical methods is a substantial experimental challenge due to their special topology, which features a closed loop. Solid-phase synthesis methods to prepare even the relatively simple 2′,5′-branched RNA are tedious (Damha and Ogilvie 1988; Damha and Zabarylo 1989; Damha et al. 1992; Ganeshan et al. 1995; Reese and Song 1999), and the closed loop of a lariat provides an added measure of complexity (Carriero and Damha 2003). Group II intron ribozymes can synthesize lariats, but numerous sequence requirements for the RNA substrates limit the applicability of this approach. We recently reported several deoxyribozymes that synthesize 2′,5′-branched RNAs (Wang and Silverman 2003a, b, Wang and Silverman 2005a; Coppins and Silverman 2004 Coppins and Silverman 2005; Pratico et al. 2005). Although some of these branch-forming deoxyribozymes can also synthesize lariat RNAs in one step from linear RNA precursors (Fig. 1A ▶), in many cases the yield of lariat is low. Presumably this is because the deoxyribozyme and substrate interfere with each other, although the structural basis of such interference is not yet known. In this article, we describe an efficient two-step strategy that sequentially applies a deoxyribozyme and then a protein enzyme (T4 RNA ligase) to construct lariat RNAs (Fig. 1B ▶). This general approach allows great flexibility in lariat RNA synthesis and also permits the controlled preparation of unnatural lariat isomers that have an alternative connectivity among their three nucleic acid arms.
FIGURE 1.
The one-step and two-step strategies for synthesis of lariat RNA. (A) One-step lariat RNA synthesis using a deoxyribozyme to mediate attack of an internal 2′-hydroxyl group on the 5′-triphosphate of the same RNA substrate strand. (B) Two-step lariat RNA synthesis, in which a deoxyribozyme first synthesizes 2′,5′-branched RNA and the loop is then closed using T4 RNA ligase. In the first step, L and R denote the left-hand and right-hand RNA substrates. The inset shows the optimal stem-loop structure near the ligation site during loop closure (see text).
RESULTS
RNA circularization efficiency depends on structure near ligation site
The deoxyribozyme-catalyzed synthesis of 2′,5′-branched RNA has been established recently in our laboratory (Wang and Silverman 2003a, b, Wang and Silverman 2005a; Coppins and Silverman 2004 Coppins and Silverman 2005; Pratico et al. 2005). Branched RNA formation by a deoxyribozyme is the first step of the two-step lariat RNA synthesis strategy (Fig. 1 ▶). The second step of the strategy—loop closure using T4 RNA ligase—has been used for RNA circularization (Silber et al. 1972; Kaufmann et al. 1974), but the substrate preferences in terms of secondary structure have not been investigated systematically (Pan et al. 1991; Dolinnaya et al. 1993). We would like to make informed choices of sites for T4 RNA ligase-mediated loop closure. Therefore, we briefly examined the role of RNA secondary structure on the efficiency of this circularization reaction, using an 80-nt linear RNA substrate with 5′-monophosphate and 3′-hydroxyl groups. When this linear substrate had no predicted secondary structure elements (i.e., a structurally featureless circle), loop closure was efficiently completed in minutes, but ~10% of side products ascribed to dimerization and higher-order multimerization were evident (data not shown). Alternatively, a 5-bp stem was placed near the ligation site to hold the reactive termini together, as shown schematically in the Figure 1B ▶ inset (see Materials and Methods for sequences). In this case, loop closure was severalfold more rapid, and the extent of side product formation depended on the lengths of the two single-stranded regions between the stem and the ligation site (data not shown). Shorter single-stranded regions (e.g., 5–10 nt) substantially suppressed multimerization relative to the absence of the 5-bp stem, whereas longer single-stranded regions (11–13 nt) did not suppress multimerization. We conclude that loop closure by T4 RNA ligase is optimal when a stem is located near the ligation site and when relatively short (<10 nt) single-stranded regions are available between the stem and the ligation site.
The two-step lariat RNA synthesis approach works where the one-step approach fails
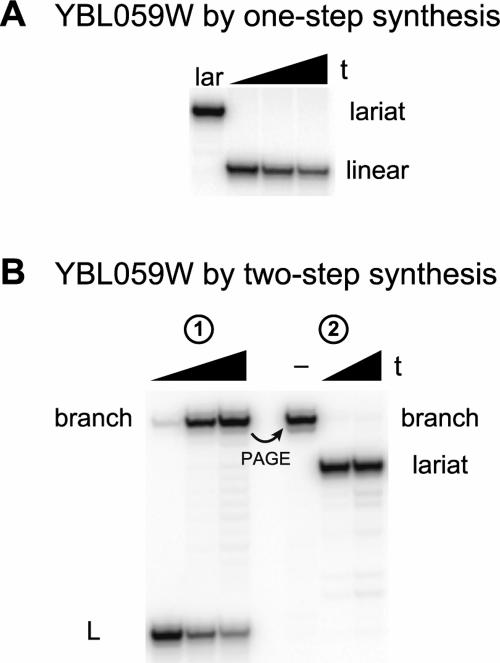
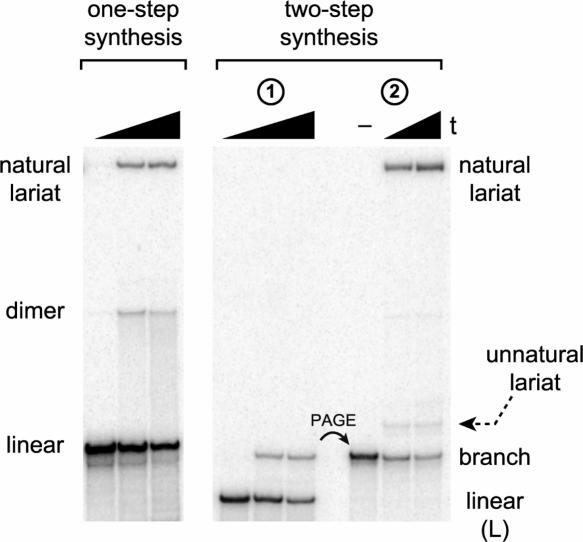
For the first test of the two-step lariat synthesis strategy, we used the small YBL059W intron lariat, which has a 51-nt loop (Davis et al. 2000; Grate and Ares 2002). The 7S11 deoxyribozyme (Coppins and Silverman 2004), which is widely applicable for branched RNA formation (Coppins and Silverman 2005), nevertheless was unable to synthesize the YBL059W lariat in one step (no detectable lariat formation; Fig. 2A ▶). Despite its inability to synthesize the YBL059W lariat in one step, 7S11 was able to synthesize the corresponding 2′,5′-branched RNA in high yield (Fig. 2B ▶, left). Completion of lariat synthesis by loop closure using T4 RNA ligase was also efficient, with <3% multi-merization side products (Fig. 2B ▶, right). For the latter reaction, the ligation site was chosen within a stem-loop structure with guidance from mfold secondary structure predictions (SantaLucia 1998; Zuker 2003).
FIGURE 2.
One-step and two-step syntheses of YBL059W lariat RNA. (A) Attempting one-step synthesis according to the strategy of Figure 1A ▶ using the 7S11 deoxyribozyme, which provides an undetectable (<1%) amount of lariat RNA (12% PAGE). The lariat standard (lar) was available from the two-step route of B. t = 0, 1.5, and 3 h. Inclusion of a disruptor DNA oligonucleotide (Wang and Silverman 2005a) did not increase the yield of lariat (data not shown). (B) Two-step synthesis according to the strategy of Figure 1B ▶, which proceeds efficiently for both steps (12% PAGE). t = 0, 0.5, and 1.5 h for branched RNA synthesis using the 7S11 deoxyribozyme, and t = 2 and 10 min for lariat synthesis using T4 RNA ligase (– denotes PAGE-purified branched RNA incubated without T4 RNA ligase). As shown in Figure 1B ▶, L denotes the left-hand RNA substrate, which contains the 32P-radiolabel.
Selectivity between 3′-ends during loop closure
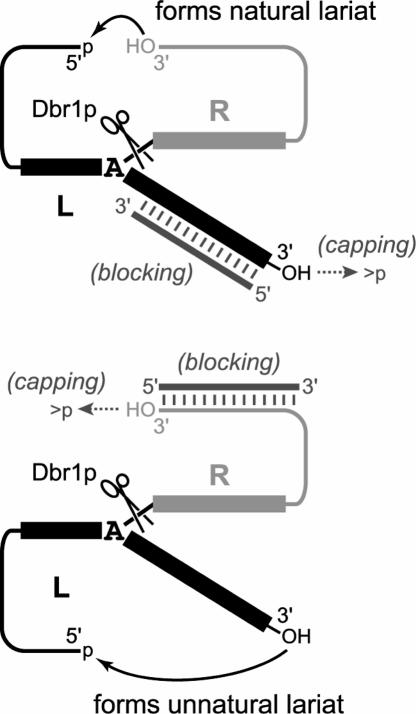
A 2′,5′-branched RNA molecule inherently has one 5′-end but two 3′-ends. In the T4 RNA ligase loop-closure step to complete lariat formation, either of these 3′-ends may become joined with the single 5′-end, forming either a “natural” lariat or its “unnatural” lariat isomer (Fig. 3 ▶). Discrimination between the two 3′-ends is expected to be low in the absence of secondary structure elements or other interventions that favor one 3′-end over the other. Choosing the ligation site near a stem element (Fig. 1B ▶, inset) inherently favors formation of one lariat isomer. However, the selectivity additionally depends on the lengths and the internal structures of the competing 3′-strands, and therefore, the selectivity must be determined experimentally for any particular substrate.
FIGURE 3.
Formation of natural and unnatural lariat RNA isomers in the T4 RNA ligase loop-closure reaction, and the blocking and capping approaches to control which isomer is formed. On each structure is marked the cleavage site for debranching enzyme Dbr1p, which is used to characterize the lariats (Fig. 4 ▶).
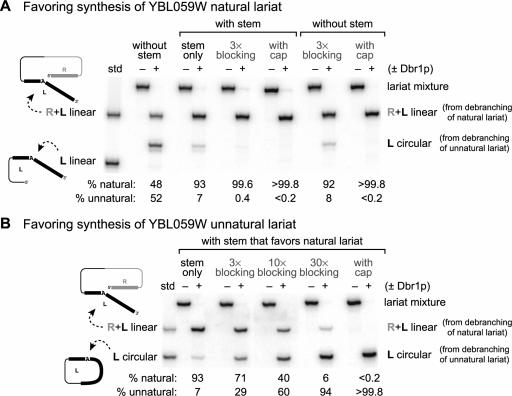
In the case of the YBL059W lariat, only one major product band (>97%) was observed during lariat formation using T4 RNA ligase (Fig. 2 ▶). However, if the natural and unnatural lariat isomers co-migrate on PAGE, the observed product would be a mixture of the two isomers. This possibility was assessed by treating the PAGE-purified lariat product with yeast debranching enzyme Dbr1p (Nam et al. 1994; Ooi et al. 2001), which is expected to debranch the natural lariat isomer to a 69-nt linear RNA but cleave the unnatural lariat isomer into a 52-nt circular RNA and a separate 17-nt linear fragment. Treatment of the YBL059W lariat product with Dbr1p showed clearly that both lariat isomers were indeed present (Fig. 4A ▶). When the lariat was synthesized with no stem at all in the T4 RNA ligase substrate, Dbr1p treatment revealed that the natural and unnatural lariat isomers were formed in ~1:1 ratio, as expected for essentially no selectivity between the 3′-ends (which are of similar length). In contrast, when a stem was present to hold the proper 3′-end near the 5′-end, the natural lariat isomer was substantially favored, but ~7% of the product was still the unnatural lariat isomer. For some applications and also from the viewpoint of biochemical purity, we sought to control lariat formation more precisely.
FIGURE 4.
Characterization of the natural and unnatural YBL059W lariat isomer ratio in lariat RNA synthesized by the variations of the two-step strategy shown in Figure 3 ▶. Each pair of lanes shows lariat RNA without or with debranching enzyme treatment (±Dbr1p). The 32P-radiolabel was in the L portion of each lariat product (12% PAGE). The ratio of the two cleavage bands is a direct measure of the ratio between natural and unnatural lariat isomers, because each isomer gives a distinct Dbr1p cleavage band as indicated. In these experiments, the stem secondary structure element (when present) favors formation of the natural lariat isomer. (A) Blocking and capping approaches to favor the natural lariat isomer. (B) Blocking and capping approaches to favor the unnatural lariat isomer. In B, these approaches must work against the natural lariat preference provided by the stem.
The two approaches developed for this purpose, termed “blocking” and “capping,” are shown in Figure 3 ▶. In the blocking approach, an excess is added of a short DNA oligonucleotide complementary to the RNA 3′-end that would otherwise react to form the unnatural lariat isomer. Because T4 RNA ligase needs a single-stranded 3′ terminus on its RNA substrate (Silverman 2004), the blocking oligo should suppress reactivity of the blocked RNA 3′-end. For lariat synthesis without a stem to direct the reactivity, inclusion of the blocking DNA oligonucleotide increased the ratio of natural to unnatural lariat isomer from ~1:1 to 92:8 (Fig. 4A ▶). For lariat synthesis when the stem element was present, the percentage of unnatural lariat isomer fell from 7% to only 0.4%, which was barely above the detection limit. In the alternative capping approach, the RNA substrate was synthesized with a 2′,3′-cyclic phosphate cap on its 3′-end, such that the 3′-hydroxyl necessary for loop closure was absent. Regardless of whether the stem was absent or present, the capping approach was entirely effective at preventing unnatural lariat isomer formation, and no unnatural lariat was detected (<0.2%).
Both the blocking and capping approaches could instead be applied to synthesize the unnatural lariat isomer. For these experiments we used the RNA substrate with the stem that favors natural lariat isomer formation, such that unnatural lariat formation must overcome the inherent 93:7 bias towards creating the natural isomer (as shown in Fig. 4A ▶). Because of this bias, it was unsurprising that the blocking approach is largely but not completely effective at inverting the selectivity to favor the unnatural lariat isomer, particularly when a large excess of blocking oligonucleotide was added (Fig. 4B ▶). The capping approach is again fully effective at controlling which lariat isomer is formed (Fig. 4B ▶).
Synthesis of the ai5γ intron lariat
We applied the two-step lariat synthesis procedure to the ai5γ group II intron, which is a commonly studied model system for group II intron splicing (Fedorova et al. 2002). Direct one-step synthesis of the ai5γ lariat using the 7S11 deoxyribozyme does proceed with modest yield for this particular substrate, and some dimer is formed as well (Fig. 5 ▶). Here we simply wished to determine if the two-step approach was applicable to a larger (~800-nt loop) ribozyme for which we could directly assay the functional competence of the synthetic lariat. Using the 7S11-synthesized ai5γ branched RNA (Coppins and Silverman 2005), loop closure using T4 RNA ligase proceeded well to form the natural lariat when a stem element was present to direct the reaction (Fig. 5 ▶). In this case, the small amount of unnatural lariat migrates differently on PAGE (~6% yield of unnatural lariat; Fig. 5 ▶, dashed arrow) and would be separable. When the stem was absent during the reaction, the unnatural lariat was the predominant product (~70%; data not shown). Evidence for both the natural and unnatural lariat structures was provided by treatment with de-branching enzyme (data not shown).
FIGURE 5.
Syntheses of ai5γ group II intron lariat RNA. The two-step RNA substrates include a stem to favor natural lariat formation. For the one-step synthesis and for the first step of the two-step synthesis, t = 0, 0.5, and 1.5 h using the 7S11 deoxyribozyme. For the second step of the two-step synthesis, t = 10 and 60 min using T4 RNA ligase (– denotes PAGE-purified branched RNA incubated without T4 RNA ligase). Samples were analyzed by 5% PAGE.
To verify the structural integrity of the natural ai5γ lariat RNA, we examined its splicing reactions (Nolte et al. 1998). In a direct comparison with the ai5γ lariat prepared independently by self-splicing of the intron, the natural isomer of the synthetic lariat was equally competent in the reverse of the first step of splicing (data not shown). The 3′-exon was not included on the lariats, or the second step of splicing would have occurred instead of the reverse of the first step. Similarly, the synthetic lariat/3′-exon intermediate was able to proceed forward through the second step of splicing when the 5′-exon was added (data not shown).
DISCUSSION
Applying deoxyribozymes to lariat RNA synthesis: One-step and two-step strategies
We now have two ways to use deoxyribozymes for lariat RNA synthesis: the one-step and two-step strategies (Fig. 1 ▶). In most cases, the one-step strategy is likely the preferable route, provided that the lengthy linear RNA substrate can be prepared readily and that lariat formation is reasonably efficient. For example, the 6BX22 deoxyribozyme is capable of efficient one-step lariat RNA synthesis (Wang and Silverman 2005a). However, we do not yet understand why 6BX22 has the unique ability among our branch-forming deoxyribozymes to synthesize lariats in one step for a wide range of RNA substrate sequences. Indeed, many of our other branch-forming deoxyribozymes, such as 7S11, 9F7 (Wang and Silverman 2003a), and 6BX34 (Wang and Silverman 2005b), are often unable to synthesize lariats efficiently in one step (7S11 with the ai5γ substrate is an exception in this regard). For deoxyribozymes such as these, the new two-step strategy for lariat RNA synthesis will be a valuable alternative. The case of YBL059W and 7S11 (Fig. 2 ▶) directly demonstrates that one-step lariat synthesis is not always feasible even when the two-step strategy works well. More generally, as we continue to develop new deoxyribozymes that collectively synthesize branched RNA of any possible sequence (Pratico et al. 2005), each deoxyribozyme will have to be examined separately for its ability to form lariat RNA in one step. For those deoxyribozymes that cannot do so efficiently and reliably, which the available data suggest is indeed difficult, the new two-step strategy will be important. The two-step strategy offers an additional advantage because the two substrates used for branched RNA formation are prepared independently, which may be valuable for incorporating site-specific modifications. In such cases, the single precursor for the onestep route would have to be prepared using one or more RNA ligation reactions, and the two-step strategy would not require a greater number of reaction steps.
To enable the most efficient application of the two-step strategy, a ligation site for the loop-closure step can generally be found with a stem-loop structure as shown in the Figure 1B ▶ inset. Although T4 RNA ligase is known to tolerate a relatively broad range of ligation substrate sequences, certain nucleotide preferences have been identified. For example, uridine in the acceptor substrate, which reacts directly with the 5′-monophosphorylated nucleotide, is detrimental to ligation activity (Ohtsuka et al. 1977; England and Uhlenbeck 1978; Romaniuk et al. 1982). Whenever possible, the ligation site for the T4 RNA ligase loop-closure step should be chosen with such preferences in mind. For practical application of this strategy, a His6-tagged version of T4 RNA ligase has been described (Wang and Unrau 2002), in addition to commercial availability of the enzyme.
Most synthetic approaches to lariat RNA are motivated by the natural occurrence of lariats in biological splicing. Indeed, natural biochemical pathways constitute one preparative route to lariat RNAs, but these reactions are limited by sequence requirements for their substrates and also by inherent reactivity of the products. For example, natural splicing reactions cannot generally be used to synthesize a lariat/3′-exon intermediate that is competent for the second step of splicing, because the intermediate will proceed rapidly through exon ligation by nucleophilic attack of the just-released 5′-exon. Therefore, lariat mutants that are competent for the second step cannot usually be synthesized by this route in more than trace amounts. In contrast, our synthetic strategy that uses a deoxyribozyme to synthesize the branched core of the lariat/3′-exon intermediate in the absence of the 5′-exon avoids this concern entirely. Closure of the loop with T4 RNA ligase in the absence of the 5′-exon allows the otherwise reactive lariat/3′-exon RNA intermediate to be prepared efficiently.
Controlling site-selectivity during loop closure by T4 RNA ligase
The intrinsic nonselectivity of T4 RNA ligase leads directly to a mixture of lariat isomers during the loop-closure reaction (Fig. 3 ▶). Inclusion of a stem near the desired ligation site is helpful but does not provide full control over the selectivity (Fig. 4 ▶). The blocking and capping approaches of Figure 3 ▶ were developed to address this practical concern. The YBL059W intron provides a quantitative experimental test for controlling the site-selectivity during loop closure. For this intron, the 2′-arm and 3′-arm extending from the branch-site adenosine are approximately the same length (17 and 18 nt), and the absence of structural features or interventions such as blocking or capping means that little selectivity in lariat formation is expected. As predicted, inclusion of a stem according to the Figure 1B ▶ inset or application of the blocking and capping strategies according to Figure 3 ▶ were successful in controlling lariat formation (Fig. 4 ▶).
The capping strategy completely suppresses formation of the undesired lariat isomer, but for this strategy the 2′,3′-cyclic phosphate must be included in the substrate. Although this adds a synthetic constraint, such cyclic phosphate termini are readily introduced. In particular, cleavage of a 3′-extended RNA substrate precursor with a ribozyme (either in cis or in trans; Grosshans and Cech 1991; Ferré-D’Amaré and Doudna 1996), with a deoxyribozyme (Santoro and Joyce 1997; Pyle et al. 2000; Cruz et al. 2004), or by other means (Lapham and Crothers 1996) is a useful way to introduce a 2′,3′-cyclic phosphate. Cyclic phosphate termini are also readily removed after the reaction if a free 2′,3′-diol is desired (Schürer et al. 2002). As an alternative to capping with a 2′,3′-cyclic phosphate, the 3′-terminal ribose moiety of the substrate could be oxidized with sodium periodate (Proudnikov and Mirzabekov 1996). Such periodate oxidation would be an acceptable capping approach if the resulting 3′-terminal aldehyde functional groups do not interfere with subsequent applications of the RNA.
For synthesis of any particular lariat according to the two-step strategy, the natural and the unnatural isomers may migrate differently on PAGE and may thus be separable during routine purification. The details of PAGE migration will depend sensitively on the details of the substrates. For example, when a 13-nt 3′-exon was included with the YBL059W substrate, lariat formation provided two readily separable PAGE bands that were assigned to the natural and unnatural lariat isomers (data not shown), instead of overlapping bands for the two isomers that were observed when the 3′-exon was absent (Fig. 4 ▶). In this case, the increased selectivity achieved via the blocking approach, combined with the ability to separate the isomers by PAGE, means that either the natural or the unnatural lariat isomer can be obtained without contamination by the undesired isomer. A similar situation exists for the ai5γ lariat (Fig. 5 ▶). In this case, the data also emphasize the importance of the stem element to direct proper loop closure, because formation of the unnatural lariat is favored unless the stem is present to guide formation of the natural lariat. To explain this inherent bias in favor of the unnatural lariat, we note that the two possible loop-closure reactions form substantially different sizes of ring (776 nt vs. 98 nt for natural vs. unnatural lariat), and it is well known that larger rings are more difficult to create by macrocyclization.
An alternative approach to lariat RNA synthesis was recently reported, in which chemical ligation with cyanogen bromide is used to close the loop of a branched nucleic acid (Carriero and Damha 2003). It is likely that our blocking and capping strategies would be applicable to loop closure using cyanogen bromide instead of T4 RNA ligase. However, it does not appear that the chemical reagent offers any particular advantages over the protein enzyme for this purpose. Indeed, the chemical reagent does not provide any control over site-selectivity during loop closure, and mixtures of 2′–5′ and 3′–5′ linkages will likely be formed. In contrast, T4 RNA ligase forms only native 3′–5′ linkages.
MATERIALS AND METHODS
RNA and DNA oligonucleotides
RNA oligonucleotides were prepared by in vitro transcription using T7 RNA polymerase. The template was either double-stranded DNA prepared by annealing two oligonucleotides (Milligan et al. 1987; Milligan and Uhlenbeck 1989), double-stranded DNA prepared by PCR, or a linearized plasmid. When DNA oligonucleotides were used as the template, the first two nucleotides at the 5′-end of the noncoding strand were 2′-methoxy derivatives to suppress heterogeneous 3′-end formation (Kao et al. 1999). DNA oligonucleotides were prepared at IDT. The sequence of the 7S11 deoxyribozyme was from our previous reports (Coppins and Silverman 2004, 2005), with binding arms complementary to the appropriate portions of the RNA substrates. All RNA and DNA oligonucleotides were purified by denaturing PAGE with running buffer 1× TBE (89 mM each Tris and boric acid, 2 mM EDTA at pH 8.3) as described previously (Flynn-Charlebois et al. 2003; Wang and Silverman 2003b).
RNA circularization assays
The 80-nt RNA substrates were prepared by transcription from a double-stranded DNA template prepared by annealing two oligonucleotides; dephosphorylated with calf intestinal phosphatase (CIP); and 5′-monophosphorylated with T4 polynucleotide kinase (T4 PNK). The RNA sequences were as follows (stem nucleotides underlined): no stem, 5′-GGAGAAGGACUACAAUCACAAACACAAACAACAAAAACAAACAACAAACAACACAAACAAAACACGACGCGCUAUAAACA-3′; stem + 5-/7-nt single-stranded regions, 5′-GGAGAAGGCCUACAAUCACAAACACAAACAACAAAAACAAACAACAAACAACACAAACAAAACACGACGGCCUAUAAACA-3′; stem + 8-/10-nt single-stranded regions, 5′-GGAGAAGGAGGCCAAUCACAAACACAAACAACAAAAACAAACAACAAACAACACAAACAAAACACGGCCUGAUAUAAACA-3′; stem + 11-/13-nt single-stranded regions, 5′-GGAGAAGGACUAGGCCCACAAACACAAACAACAAAAACAAACAACAAACAACACAAACAAAAGGCCUCGCGAUAUAAACA-3′. The assays were conducted as follows: 5′-Monophosphorylated linear RNA (10 pmol) was annealed in 7.5 μL of 5 mM HEPES (pH 7.5), 15 mM NaCl, and 0.1 mM EDTA by heating at 95 °C for 3 min and cooling on ice for 5 min. Portions of 5× ligation buffer and T4 RNA ligase (Fermentas) were added, bringing the final conditions to 50 mM HEPES (pH 7.5), 10 mM MgCl2, 10 mM DTT, and 50 μM ATP with 1 U/μL of T4 RNA ligase in 10 μL total volume. The sample with final RNA concentration of 1 μM was incubated at 37°C. Aliquots were withdrawn at desired time points (0.5, 10, 30, and 60 min) and quenched onto stop solution (80% formamide, 1× TB, 50 mM EDTA, 0.025% each bromophenol blue and xylene cyanol), followed by PAGE and exposure to a Phosphor-Imager screen.
One-step and two-step lariat RNA synthesis for YBL059W (Fig. 2 ▶)
For deoxyribozyme-catalyzed one-step YBL059W lariat RNA synthesis (Fig. 2A ▶), the linear RNA substrate was 5′-GUAUGCAUAGGCAAUAACUUCGGCCUCAUACUCAAAGAACACGUUUACUAACAUAACUUAUUUACAUAG-3′. The branch-site adenosine is underlined, and no 3′-exon is included. This RNA was prepared by transcription with α-32P-CTP, such that the product was internally radiolabeled. The RNA substrate and deoxyribozyme (each 5 pmol) were annealed in 5.9 μL of 5 mM HEPES (pH 7.5), 15 mM NaCl, and 0.1 mM EDTA by heating at 95°C for 3 min and cooling on ice for 5 min. The sample volume was raised to 10 μL with final concentrations of 50 mM EPPS (pH 8.5), 150 mM NaCl, 2 mM KCl, and 40 mM MgCl2. The sample was incubated at 37°C. Aliquots were quenched and assayed as described above.
For two-step YBL059W lariat RNA synthesis (Fig. 2B ▶), the left-hand (L) RNA substrate was prepared by taking the 69-nt transcript whose sequence is given above and cleaving with a 10–23 deoxyribozyme (5′-GTATGAGGCCGAAGGGCTAGCTACAACGATATTGCCTATGCAT-3′) between nucleotides 17 and 18, providing L as the 3′-fragment. The 52-nt product was purified by PAGE and 5′-32P-radiolabeled using γ-32P-ATP and T4 polynucleotide kinase (T4 PNK). Another portion of L was 5′-monophosphorylated using unradiolabeled ATP and T4 PNK. For deoxyribozyme-catalyzed branched RNA formation (step 1), the L substrate (5 pmol, of which 0.5 pmol was 5′-32P-radiolabeled), deoxyribozyme (15 pmol), and R substrate (30 pmol; nucleotides 1–17 of the sequence given above prepared as an independent unradiolabeled transcript) were annealed in 5.9 μL of 5 mM HEPES (pH 7.5), 15 mM NaCl, and 0.1 mM EDTA by heating at 95°C for 3 min and cooling on ice for 5 min. The sample volume was raised to 10 μL with final concentrations of 50 mM EPPS (pH 8.5), 150 mM NaCl, 2 mM KCl, and 40 mM MgCl2. The sample was incubated at 37°C; aliquots were quenched and assayed as described above.
For loop closure catalyzed by T4 RNA ligase (step 2), the branched product from step 1 was prepared by a modification of the above procedure. The L substrate (50 pmol, of which 10 pmol was 5′-32P-radiolabeled), deoxyribozyme (100 pmol), and R substrate (200 pmol) were annealed in 29.5 μL of 5 mM HEPES (pH 7.5), 15 mM NaCl, and 0.1 mM EDTA by heating at 95°C for 3 min and cooling on ice for 5 min. The sample volume was raised to 50 μL with final concentrations of 50 mM EPPS (pH 8.5), 150 mM NaCl, 2 mM KCl, and 40 mM MgCl2. The sample was incubated at 37°C for 1.5 h and quenched with 60 μL of stop solution. The branched RNA product was purified by 12% PAGE. Then, loop closure catalyzed by T4 RNA ligase was performed as follows: Branched RNA (5 pmol) was annealed in 7.5 μL of 5 mM HEPES (pH 7.5), 15 mM NaCl, and 0.1 mM EDTA by heating at 95°C for 3 min and cooling on ice for 5 min. Portions of 5× ligation buffer and T4 RNA ligase (Fermentas) were added, bringing the final conditions to 50 mM HEPES (pH 7.5), 10 mM MgCl2, 10 mM DTT, and 50μM ATP with 1 U/μL of T4 RNA ligase in 10 μL total volume. The sample was incubated at 37°C. Aliquots were quenched and assayed as described above. To prepare the lariat RNA samples for the assays of Figure 4 ▶, the entire sample was quenched after 15 min with 20 μL of stop solution, and the lariat RNA product was purified by 12% PAGE.
Controlling and assaying natural and unnatural lariat RNA formation for YBL059W (Fig. 4 ▶)
For application of the blocking approach, an appropriate DNA oligonucleotide designed as shown in Figure 3 ▶ was included during the loop closure reaction. For YBL059W natural lariat formation, the blocking oligo sequence was 5′-CTATGTAAATAAGTT ATG-3′; for YBL059W unnatural lariat formation, the blocking oligo sequence was 5′-TTATTGCCTATGCATAC-3′. The blocking oligo was added in 3- to 30-fold excess relative to the branched RNA. For application of the capping approach, either the L or the R RNA substrate was prepared with a 2′,3′-cyclic phosphate cap as appropriate. The 52-nt capped L substrate was prepared by sequentially cleaving an 82-nt transcript (equivalent to the 69-nt transcript described above with a 3′-extension) with two 10–23 deoxyribozymes. The first cleaves 17 nt from the 5′ terminus and was the same deoxyribozyme as used to prepare uncapped L above. The second was 5′-GTGTTCAATGGCAGGCTAGCTA CAACGATATGTAAATAAGTTATGTTAG-3′, which cleaves 13 nt from the 3′ terminus and provides the 2′,3′-cyclic phosphate. The 17-nt capped R substrate was prepared by cleavage of the 69-nt transcript with the same 10–23 deoxyribozyme as used to prepare L as the 52-nt 3′-fragment of the 69-nt transcript above. The sequences of the YBL059W substrates for step 2 without a stem near the ligation site was derived from the sequences given above by leaving L unchanged and by altering R to 5′-GUGACAAUACUCAAUAA-3′, where the six underlined nucleotides have been changed.
For the debranching enzyme assays, 0.2–0.5 pmol of PAGE-purified lariat RNA was incubated with 1.5 pmol (75 ng) Dbr1p and 16 U of RNase inhibitor (Boehringer Mannheim) in 10 μL of 20 mM HEPES (pH 7.5), 0.5 mM MgCl2, 125 mM KCl, and 1 mM DTT for 15 min at 30°C. Aliquots of 4 μL were quenched onto 5 μL of stop solution, followed by PAGE and exposure to a PhosphorImager screen. The linear RNA standards were prepared by 5′-32P-radiolabeling of appropriate substrates. The circular RNA standard was prepared by circularization of the linear standard with T4 RNA ligase; its structure was verified by partial alkaline hydrolysis, which gave a linear product that co-migrated with the linear standard (data not shown).
One-step and two-step lariat RNA synthesis for ai5γ (Fig. 5 ▶)
For one-step ai5γ lariat RNA synthesis, the 783-nt linear RNA substrate was prepared by ligation of two transcripts to form a single RNA substrate that contains both domains 1–2-3 (D123) (691 nt) and domains 5–6 (D56) (92 nt without the 3′-exon or 98 nt with the 3′-exon) connected via a small stem-loop (not shown) that replaces the full domain 4 of the natural RNA. The D123 with a 5′-triphosphate was prepared by transcription using α-32P-CTP, such that the product was internally radiolabeled. The D56 was prepared without a radiolabel; the transcript was dephosphorylated with CIP and 5′-monophosphorylated with unradiolabeled ATP and T4 PNK before ligation with D123. The D123 and D56 were joined using T4 RNA ligase. For this reaction, 100 pmol of D123 and 300 pmol of D56 (along with 600 pmol of a DNA oligonucleotide complementary to the 3′ terminus of D56, to suppress D56 circularization; 5′-ATCCCGATAGGTAGACCTT TACA-3′) were annealed in 21 μL of 5 mM HEPES (pH 7.5), 15 mM NaCl, and 0.1 mM EDTA by heating at 95°C for 3 min and cooling on ice for 5 min. Portions of 5× ligation buffer and T4 RNA ligase (Fermentas) were added, bringing the final conditions to 50 mM HEPES (pH 7.5), 10 mM MgCl2, 10 mM DTT, and 50 μM ATP with 1 U/μL of T4 RNA ligase in 30 μL total volume. The sample was incubated at 37°C for 3 h and quenched with 30 μL of stop solution. The 783-nt linear RNA substrate was purified by 5% PAGE. One-step lariat synthesis was performed using 5 pmol of the linear substrate as described above for the YBL059W lariat. Two disruptor oligonucleotides (50 pmol each) were required, one each for the D123 and D56 portions of the substrate (data not shown).
For two-step ai5γ lariat RNA synthesis, the L substrate (D123) was prepared as above with 5′-triphosphate and internal 32P-radio-labels, and the R substrate (D56) was also prepared as above with 5′-monophosphate. For deoxyribozyme-catalyzed branched RNA formation (step 1), the R substrate (D123) (5 pmol), deoxyribozyme (15 pmol), and L substrate (D56) (30 pmol) were annealed in 5.9 μL of 5 mM HEPES (pH 7.5), 15 mM NaCl, and 0.1 mM EDTA by heating at 95°C for 3 min and cooling on ice for 5 min. The sample volume was raised to 10 μL, with final concentrations of 50 mM EPPS (pH 8.5), 150 mM NaCl, 2 mM KCl, and 40 mM MgCl2. The sample was incubated at 37°C; aliquots were quenched and assayed as described for the YBL059W lariat. For two-step ai5γ lariat RNA synthesis without a stem-loop to control reactivity during step 2, D123 was shortened by 16 nt, such that no stem-loop was present. The remainder of the reaction was performed as described here for step 1.
For loop closure catalyzed by T4 RNA ligase (step 2), the branched product from step 1 was prepared by a modification of the above procedure. The R substrate (D123; 100 pmol), deoxyribozyme (150 pmol), and L substrate (D56; 250 pmol) were annealed in 37.5 μL of 5 mM HEPES (pH 7.5), 15 mM NaCl, and 0.1 mM EDTA by heating at 95°C for 3 min and cooling on ice for 5 min. The sample volume was raised to 50 μL with final concentrations of 50 mM EPPS (pH 8.5), 150 mM NaCl, 2 mM KCl, and 40 mM MgCl2. The sample was incubated at 37°C for 1.5 h and quenched with 50 μL of stop solution. The branched RNA product was purified by 5% PAGE. Then, loop closure catalyzed by T4 RNA ligase was performed as described above for the second step of the two-step YBL059W lariat synthesis. To prepare the lariat RNA samples for the debranching enzyme assays, the entire 10-μL sample was quenched after 1 h with 20 μL of stop solution, and the lariat RNA product was purified by 5% PAGE.
Acknowledgments
This research was supported by the Burroughs Wellcome Fund (New Investigator Award in the Basic Pharmacological Sciences), the March of Dimes Birth Defects Foundation (Research Grant No. 5-FY02-271), the National Institutes of Health (GM-65966), the American Chemical Society Petroleum Research Fund (38803-G4), and the UIUC Department of Chemistry (all to S.K.S.). S.K.S. is the recipient of a fellowship from the David and Lucile Packard Foundation. We are grateful to Scott W. Stevens (University of Texas at Austin) for the generous gift of purified yeast debranching enzyme Dbr1p. We also thank members of the Silverman lab for discussions.
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.2259406.
REFERENCES
- Burge, C.B., Tuschl, T., and Sharp, P.A. 1999. Splicing of precursors to mRNAs by the spliceosomes. In The RNA world (eds. R.F. Gesteland et al.), pp. 525–560. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Carriero, S. and Damha, M.J. 2003. Template-mediated synthesis of lariat RNA and DNA. J. Org. Chem. 68: 8328–8338. [DOI] [PubMed] [Google Scholar]
- Coppins, R.L. and Silverman, S.K. 2004. A DNA enzyme that mimics the first step of RNA splicing. Nat. Struct. Mol. Biol. 11: 270–274. [DOI] [PubMed] [Google Scholar]
- ———. 2005. A deoxyribozyme that forms a three-helix-junction complex with its RNA substrates and has general RNA branch-forming activity. J. Am. Chem. Soc. 127: 2900–2907. [DOI] [PubMed] [Google Scholar]
- Cruz, R.P.G., Withers, J.B., and Li, Y. 2004. Dinucleotide junction cleavage versatility of 8–17 deoxyribozyme. Chem. Biol. 11: 57–67. [DOI] [PubMed] [Google Scholar]
- Damha, M.J. and Ogilvie, K.K. 1988. Synthesis and spectroscopic analysis of branched RNA fragments: Messenger RNA splicing intermediates. J. Org. Chem. 53: 3710–3722. [Google Scholar]
- Damha, M.J. and Zabarylo, S. 1989. Automated solid-phase synthesis of branched oligonucleotides. Tetrahedron Lett. 30: 6295–6298. [Google Scholar]
- Damha, M.J., Ganeshan, K., Hudson, R.H., and Zabarylo, S.V. 1992. Solid-phase synthesis of branched oligoribonucleotides related to messenger RNA splicing intermediates. Nucleic Acids Res. 20: 6565–6573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis, C.A., Grate, L., Spingola, M., and Ares Jr., M. 2000. Test of intron predictions reveals novel splice sites, alternatively spliced mRNAs and new introns in meiotically regulated genes of yeast. Nucleic Acids Res. 28: 1700–1706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolinnaya, N.G., Blumenfeld, M., Merenkova, I.N., Oretskaya, T.S., Krynetskaya, N.F., Ivanovskaya, M.G., Vasseur, M., and Shabarova, Z.A. 1993. Oligonucleotide circularization by template-directed chemical ligation. Nucleic Acids Res. 21: 5403–5407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- England, T.E. and Uhlenbeck, O.C. 1978. Enzymatic oligoribonucleotide synthesis with T4 RNA ligase. Biochemistry 17: 2069–2076. [DOI] [PubMed] [Google Scholar]
- Fedorova, O., Su, L.J., and Pyle, A.M. 2002. Group II introns: Highly specific endonucleases with modular structures and diverse catalytic functions. Methods 28: 323–335. [DOI] [PubMed] [Google Scholar]
- Ferré-D’Amaré, A.R., and Doudna, J.A. 1996. Use of cis- and trans-ribozymes to remove 5′ and 3′ heterogeneities from milligrams of in vitro transcribed RNA. Nucleic Acids Res. 24: 977–978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flynn-Charlebois, A., Wang, Y., Prior, T.K., Rashid, I., Hoadley, K.A., Coppins, R.L., Wolf, A.C., and Silverman, S.K. 2003. Deoxyribozymes with 2′-5′ RNA ligase activity. J. Am. Chem. Soc. 125: 2444–2454. [DOI] [PubMed] [Google Scholar]
- Ganeshan, K., Tadey, T., Nam, K., Braich, R., Purdy, W.C., Boeke, J.D., and Damha, M.J. 1995. Novel approaches to the synthesis and analysis of branched RNA. Nucleosides Nucleotides 14: 1009–1013. [Google Scholar]
- Grate, L. and Ares Jr., M. 2002. Searching yeast intron data at Ares lab Web site. Methods Enzymol. 350: 380–392. [DOI] [PubMed] [Google Scholar]
- Grosshans, C.A. and Cech, T.R. 1991. A hammerhead ribozyme allows synthesis of a new form of the Tetrahymena ribozyme homogeneous in length with a 3′ end blocked for transesterification. Nucleic Acids Res. 19: 3875–3880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacquier, A. 1996. Group II introns: Elaborate ribozymes. Biochimie 78: 474–487. [DOI] [PubMed] [Google Scholar]
- Kao, C., Zheng, M., and Rudisser, S. 1999. A simple and efficient method to reduce nontemplated nucleotide addition at the 3′ terminus of RNAs transcribed by T7 RNA polymerase. RNA 5: 1268–1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufmann, G., Klein, T., and Littauer, U.Z. 1974. T4 RNA ligase: Substrate chain length requirements. FEBS Lett. 46: 271–275. [DOI] [PubMed] [Google Scholar]
- Lapham, J. and Crothers, D.M. 1996. RNase H cleavage for processing of in vitro transcribed RNA for NMR studies and RNA ligation. RNA 2: 289–296. [PMC free article] [PubMed] [Google Scholar]
- Michel, F. and Ferat, J.L. 1995. Structure and activities of group II introns. Annu. Rev. Biochem. 64: 435–461. [DOI] [PubMed] [Google Scholar]
- Milligan, J.F. and Uhlenbeck, O.C. 1989. Synthesis of small RNAs using T7 RNA polymerase. Methods Enzymol. 180: 51–62. [DOI] [PubMed] [Google Scholar]
- Milligan, J.F., Groebe, D.R., Witherell, G.W., and Uhlenbeck, O.C. 1987. Oligoribonucleotide synthesis using T7 RNA polymerase and synthetic DNA templates. Nucleic Acids Res. 15: 8783–8798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nam, K., Hudson, R.H., Chapman, K.B., Ganeshan, K., Damha, M.J., and Boeke, J.D. 1994. Yeast lariat debranching enzyme. Substrate and sequence specificity. J. Biol. Chem. 269: 20613–20621. [PubMed] [Google Scholar]
- Nolte, A., Chanfreau, G., and Jacquier, A. 1998. Influence of substrate structure on in vitro ribozyme activity of a group II intron. RNA 4: 694–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohtsuka, E., Nishik, S., Fukumoto, R., Tanaka, S., Markham, A.F., Ikehara, M., and Sugiura, M. 1977. Joining of synthetic ribotrinucleotides with defined sequences catalyzed by T4 RNA Ligase. Eur. J. Biochem. 81: 285–291. [DOI] [PubMed] [Google Scholar]
- Ooi, S.L., Dann 3rd, C., Nam, K., Leahy, D.J., Damha, M.J., and Boeke, J.D. 2001. RNA lariat debranching enzyme. Methods Enzymol. 342: 233–248. [DOI] [PubMed] [Google Scholar]
- Pan, T., Gutell, R.R., and Uhlenbeck, O.C. 1991. Folding of circularly permuted transfer RNAs. Science 254: 1361–1364. [DOI] [PubMed] [Google Scholar]
- Pratico, E.D., Wang, Y., and Silverman, S.K. 2005. A deoxyribozyme that synthesizes 2′,5′-branched RNA with any branch-site nucleotide. Nucleic Acids Res. 33: 3503–3512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proudnikov, D. and Mirzabekov, A. 1996. Chemical methods of DNA and RNA fluorescent labeling. Nucleic Acids Res. 24: 4535–4542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyle, A.M., Chu, V.T., Jankowsky, E., and Boudvillain, M. 2000. Using DNAzymes to cut, process, and map RNA molecules for structural studies or modification. Methods Enzymol. 317: 140–146. [DOI] [PubMed] [Google Scholar]
- Reese, C.B. and Song, Q. 1999. A new approach to the synthesis of branched and branched cyclic oligoribonucleotides. Nucleic Acids Res. 27: 2672–2681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romaniuk, E., McLaughlin, L.W., Neilson, T., and Romaniuk, P.J. 1982. The effect of acceptor oligoribonucleotide sequence on the T4 RNA ligase reaction. Eur. J. Biochem. 125: 639–643. [DOI] [PubMed] [Google Scholar]
- Saldanha, R., Mohr, G., Belfort, M., and Lambowitz, A.M. 1993. Group I and group II introns. FASEB J. 7: 15–24. [DOI] [PubMed] [Google Scholar]
- SantaLucia Jr., J. 1998. A unified view of polymer, dumbbell, and oligonucleotide DNA nearest-neighbor thermodynamics. Proc. Natl. Acad. Sci. 95: 1460–1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santoro, S.W. and Joyce, G.F. 1997. A general purpose RNA-cleaving DNA enzyme. Proc. Natl. Acad. Sci. 94: 4262–4266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schürer, H., Lang, K., Schuster, J., and Mörl, M. 2002. A universal method to produce in vitro transcripts with homogeneous 3′-ends. Nucleic Acids Res. 30: e56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silber, R., Malathi, V.G., and Hurwitz, J. 1972. Purification and properties of bacteriophage T4-induced RNA ligase. Proc. Natl. Acad. Sci. 69: 3009–3013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverman, S.K. 2004. Practical and general synthesis of 5′-adenylated RNA (5′-AppRNA). RNA 10: 731–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Y. and Silverman, S.K. 2003a. Deoxyribozymes that synthesize branched and lariat RNA. J. Am. Chem. Soc. 125: 6880–6881. [DOI] [PubMed] [Google Scholar]
- ———. 2003b. Characterization of deoxyribozymes that synthesize branched RNA. Biochemistry 42: 15252–15263. [DOI] [PubMed] [Google Scholar]
- ———. 2005a. Efficient one-step synthesis of biologically related lariat RNAs by a deoxyribozyme. Angew. Chem. Int. Ed. 44: 5863–5866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ———. 2005b. Directing the outcome of deoxyribozyme selections to favor native 3′-5′ RNA ligation. Biochemistry 44: 3017–3023. [DOI] [PubMed] [Google Scholar]
- Wang, Q.S. and Unrau, P.J. 2002. Purification of histidine-tagged T4 RNA ligase from E. coli. BioTechniques 33: 1256–1260. [DOI] [PubMed] [Google Scholar]
- Zuker, M. 2003. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 31: 3406–3415. [DOI] [PMC free article] [PubMed] [Google Scholar]