Abstract
RNA folding into stable tertiary structures is remarkably sensitive to the concentrations and types of cations present; an understanding of the physical basis of ion-RNA interactions is therefore a prerequisite for a quantitative accounting of RNA stability. This article summarizes the energetic factors that must be considered when ions interact with two different RNA environments. “Diffuse ions” accumulate near the RNA because of the RNA electrostatic field and remain largely hydrated. A “chelated” ion directly contacts a specific location on the RNA surface and is held in place by electrostatic forces. Energetic costs of ion chelation include displacement of some of the waters of hydration by the RNA surface and repulsion of diffuse ions. Methods are discussed for computing both the free energy of the set of diffuse ions associated with an RNA and the binding free energies of individual chelated ions. Such calculations quantitatively account for the effects of Mg2+ on RNA stability where experimental data are available. An important conclusion is that diffuse ions are a major factor in the stabilization of RNA tertiary structures.
Keywords: RNA folding, electrostatics, diffuse ions, Mg2+, K+, non-linear Poisson-Boltzmann equation
INTRODUCTION
The high negative charge of an RNA molecule works against its folding into a compact structure. Positive ions promote folding by reducing the repulsion between RNA phosphates, but some ions are much more effective than others at this task. A dramatic example is the ability of millimolar Mg2+ concentrations to stabilize RNA tertiary structures that are otherwise only marginally stable in the presence of high monovalent cation concentrations. This surprising phenomenon was recognized in studies of tRNA folding 30 years ago (Cole et al. 1972; Römer and Hach 1975; Stein and Crothers 1976a), and has motivated much of the subsequent work on ion-RNA interactions. The possibility that Mg2+ also participates directly in the catalytic mechanisms of ribozymes (Cech and Bass 1986; Dahm and Uhlenbeck 1991) has added impetus to the study of ion interactions with RNA.
Although a large literature on ion-RNA interactions has accumulated over the years, the physical basis for Mg2+ stabilizing tertiary structures much more effectively than monovalent ions has not been well understood. To answer questions about the effects of ions on RNA stability, a coherent framework based on the physical properties of ions and nucleic acids is needed. Models for the behavior of ions near DNA have been discussed for years and can quantitatively account for the variation of DNA duplex stability with added mono- or divalent cations (Record 1975; Manning 1978; Anderson and Record 1995). Until recently, there has been no similarly useful model for the analogous problem of RNA tertiary structure stability.
A major difficulty in devising models of ion-RNA interactions stems from the irregularity of RNA tertiary structure, which provides a variety of ion environments. Drawings in Figure 1 ▶ illustrate the problem. At the right, a fully hydrated ion is held in the vicinity of the RNA by the RNA electrostatic field. On the left is an ion that is chelated by RNA groups such as phosphate oxygen. (“Chelate” is used here in its usual chemical sense of an ion making two or more direct contacts.) Although this ion is also held in place by electrostatic forces, the displacement of some of its hydrating water by the RNA is an important energetic consideration. Environments with intermediate levels of ion hydration can be imagined; the middle drawing shows an ion with a first shell of bound waters making hydrogen bonds to acceptor groups on the RNA surface. The energetics of placing an ion in each of these environments depends on different factors, and all of the environments could be energetically important for folding a given RNA structure. Most RNA experiments are done in the presence of both monovalent and divalent ions, which adds to the difficulty of the problem. The challenge has been to devise a model of ion-RNA interactions that captures the full range of energetically important interactions but is simple enough that quantitative calculations of ion-dependent phenomena are feasible.
FIGURE 1.
Potential ion environments in an RNA. Represented are a Mg2+ ion (green dot), nonbridging phosphate oxygens (red circles at the hatched RNA surface), and water molecules (red and gray). At the left, Mg2+ is chelated and partially dehydrated by RNA phosphates; at the right, the Mg2+ remains hydrated but still interacting with the RNA electrostatic field. A possible intermediate situation, in which Mg2+ retains one layer of hydrating water molecules that in turn interact with the RNA surface, is shown in the middle.
There is a well-developed formalism for describing the interactions of small molecules with a fixed number of sites on a macromolecule; many readers will have encountered it in the classic treatment of hemoglobin-O2 binding curves as developed by Wyman and others (Wyman and Gill 1990). Central to this “ligand binding” approach is the binding polynomial, an equation which contains terms characterizing all of the possible small molecule binding sites and the interactions among them. An early use of binding polynomials was in the analysis of ion binding by proteins (Edsall and Wyman 1958), and it was natural that this approach was used in analysis of the first studies of Mg2+-transfer RNA interactions (Stein and Crothers 1976b; Schimmel and Redfield 1980); I have perpetrated its use in this context as well (Draper 1985; Laing et al. 1994). However, ions under the influence of long-range electrostatic forces do not conform to the mass action laws on which binding polynomials depend.1 As a result, the ion “sites” described by a ligand binding analysis are mathematical constructs that do not necessarily correspond to physical reality. For a deeper understanding of ion-RNA interactions, an approach rooted in electrostatic theory is needed.
The purpose of this article is to describe the factors that must be considered in the energetics of ion-RNA interactions for two of the ion environments of Figure 1 ▶, chelated ions and fully hydrated ions. In doing so, the limitations of using a ligand binding formalism to describe ion behavior will become evident, as will the surprising importance of fully hydrated ions in stabilizing RNA tertiary structure and linking together the behavior of ions in all environments. The article concludes with a model of ion-RNA interactions based on chelated and fully hydrated ions; the model has been remarkably successful at predicting the effects of Mg2+ on RNA stability, and can serve as a starting point for considering a wide range of ion-RNA problems.
Characteristics of K+ and Mg2+ ions
The following discussion considers only K+ and Mg2+, the two most common inorganic cations in vivo. Because both ions have a closed shell electronic structure, they interact with other atoms or molecules primarily through electrostatic forces. In contrast, metal ions commonly used as analogs of Mg2+ or K+ in RNA folding experiments (such as Mn2+, Cd2+, Pb2+, Eu3+, and Tl+) have d electrons and/or high polarizability which promote covalent bonding. These metals tend to bind nucleic acids more strongly than do Mg2+ or K+; much of the added energy comes from bonding to base nucleophiles (i.e., purine N7).
Ions in solution interact strongly with the partial negative charge of water oxygen atoms. This electrostatic interaction becomes stronger in ions with higher charge density at the ion surface; either higher charge or smaller ion radius increases charge density. Mg2+ is a small ion with an ionic radius of ~0.65 Å (Pauling 1960). (For comparison, Li+ is only slightly smaller, ~0.60 Å.) Thus, the electrostatic interaction between Mg2+ and water is extremely strong; as a result, the free energy cost of moving Mg2+ from pure water to a less polar solvent such as ethanol is on the order of 250 kcal/mole (Draper and Misra 1998). Detailed calculations have shown that the strength of the Mg2+ electrostatic interaction with water dipoles keeps six water molecules (the number that can pack around a sphere the size of Mg2+) ordered in a characteristic octahedral arrangement (Markham et al. 2000). K+, with a larger radius (~1.3 Å) and only +1 charge, interacts less strongly with water, and the water molecules are not as well ordered. Nevertheless, moving K+ from water to ethanol still involves energies on the order of tens of kcal/mole.
Ion interactions with RNA negative charges
Each K+ in a KCl crystal has as its nearest neighbors Cl− ions; likewise, each Cl− ion is surrounded by K+ ions. In water, the same ions adopt a similar, though far less ordered, arrangement: in the vicinity of each cation is an excess of anions and a deficiency of cations. This spatial arrangement maximizes favorable interactions between opposite charges while minimizing repulsion between like charges. The same general principles apply when ions interact with the negative charges of RNA or DNA, but with important differences because the distances between polynucleotide phosphates are fixed. To visualize the consequences of this constraint, imagine that the potassium salt of an RNA is dissolved in a dilute KCl solution and dialyzed against the same concentration of KCl (Fig. 2A ▶). The initial concentration of K+ inside the dialysis bag exceeds the outside concentration by the number of RNA phosphates (Fig. 2B ▶). For an “ideal” (and imaginary) polynucleotide in which the phosphate charges are very far apart, half of the excess K+ inside the bag diffuses out, and (to maintain charge neutrality) an equal number of Cl− also leave the bag (Fig. 2C ▶). The phosphate charges thus maintain both an excess of K+ and an equivalent deficiency of Cl− inside the dialysis bag; this symmetry might be anticipated, because an isolated phosphate negative charge repels Cl− as effectively as it attracts K+. However, when phosphates are brought close together by the RNA structure, the retention of cations by the RNA becomes larger and the exclusion of anions is correspondingly smaller (Fig. 2 ▶, cf. D,E and C). The underlying reason for this asymmetry is the large electrostatic repulsion between closely spaced phosphates (called the electrostatic “stress”). The stress is greatly relieved by K+ ions accumulated nearby, but a reduced concentration of Cl− near the phosphates is not as effective in reducing the phosphate repulsion. (The preceding statements strictly apply at low KCl concentrations, but the extent of K+ retention by the nucleic acid is approximately constant into the physiological salt concentration range.)
FIGURE 2.
Distribution of ions in equilibrium dialysis experiments with RNA. (A) When an RNA is dialyzed against a KCl solution, K+ (blue) accumulates inside the dialysis bag and Cl− (yellow) is excluded from the bag, relative to the concentrations of the ions outside the bag. Quantitation of the retained and excluded ions is shown in the series of histograms B–E (y-axis: K+, Cl−, and phosphate ion concentrations inside or outside the dialysis bag). (B) “Initial” shows the concentrations of ions after the RNA is placed in the dialysis bag but before dialysis begins. (C) “Ideal” shows the equilibrium concentrations that would be reached if the phosphate charges were infinitely far apart. (D,E) Show the ion concentrations for poly(U) (0.84 K+ retained per phosphate) or poly(U)•poly(A) (0.94 K+ retained per phosphate). The quantitations are based on polyelectrolyte theory and are valid in the limit of low salt concentration, although the cited numbers are relatively independent of the “outside” KCl concentration over a wide range. This diagram and the quantitations of ion retentions are adapted from Record and Richey (1988).
An important principle illustrated by the Figure 2 ▶ equilibrium dialysis experiments is that the number of retained K+ ions depends on how closely the phosphates are spaced (the RNA charge density): the increase from 0.84 K+ per phosphate to 0.94 K+ per phosphate in going from single-stranded poly(U) to a double-helical RNA is because of the closer phosphate–phosphate spacing in the latter molecule. Likewise, any formation of RNA tertiary structure which brings phosphates yet closer together will be accompanied by an increase in the number of associated K+. The principle that the stoichiometry of cation association with an RNA depends on the RNA charge density is quite different from the prevalent notion (based on ligand-binding analysis) that each RNA phosphate constitutes a potential ion binding “site.”
The same principles that govern K+ retention by an RNA also apply to Mg2+, although of course only about half as many Mg2+ ions as K+ are retained by a given RNA [~0.49 Mg2+ per phosphate in poly(A)-poly(U)]. Mixtures of K+ and Mg2+ are more complicated to deal with, but the basic principle remains the same: the fraction of the nucleic acid charge neutralized by associated cations increases with the overall charge density of an RNA. A more rigorous but still accessible discussion of the concepts behind the dialysis experiment described here is given by Record and Richey (1988).
Diffuse ions
There is persuasive NMR evidence that group I monovalent ions, Na+ and K+ in particular, remain well hydrated in the presence of duplex DNA (Braunlin 1995). This is true for nearly all single-stranded and duplex RNAs as well. These ions, shown on the right-hand side of Figure 1 ▶, are variously called “diffuse ions” or the “ion atmosphere.” “Diffuse” ions are defined as the set of ions whose collective behavior is overwhelmingly determined by their long-range electrostatic interactions with the RNA and with each other; within the group, energies associated with ion dehydration or direct contacts with the nucleic acid surface are, by comparison, insignificant. Because diffuse ions follow the electrostatic field of the RNA, the concentration of diffuse ions in a particular region is proportional to the magnitude of the electrostatic potential there. This is illustrated by a cross-section of an RNA hairpin in Figure 3 ▶; the negative potential of the RNA appears as red shading, and the concentrations of monovalent cations are shown by contour lines. The highest potential is deep in the major groove; the highest concentration of cations (more than one molar) is in the same region.
FIGURE 3.

The ion atmosphere around an RNA helix. Red shading, the electrostatic potential; the contour lines map the concentration of cations. Contour lines range from 1.6 M (brown) to 0.2 M (magenta) with intermediate concentrations of 0.8 M and 0.4 M. The bulk monovalent salt concentration is 0.1 M. The highest concentration of cations is found in the major groove, where the electrostatic potential is also highest. Based on Figure 6 of García-García and Draper (2003).
Mg2+ ions, when present in solution, also participate in the diffuse ion atmosphere of a nucleic acid. Although there is evidence that the hydration of Mg2+ associated with a DNA duplex is affected to a small degree (Kankia and Marky 1999), there does not appear to be direct contact between Mg2+ and duplex nucleic acids (Duguid et al. 1993). Thus, under typical conditions used to study RNAs with tertiary structure, the diffuse ion atmosphere will be composed of both monovalent and divalent ions.
The electrostatic potential and ion distribution in Figure 3 ▶ were calculated using the nonlinear Poisson-Boltzmann (NLPB) equation, which finds an overall extent of K+ and Mg2+ ion association with RNA or DNA in good agreement with experimental data (Hecht et al. 1995; Misra and Draper 2000; Das et al. 2003; García-García and Draper 2003). Several groups have used similar calculations to visualize the electrostatic potential near RNA surfaces or to predict the movement of ions under the influence of the RNA electrostatic potential; illustrations can be found in a number of references (Sharp et al. 1990; Hermann and Westhof 1998; Chin et al. 1999; Colmenarejo and Tinoco 1999; Misra and Draper 2000).
Diffuse ions and tRNA stability
The native tertiary structure of an RNA is more compact (and thus has a higher charge density) than partially unfolded forms of the RNA containing only secondary structure. Therefore diffuse ions, either monovalent or divalent, should preferentially stabilize the native structure. In classic studies of tRNA folding, it was found that native tRNA structure becomes more stable as the concentration of Na+ or other group I cations is increased (Cole et al. 1972; Urbanke et al. 1975), and, as noted in the Introduction, tRNA stability increases dramatically with addition of Mg2+. The question is whether the observed tertiary structure stabilization is fully determined by the diffuse ion atmosphere, or whether individual ions that make strong local interactions with special regions of the tertiary structure are important contributors.
To answer this question, comparisons have been made between the results of NLPB-based calculations and measurements of either the extent of Mg2+ association with tRNAPhe (Misra and Draper 2000) or the Mg2+-induced stabilization of tRNAPhe tertiary structure (Misra and Draper 2002). The correspondence between theory and experiment is quite good in both cases. These results show that the effect of Mg2+ on tRNA tertiary structure can be explained solely in terms of the interaction of diffuse ions with the higher charge density of the native tRNA structure, and suggest that other possible modes by which Mg2+ might be interacting with tRNA are not energetically important.2 The idea that diffuse ions have a dominant role in tRNA folding runs counter to the long-standing argument that a small number of “strong” Mg2+ ion binding sites are responsible for the dramatic stabilization of tRNA tertiary structure by Mg2+ (Schimmel and Redfield 1980).
There is a simple entropic reason, first suggested more than 25 years ago by Manning (1978), as to why diffuse Mg2+ ions are so effective in stabilizing tRNA structure. One Mg2+ ion alleviates the electrostatic stress arising from closely packed phosphates to about the same extent as two K+ ions, but at a lower entropic cost because fewer ions are confined near the RNA. The higher the charge density of the RNA, the higher the concentration of ions near the surface and the greater the entropic advantage in using divalent ions to relieve the electrostatic stress. Thus, Mg2+ is far more effective than K+ in stabilizing the high charge density of tRNA tertiary structure, but has a less dramatic effect on secondary structure stability.
Chelated ions
Chelation of an ion by a set of RNA contacts, as illustrated on the left side of Figure 1 ▶, involves three kinds of interactions that contribute to the free energy of ion binding. These energetic factors are diagrammed in Figure 4 ▶ for chelation of a Mg2+ ion:
FIGURE 4.
Energetics of placing a Mg2+ ion in an RNA chelation site. (A,B) The major unfavorable energetic factors. The ion must be partially dehydrated to make direct contact with the RNA surface (ΔGhyd, panel A). The chelated ion displaces ~two diffuse ions from the ion atmosphere, removing their favorable interactions with the RNA, and has repulsive interactions (gray arrows) with the remaining diffuse ions (ΔGMg-diffuse, panel B). Mg2+ interactions with the RNA are favorable (double-headed arrows) and almost entirely electrostatic (ΔGMg-RNA, panel C). Only in unusual chelation sites is ΔGMg-RNA large enough to outweigh the other two unfavorable factors.
Hydration (ΔGhyd)
The close proximity of a chelated Mg2+ to the RNA surface requires partial dehydration of the ion (Fig. 4A ▶); as remarked already, this process is energetically costly. Because the influence of the ionic charge extends to two or more layers of water molecules, the dehydration cost depends on the shape and size of the chelation site as well as the number of direct Mg2+-RNA contacts. There is also a free energy cost for removing water from the surface of the RNA chelation site (not shown), although it tends to be much smaller than the ion dehydration energy.
Interactions with diffuse ions (ΔGMg-diffuse)
Electrostatic interactions between a chelated Mg2+ ion and the surrounding diffuse ion atmosphere are energetically unfavorable for two reasons (Fig. 4B ▶). First, neutralization of RNA phosphate charges by the bound Mg2+ causes a reduction in the average number of diffuse ions associated with the RNA. Although displacement of the ions is entropically favorable, the loss of electrostatic interactions between the displaced ions and the RNA results in a net unfavorable free energy. Second, there are repulsive electrostatic forces between the chelated Mg2+ and the remaining diffuse cations. The overall unfavorable energetic cost of these interactions is of the same order of magnitude as the dehydration penalty, tens of kcals/mole.
RNA interactions (ΔGMg-RNA)
The large unfavorable free energies are offset by strong interactions between the chelated Mg2+ and the RNA (Fig. 4C ▶). By far the most important component of this free energy is the simple electrostatic interaction between the Mg2+ ionic charge and the RNA electrostatic field. There are other possible considerations, such as the polarizability of the ion or Van der Waals contacts, but for chelated group I and group II ions these are small factors compared to the electrostatic attraction. (Not explicitly shown in Fig. 4 ▶ is the reduction in entropy when Mg2+ is confined to a specific location, which amounts to about 3 kcal/mole; Amzel 1997.)
All of the free energy changes associated with chelation can be estimated by widely used methods (Misra and Draper 2001). Of 11 crystallographically observed Mg2+ ions in the crystal structure of a 58mer rRNA fragment with extensive tertiary structure, only two are chelated by the RNA structure, and only one of these has an overall favorable binding free energy when all of the above energetic factors are accounted for. This ion is in a deep pocket where the electrostatic potential is nearly an order of magnitude more intense than that found at the location of any metal ion in tRNAPhe crystals (Misra and Draper 2001). The very favorable electrostatic interaction at this site (~ −110 kcal/mole) overcomes the large penalties for dehydration (~60 kcal/mole) and repulsion of diffuse ions (~40 kcal/mole).
Interdependence of RNA-associated ions
An important principle is that none of the ions associated with an RNA behave independently. For instance, association of a Mg2+ ion with an RNA is accompanied by the displacement of ~two K+ ions, whether the Mg2+ behaves as a chelated ion (Fig. 4 ▶) or a hydrated “diffuse” ion. In a reciprocal fashion, raising the concentration of K+ displaces either diffuse or chelated Mg2+ from the RNA; Mg2+ and diffuse K+ ions therefore compete for interaction with the RNA. In a typical ligand-binding model for ion interactions, competition between monovalent and divalent ions would mean that the two kinds of ions compete for the same discrete binding “sites”. However, the Mg2+–K+ competition just described does not take place because the ions are directly contacting the same RNA groups; rather, the ions respond to each other’s presence through long-range electrostatic interactions. This kind of competition includes any charged molecules, from simple ions to proteins (Misra et al. 1994; García-García and Draper 2003).
A common experiment is to detect the binding of a reporter ion in a specific region of RNA and assume, because adding Mg2+ competes away the signal of the bound ion, that Mg2+ occupies the same “site”. It should be apparent that the observed competition is an inevitable feature of the electrostatic nature of the interaction. Mg2+, as a diffuse ion, will displace any other RNA-associated ions regardless of whether Mg2+ can occupy the identical environment or “site”.
Quantitative calculations of ion-RNA interactions
The foregoing sections have outlined the thermodynamic characteristics of ions in two RNA environments, “diffuse” and “chelated”. A key point of these discussions is that no ion or set of ions can be considered in isolation. Building on these considerations, this section outlines a simple approach by which the extent and energetics of ion association with an RNA may be calculated.
The model divides ions around the RNA into two classes: a large number of diffuse ions that are treated as a group or “ensemble,” and a small number of “explicit” ions that are energetically distinct and treated individually (Misra et al. 2003). Because the properties of both classes of ions depend on the three-dimensional shape and charge distribution of the RNA, the starting point of the calculations is an atomic resolution structure of the RNA in question. The first step is to calculate the properties of the surrounding ensemble of diffuse mono- and divalent ions for a specific set of solution conditions (ion concentrations and temperature). The NLPB equation, mentioned above in reference to calculations on tRNA, has been shown to work very well for this purpose (Misra and Draper 2000). The ensemble description implicitly takes into account not only the interactions of the ions with the RNA, but also the strong interactions between ions.
The second step is to screen for individual ions whose energetics are strongly influenced by additional factors besides electrostatic interactions with the RNA; such factors include dehydration (as described for chelated ions) or, in the case of transition metals, covalent bonding. Ions identified in an RNA crystal structure may suggest locations with such special energetic properties. As described above for chelated ions, the energetics of these explicitly considered ions must include the strongly unfavorable interactions of the ion with the diffuse ion atmosphere.
Although the calculations described here are not trivial, recent numerical implementations of the NLPB equation are capable of all the necessary computations in mixed mono- and divalent salt solutions (Rocchia et al. 2001); the model presented here and the corresponding calculations are described in more detail elsewhere (Misra and Draper 2001; Misra et al. 2003). For the purposes of this article, the important point is that the conceptual distinction between diffuse and explicit ions provides a rigorous basis for calculations. More sophisticated methods may be developed in the future to explore other ion environments which are not chelates but may have characteristics different from those of diffuse ions. For example, the energetic characteristics of a Mg2+ hexahydrate-RNA complex (Fig. 1 ▶, middle panel) are currently unknown. In some situations, calculations with low-resolution models (e.g., cylinders for helices) may also provide useful insights into ion behavior (Bokinsky et al. 2003).
Mg2+-induced RNA stabilization
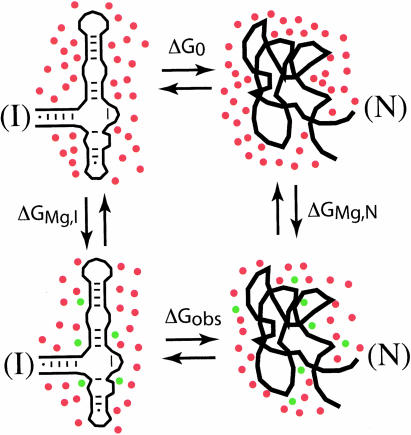
A goal of the calculations described here is to predict how much a change in MgCl2 concentration will affect the stability of an RNA tertiary structure. The way this question is approached is illustrated in Figure 5 ▶. Two forms of the RNA are shown, a partially folded conformation containing only secondary structure (usually referred to as an intermediate, or I form; Fang et al. 1999) and the completely folded tertiary structure (N form). As indicated by the vertical arrows, added Mg2+ interacts with both I and N forms of the RNA. The Mg2+-induced stabilization of the RNA is the difference between the stability of the RNA tertiary structure in the presence or absence of a given Mg2+ concentration, ΔGobs − ΔG0. These two free energies can be experimentally measured, for instance by thermal denaturation (Bukhman and Draper 1997).
FIGURE 5.
Thermodynamic cycle used to relate the measurement of Mg2+-induced stabilization of an RNA tertiary structure to calculations of Mg2+ interactions with RNA. The two left-hand diagrams represent an rRNA fragment with only secondary structure present (the partially folded I state); the right-hand structures are the same RNA in its native tertiary structure (N state). Red dots represent K+ accumulated by the RNA in excess of the K+ concentration in the bulk solution; green dots are Mg2+ added in the reactions depicted by the vertical arrows. (The depletion of anions in the vicinity of the RNA is not illustrated.) The overall free energy in going from I state RNA in the absence of Mg2+ (upper left) to the native RNA with Mg2+ (lower right) is the same whichever pathway is taken; thus ΔG0 + ΔGMg,N = ΔGMg,I + ΔGobs. Mg2+-induced stabilization of the RNA tertiary structure is the difference between the stabilities in the presence and absence of Mg2+, ΔGobs − ΔG0. By the foregoing constraint, this must be equal to ΔGMg,N − ΔGMg,I, a quantity that can be calculated by the approaches described in the text.
The RNA conformational changes associated with ΔGobs and ΔG0 are complex and prevent reliable computation of these free energies. The free energies of Mg2+ association with either RNA form, ΔGMg,I and ΔGMg,N (Fig. 5 ▶, vertical arrows), do not incorporate changes in the RNA structure and can be calculated by the methods outlined in the preceding sections. The I form of the RNA is somewhat problematic, as it is in reality a poorly defined mixture of conformations, but it appears that simple models may imitate the average extent of ion interactions with the partially unfolded RNA (Misra and Draper 2002). A consequence of the way the four equilibria of Figure 5 ▶ are arranged in a square is that the Mg2+-induced stabilization, ΔGobs − ΔG0, is identical to the difference between the Mg2+-RNA association energies, ΔGMg,N − ΔGMg,I (see Fig. 5 ▶ legend). This correspondence allows the calculation of Mg2+-induced RNA stabilization for direct comparison with experiments.
Calculated Mg2+-induced stabilization free energies are in good agreement with experimental values for both tRNAPhe and a 58-nt rRNA fragment (Misra and Draper 2002). As already mentioned, diffuse ions alone can account for tRNAPhe folding; one explicit Mg2+ ion must be included in calculations with the rRNA fragment. To date, these are the only two RNAs for which there are experimental data to compare with calculations. (Few RNA tertiary structures fold completely with only monovalent ions present, which limits the number of RNAs for which ΔG0 can be directly measured.) Nevertheless, on the basis of this limited number of tests, it seems likely that diffuse Mg2+ ions do a large fraction of the work in stabilizing RNA tertiary structures. Thus the tendency in the RNA literature to focus on chelated ions to the exclusion of diffuse ions does not give a balanced picture of folding energetics.
Practical considerations
Ions and “sites”
In reference to small molecules interacting with a macromolecule, the terms “site” and “binding” describe interactions that involve defined sets of contacts, saturate at integral stoichiometries, and follow the mass action law. Chelated ions satisfy these criteria: they make sets of specific contacts with the RNA, and it is meaningful to estimate their individual binding free energies (Fig. 4 ▶). Diffuse ions are constrained only by their long-range interactions with the RNA electrostatic field and not by direct RNA contacts. There may be many spatial arrangements of diffuse ions which satisfy the overall electrostatic constraints imposed by the RNA; the energetics of diffuse ions can only be calculated as an average of all such allowable arrangements for the entire set of ions. For these reasons, it does not make sense to refer to diffuse ion “sites,” and it can be misleading even to speak of diffuse ions “binding” an RNA. To avoid ambiguity, this article has referred to ions “associated” with an RNA; “binding” and “site” have been used only in reference to ion-RNA chelates.
Hill plot analysis of Mg2+-RNA titrations
A common experiment is to follow some measure of RNA tertiary folding (e.g., protections from hydroxyl radical reaction) as a function of Mg2+ ion concentration and fit the resulting curve to the Hill equation (Schimmel and Redfield 1980; Fang et al. 1999; Silverman and Cech 1999). Although the equation usually fits the data moderately well, the fitting parameters are based on a simplified ligand-binding model of Mg2+-RNA association and lack physical relevance. The basic problem is the assumption that the native RNA structure always has n more Mg2+ associated with it than does the partially unfolded form; in reality, this number reaches a maximum near the point where half of the molecules are folded (Misra and Draper 2002). The consequence of this simplification is that free energies derived from the Hill equation do not correspond to actual folding free energies, which are instead comprised of the “intrinsic” free energy of RNA folding (ΔG0) and the free energies of Mg2+ association with both RNA forms (ΔGMg,I and ΔGMg,N in Fig. 5 ▶). These problems are discussed more fully by Misra and Draper (2002).
Detecting partially dehydrated ions
There is no simple test that reliably tells whether the stability of an RNA tertiary structure is derived solely from diffuse ions or has some contribution from chelated ions. (A previous suggestion that the Mg2+ dependence of the Tm of an RNA structure might be used for this purpose neglected the strong interactions between chelated and diffuse ions; Laing et al. 1994.) However, a hint that chelated ions participate in RNA folding can follow from a series of experiments using group I (Li+ to Cs+) or group II (Mg2+ to Ba2+) ions. The reasoning is based on the fact that ion hydration energies strongly depend on the ion radius (Draper and Misra 1998). Thus, the effects of diffuse ions (which remain highly hydrated) on RNA stability should be relatively independent of their size, but interactions of chelated ions, for which ΔGhyd is significant, should vary in strength with ion radius. These kinds of experiments with a 58-nt rRNA fragment suggested the presence of a single chelated K+ (Shiman and Draper 2000) and a chelated Mg2+ (Bukhman and Draper 1997) in the folded structure; the existence and importance of such chelated ions were later corroborated by x-ray crystallography and calculations (Misra and Draper 2001; Conn et al. 2002).
Mn2+ rescue experiments
“Mn2+ rescue” experiments were devised to identify phosphates that must be contacting Mg2+ for an RNA to fold into a stable tertiary structure (Christian and Yarus 1993). The premises of the experiment are that (1) proper RNA folding requires occupancy of a minimum number of defined Mg2+ ion binding sites containing nonbridging phosphate oxygen contacts; (2) simple substitution of phosphate oxygen with sulfur prevents Mg2+ interaction with a site; and (3) occupancy of a disrupted site can be subsequently restored by adding a “soft” divalent metal that tightly binds sulfur. These premises become problematic when long-range electrostatic forces are viewed as the primary determinant of ion interactions. In addition, thiophosphate substitutions are potentially disruptive to RNA structure in two ways. First, the larger and less electronegative sulfur redistributes negative charge onto the nonbridging oxygen in a thiophosphate, which alters the RNA electrostatic field and potentially disfavors the native conformation. Second, the sulfur extends ~0.8 Å further than an oxygen, which may cause steric clashes or disrupt hydrogen bonds (Saenger 1983). Thus, thiophosphate destabilization of an RNA structure may stem from a combination of electrostatic and steric consequences of the sulfur substitution. In support of this view, the disruptive influence of an R oxygen thiophosphate substitution in the hammerhead ribozyme was partially compensated when the S oxygen was also substituted (Derrick et al. 2000), the anticipated result if charge redistribution disrupts the hammerhead structure. The strong interaction of a transition metal with the thiophosphate may well alleviate electrostatic problems created by charge redistribution, but interpretation of such a “rescue” in terms of a Mg2+-phosphate contact in the native structure is problematic. Mn2+ rescue experiments on the group I intron P4–P6 domain, which has three chelated Mg2+ ions in its crystal structure, identify one of the chelated ions but also give “nonspecific” rescues that are not easily interpreted (Basu and Strobel 1999).
Concluding remarks
Biochemical interactions such as enzyme-substrate complexes depend to some extent on short-range forces (e.g., Van der Waals interactions or hydrogen bonds); the direct contacts between the molecules are the basis for defining a binding site, binding stoichiometry, and site-specific equilibrium constant. These concepts do not generally apply to ions whose interactions with RNA are mainly electrostatic, as the interactions may extend over distances many times the diameter of a simple ion (García-García and Draper 2003). For this reason, models of ion-RNA interactions based entirely on discrete and independent “sites” must be inadequate. This article has attempted to show how a consideration of the polyelectrolyte properties of an RNA can lead to a quantitative accounting of the effects of ions on RNA folding. The most important conclusion is that diffuse ions cannot be overlooked in any discussion of ions and RNA: they probably do most of the work in stabilizing RNA tertiary structures, and they link (in an energetic sense) all of the ions associated with an RNA.
Acknowledgments
I thank a number of people for helpful comments on drafts of this paper, including Marty Fedor, Dan Grilley, and Ana Maria Soto. I am especially indebted to Vinod Misra and Ross Shiman for their insights into electrostatic theory and ion-RNA interactions over the last few years and for their extensive criticisms of this paper; it is of course my responsibility, and not lack of effort on their part, if any ideas remain erroneously or poorly expressed here. Work on ion-RNA interactions in my lab has been supported by NIH grant R01 GM58545.
Article and publication are at http://www.rnajournal.org/cgi/doi/10.1261/rna.5205404.
Footnotes
Wyman and Gill (1990), on page 64 of their classic book, cautioned against the use of a binding polynomial approach to treat salt- or solvent-dependent phenomena. A general discussion of Wyman’s linkage relations and charged molecules is in section II.A of M.T. Record’s comprehensive review of salt effects on biopolymers (Record et al. 1998).
The conclusion that tRNA interactions with ions are dominated by the polyelectrolyte character of RNA, rather than by a small number of special binding sites, was clearly stated by M. Guéron and J.L. Leroy more than 20 years ago (Leroy and Guéron 1977; Guéron and Leroy 1982).
REFERENCES
- Amzel, L.M. 1997. Loss of translational entropy in binding, folding, and catalysis. Proteins 28: 144–149. [PubMed] [Google Scholar]
- Anderson, C.F. and Record Jr., M.T. 1995. Salt-nucleic acid interactions. Annu. Rev. Phys. Chem. 46: 657–700. [DOI] [PubMed] [Google Scholar]
- Basu, S. and Strobel, S.A. 1999. Thiophilic metal ion rescue of phosphorothioate interference within the Tetrahymena ribozyme P4–P6 domain. RNA 5: 1399–1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bokinsky, G., Rueda, D., Misra, V.K., Rhodes, M.M., Gordus, A., Babcock, H.P., Walter, N.G., and Zhuang, X. 2003. Single-molecule transition-state analysis of RNA folding. Proc. Natl. Acad. Sci. 100: 9302–9307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braunlin, W.H. 1995. NMR Studies of cation-binding environments on nucleic acids. Adv. Biophys. Chem. 5: 89–139. [Google Scholar]
- Bukhman, Y.V. and Draper, D.E. 1997. Affinities and selectivities of divalent cation binding sites within an RNA tertiary structure. J. Mol. Biol. 274: 1020–1031. [DOI] [PubMed] [Google Scholar]
- Cech, T.R. and Bass, B.L. 1986. Biological catalysis by RNA. Annu. Rev. Biochem. 55: 599–630. [DOI] [PubMed] [Google Scholar]
- Chin, K., Sharp, K.A., Honig, B., and Pyle, A.M. 1999. Calculating the electrostatic properties of RNA provides new insights into molecular interactions and function. Nat. Struct. Biol. 6: 1055–1061. [DOI] [PubMed] [Google Scholar]
- Christian, E.L. and Yarus, M. 1993. Metal coordination sites that contribute to structure and catalysis in the group I intron from Tetrahymena. Biochemistry 32: 4475–4480. [DOI] [PubMed] [Google Scholar]
- Cole, P.E., Yang, S.K., and Crothers, D.M. 1972. Conformational changes of transfer ribonucleic acid. Equilibrium phase diagrams. Biochemistry 11: 4358–4368. [DOI] [PubMed] [Google Scholar]
- Colmenarejo, G. and Tinoco Jr., I. 1999. Structure and thermodynamics of metal binding in the P5 helix of a group I intron ribozyme. J. Mol. Biol. 290: 119–135. [DOI] [PubMed] [Google Scholar]
- Conn, G.L., Gittis, A.G., Lattman, E.E., Misra, V.K., and Draper, D.E. 2002. A compact RNA tertiary structure contains a buried backbone-K+ complex. J. Mol. Biol. 318: 963–973. [DOI] [PubMed] [Google Scholar]
- Dahm, S.C. and Uhlenbeck, O.C. 1991. Role of divalent metal ions in the hammerhead RNA cleavage reaction. Biochemistry 30: 9464–9469. [DOI] [PubMed] [Google Scholar]
- Das, R., Mills, T.T., Kwok, L.W., Maskel, G.S., Millett, I.S., Doniach, S., Finkelstein, K.D., Herschlag, D., and Pollack, L. 2003. Counterion distribution around DNA probed by solution X-ray scattering. Phys. Rev. Lett. 90: 188103. [DOI] [PubMed] [Google Scholar]
- Derrick, W.B., Greef, C.H., Caruthers, M.H., and Uhlenbeck, O.C. 2000. Hammerhead cleavage of the phosphorodithioate linkage. Biochemistry 39: 4947–4954. [DOI] [PubMed] [Google Scholar]
- Draper, D.E. 1985. On the coordination properties of Eu3+ bound to tRNA. Biophys. Chem. 21: 91–101. [DOI] [PubMed] [Google Scholar]
- Draper, D.E. and Misra, V.K. 1998. RNA shows its metal. Nat. Struct. Biol. 5: 927–930. [DOI] [PubMed] [Google Scholar]
- Duguid, J.G., Bloomfield, V.A., Benevides, J.M., Thomas Jr., G.J. 1993. Raman spectroscopy of DNA-metal complexes. I. Interactions and conformational effects of the divalent cations: Mg, Ca, Sr, Ba, Mn, Co, Ni, Cu, Pd, and Cd. Biophys. J. 65: 1916–1928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edsall, J.T. and Wyman, J. 1958. Biophysical chemistry. Academic Press, New York.
- Fang, X., Pan, T. and Sosnick, T.R. 1999. A thermodynamic framework and cooperativity in the tertiary folding of a Mg2+-dependent ribozyme. Biochemistry 38: 16840–16846. [DOI] [PubMed] [Google Scholar]
- García-García, C. and Draper, D.E. 2003. Electrostatic interactions in a peptide-RNA complex. J. Mol. Biol. 331: 75–88. [DOI] [PubMed] [Google Scholar]
- Guéron, M. and Leroy, J.L. 1982. Significance and mechanism of divalent-ion binding to transfer RNA. Biophys. J. 38: 231–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hecht, J.L., Honig, B., Shin, Y.-K., and Hubbell, W.L. 1995. Electrostatic potentials near the surface of DNA: Comparing theory and experiment. J. Phys. Chem. 99: 7782–7786. [Google Scholar]
- Hermann, T. and Westhof, E. 1998. Exploration of metal ion binding sites in RNA folds by brownian-dynamics simulations. Structure 6: 1303–1314. [DOI] [PubMed] [Google Scholar]
- Kankia, B.I. and Marky, L.A. 1999. DNA, RNA, and RNA/RNA oligomer duplexes: A comparative study of their stability, heat, hydration, and Mg2+ binding properties. J. Phys. Chem. B 103: 8759–8767. [Google Scholar]
- Laing, L.G., Gluick, T.C., and Draper, D.E. 1994. Stabilization of RNA structure by Mg2+ Ion: Specific and nonspecific effects. J. Mol. Biol. 237: 577–587. [DOI] [PubMed] [Google Scholar]
- Leroy, J.L. and Guéron, M. 1977. Electrostatic effects in divalent ion binding to tRNA. Biopolymers 16: 2429–2446. [DOI] [PubMed] [Google Scholar]
- Manning, G.S. 1978. The molecular theory of polyelectrolyte solutions with applications to the electrostatic properties of polynucleotides. Q. Rev. Biophys. 11: 179–246. [DOI] [PubMed] [Google Scholar]
- Markham, G.D., Glusker, J.P., and Bock, C.W. 2000. The arrangement of first- and second-sphere water molecules in divalent magnesium complexes: Results from molecular orbital and density functional theory and from structural crystallography. J. Phys. Chem. B 106: 5118–5134. [DOI] [PubMed] [Google Scholar]
- Misra, V.K. and Draper, D.E. 2000. Mg(2+) binding to tRNA revisited: The nonlinear Poisson-Boltzmann model. J. Mol. Biol. 299: 813–825. [DOI] [PubMed] [Google Scholar]
- Misra, V.K. and Draper, D.E. 2001. A thermodynamic framework for Mg2+ binding to RNA. Proc. Natl. Acad. Sci. 98: 12456–12461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misra, V.K. and Draper, D.E. 2002. The linkage between magnesium binding and RNA folding. J. Mol. Biol. 317: 507–521. [DOI] [PubMed] [Google Scholar]
- Misra, V.K., Sharp, K.A., Friedman, R.A., and Honig, B. 1994. Salt effects on ligand-DNA binding: Minor groove binding antibiotics. J. Mol. Biol. 238: 245–263. [DOI] [PubMed] [Google Scholar]
- Misra, V.K., Shiman, R., and Draper, D.E. 2003. A thermodynamic framework for the magnesium-dependent folding of RNA. Biopolymers 69: 118–136. [DOI] [PubMed] [Google Scholar]
- Pauling, L. 1960. The nature of the chemical bond. Cornell University Press, Ithaca, NY.
- Record MT. 1975. Effects of Na+ and Mg++ ions on the helix-coil transition of DNA. Biopolymers 14: 2137–2158. [Google Scholar]
- Record Jr., M.T. and Richey, B. 1988. Physical chemical analysis of biopolymer self-assembly interactions. In ACS sourcebook for physical chemistry instructors (ed. E.T. Lippincott), pp. 145–159. American Chemical Society, Washington, DC.
- Record Jr., M.T., Zhang, W., and Anderson, C.F. 1998. Analysis of effects of salts and uncharged solutes on protein and nucleic acid equilibria and processes: A practical guide to recognizing and interpreting polyelectrolyte effects, Hofmeister effects, and osmotic effects of salts. Adv. Protein Chem. 51: 281–353. [DOI] [PubMed] [Google Scholar]
- Rocchia, W., Alexov, E., and Honig, B. 2001. Extending the applicability of the nonlinear Poisson-Botlzmann equation: Multiple dielectric constants and multivalent ions. J. Phys. Chem. B 105: 6507–6514. [Google Scholar]
- Römer, R. and Hach, R. 1975. tRNA conformation and magnesium binding. A study of a yeast phenylalanine-specific tRNA by a fluorescent indicator and differential melting curves. Eur. J. Biochem. 55: 271–284. [DOI] [PubMed] [Google Scholar]
- Saenger, W. 1983. Principles of nucleic acid structure. Springer Verlag, New York.
- Schimmel, P.R. and Redfield, A.G. 1980. Transfer RNA in Solution: Selected topics. Annu. Rev. Biophys. Bioeng. 9: 181–221. [DOI] [PubMed] [Google Scholar]
- Shiman, R. and Draper, D.E. 2000. Stabilization of RNA tertiary structure by monovalent cations. J. Mol. Biol. 302: 79–91. [DOI] [PubMed] [Google Scholar]
- Silverman, S.K. and Cech, T.R. 1999. Energetics and cooperativity of tertiary hydrogen bonds in RNA structure. Biochemistry 38: 8691–8702. [DOI] [PubMed] [Google Scholar]
- Stein, A. and Crothers, D.M. 1976a. Conformational changes of transfer RNA. The role of magnesium(II). Biochemistry 15: 160–167. [DOI] [PubMed] [Google Scholar]
- Stein, A. and Crothers, D.M. 1976b. Equilibrium binding of magnesium(II) by Escherichia coli tRNAfMet. Biochemistry 15: 157–160. [DOI] [PubMed] [Google Scholar]
- Urbanke, C., Romer, R., and Maass, G. 1975. Tertiary structure of tRNAPhe (yeast): Kinetics and electrostatic repulsion. Eur. J. Biochem. 55: 439–444. [DOI] [PubMed] [Google Scholar]
- Wyman, J. and Gill, S. 1990. Binding and linkage. Functional chemistry of biological macromolecules. University Science Books, Mill Valley, CA.