Abstract
MicroRNAs (miRNAs) are small regulatory RNAs that control gene expression by base-pairing with their mRNA targets. miRNAs assemble into ribonucleoprotein complexes termed miRNPs. Animal miRNAs recognize their mRNA targets via partial antisense complementarity and repress mRNA translation at a step after translation initiation. How animal miRNAs recognize their mRNA targets and how they control their translation is unknown. Here we describe that in a human neuronal cell line, the miRNP proteins eIF2C2 (a member of the Argonaute family of proteins), Gemin3, and Gemin4 along with miRNAs cosediment with polyribosomes. Furthermore, we describe a physical association between a let-7b (miRNA)-containing miRNP and its putative human mRNA target in polyribosome-containing fractions. These findings suggest that miRNP proteins may play important roles in target mRNA recognition and translational repression.
Keywords: microRNA, miRNP, translation, polyribosomes, Argonaute, lin-28
INTRODUCTION
MicroRNAs (miRNAs) comprise an evolutionarily conserved class of ~22-nt RNAs that participate in genetic regulation at the posttranscriptional level (Lee et al. 1993; Wightman et al. 1993; Reinhart et al. 2000; Lagos-Quintana et al. 2001; Lau et al. 2001; Lee and Ambros 2001; Mourelatos et al. 2002; Nelson et al. 2003). miRNAs are processed by the Dicer nuclease from longer RNA precursors (pre-miRNAs; Hutvagner et al. 2001; Ketting et al. 2001; Knight and Bass 2001). Dicer also processes double-stranded RNAs into ~22-nt short interfering RNAs (siRNAs; Hamilton and Baulcombe 1999; Bernstein et al. 2001; Elbashir et al. 2001). mi/siRNAs control gene expression by base-pairing with their mRNA targets. We refer to the sequences that miRNAs recognize on their mRNA targets as miRNA recognition elements (MREs). If the complementarity between the mRNA target and its cognate mi/siRNA is extensive, the mRNA is endonucleolyticaly cleaved (Elbashir et al. 2001; Hutvagner and Zamore 2002); if it is partial, the mRNA levels are not affected but rather the accumulation of the nascent polypeptide is prevented (Olsen and Ambros 1999; Seggerson et al. 2002; Zeng et al. 2002; Doench et al. 2003). miRNAs and siRNAs assemble in multiprotein complexes termed microribonucleoproteins (miRNPs; Mourelatos et al. 2002) and RNAi induced silencing complexes (RISCs; Hammond et al. 2000), respectively. Both siRNAs and miRNAs are bound to proteins that belong to the Argonaute family (Hammond et al. 2001; Martinez et al. 2002; Mourelatos et al. 2002). The Argonaute proteins are ~100-kD proteins that contain two conserved domains of unknown biochemical function, termed the PAZ and PIWI domains. Argonaute proteins are evolutionary conserved and multiple paralogs exist in many organisms (humans have at least seven; for review, see Carmell et al. 2002). Purified RISCs from Drosophila have an apparent molecular mass of ~500 kD and contain the Argonaute protein Ago-2 (Hammond et al. 2001) as well as dFXR, the Drosophila homolog of the Fragile X mental retardation protein (Caudy et al. 2002; Ishizuka et al. 2002), the vasa intronic gene (VIG) protein (Caudy et al. 2002), and the tudor staphylococcal nuclease (Caudy et al. 2003). Affinity purified human RISCs have an apparent molecular mass of 90–160 kD and contain two Argonaute proteins, eIF2C1 and eIF2C2 (Martinez et al. 2002). Human miRNPs sediment as ~15S particles and contain in addition to miRNAs, the proteins eIF2C2, Gemin3 (a putative RNA helicase), and Gemin4 (Mourelatos et al. 2002). The Gemin3 and Gemin4 proteins are also found in the ~50S survival of motor neurons (SMN) complex, an assemblyosome of diverse RNPs (for review, see Paushkin et al. 2002). The core components of SMN complexes are the SMN protein, mutations of which cause Spinal Muscular Atrophy a pediatric neurodegenerative disease, and the Gemin2 protein (Paushkin et al. 2002). RISCs and miRNPs contain the si/miRNA-directed endonuclease, which cleave target mRNAs that bear extensive complementarity with si/miRNAs (Hammond et al. 2000; Hutvagner and Zamore 2002; Martinez et al. 2002); the identity of this endonuclease is unknown. Most plant miRNAs show near perfect complementarity with their mRNA targets and cleave them (Llave et al. 2002b; Rhoades et al. 2002; Kasschau et al. 2003; for review, see Bartel and Bartel 2003). In contrast, most animal miRNAs recognize their mRNA targets via partial antisense complementarity and repress their translation at a step after translation initiation. This novel form of gene expression regulation was first described in Caenorhabditis elegans with the demonstration that the lin-14 mRNA and its lin-4 miRNA regulator are present in translating polyribosomes (Olsen and Ambros 1999). Similarly, the translation of the C. elegans lin-28 mRNA is repressed by lin-4 and both lin-28 and lin-4 associate with translating polyribosomes (Seggerson et al. 2002). Drosophila RISCs copurify with ribosomes (Hammond et al. 2000, 2001) and a fraction of siRNAs associates with polyribosomes in Trypanosoma brucei (Djikeng et al. 2003). How animal miRNAs attenuate the translation of their mRNA targets is unknown. Uncovering the mechanism of this novel form of gene expression regulation will require knowledge of how miRNAs recognize their mRNA targets and which factors participate in these interactions.
Here we describe that the miRNP proteins eIF2C2, Gemin3, and Gemin4 along with miRNA cosediment with polyribosomes in a human neuronal cell line. We also provide evidence for a physical association between a let-7b-containing miRNP and its putative human mRNA target in polyribosome-containing fractions. These findings suggest that miRNP proteins are likely to play important roles in target mRNA recognition and translational repression and indicate that cloning of miRNA targets from polyribosome-containing miRNPs is possible.
RESULTS AND DISCUSSION
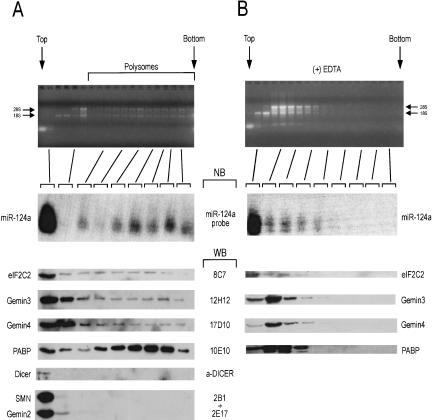
To assess the sedimentation profile of miRNA and miRNA-related proteins, lysates from Weri cells (a human retinoblastoma cell line) were subjected to sedimentation on 15%–45% sucrose gradients in the presence of cycloheximide (to preserve the association of translating ribosomes with mRNAs). The gradients were fractionated and aliquots from different fractions were analyzed as shown in Figure 1A ▶. Analysis of total RNA from each fraction indicated the position of soluble, light particles, monoribosomes, and heavier sedimenting polyribosomes (Fig. 1A, ▶top panel). We analyzed equal volume aliquots from different fractions by Western blotting with antibodies against miRNP and non-miRNP proteins. As shown in Figure 1A ▶, the poly(A)-binding protein (PABP) is enriched in polyribosomal-containing particles, as expected. The majority of the miRNP proteins eIF2C2, Gemin3, and Gemin4 are found towards the top of the gradient, consistent with their presence in a ~15S miRNP particle (Mourelatos et al. 2002). However, a portion of these proteins are also found in heavier sedimenting, monoribosome- and polyribosome-containing particles. In contrast, Dicer or the two core components of the SMN complex, the SMN and Gemin2 proteins, are found only in the top fractions. A Northern blot against miR-124a, a neuronal-specific miRNA (Lagos-Quintana et al. 2002) shows that although the majority of miR-124a is found in the top fraction, a substantial amount is also found in polyribosome-containing fractions. This result is consistent with the finding that the C. elegans miRNA lin-4 cosediments with polyribosomes (Olsen and Ambros 1999; Seggerson et al. 2002). To further substantiate these findings we treated Weri cell lysates with EDTA (to disrupt the polyribosomes) and analyzed them on a 15%–45% sucrose gradient, as performed for the experiments shown in Figure 1A ▶. As shown in Figure 1B ▶, the EDTA treatment shifts the sedimentation of ribosomal-containing particles to lighter fractions and has a similar effect on the distribution of PABP, of the miRNP proteins eIF2C2, Gemin3, and Gemin4, and of miR-124a. These results indicate that miRNP proteins and miRNAs cosediment with polyribosomes.
FIGURE 1.
miRNP proteins and miRNA cosediment with polyribosomes. Lysates from human Weri cells prepared in the presence of cycloheximide (to preserve polyribosomes; A) or EDTA (to disrupt polyribosomes; B), were sedimented on 15%–40% sucrose gradients. Fractions were collected and aliquots of equal volume were analyzed as shown. Top and bottom of gradients are shown. RNA from each fraction was resolved on agarose gels and stained with ethidium bromide to monitor the integrity of the RNA and the presence of polyribosomes; ribosomal RNAs (28S and 18S) are indicated. RNA from indicated fractions was analyzed by Northern blots (NB) using a miR-124a specific probe. Aliquots (equal volume) from indicated fractions were analyzed by Western blots (WB) with antibodies against the Argonaute protein eIF2C2 (8C7), Gemin3 (12H12), Gemin4 (17D10), poly(A) binding protein (PABP; 10E10), Dicer (α-DICER), SMN (2B1), and Gemin2 (2E17). PABP was used as a positive control to monitor the presence of mRNAs in fast-sedimenting (polyribosome-containing) fractions. SMN and Gemin2 proteins do not associate with miRNPs and are not present in polyribosome-containing fractions.
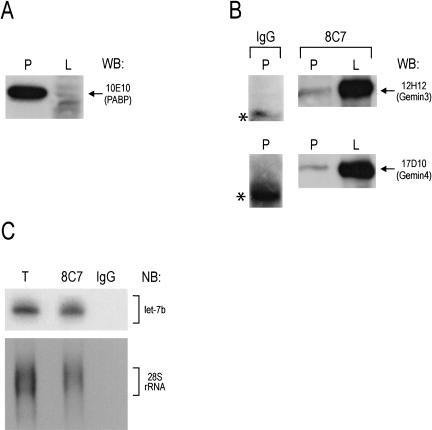
The eIF2C2, Gemin3, and Gemin4 proteins associate physically in a RNA-independent manner as a 15S miRNP (Mourelatos et al. 2002). To assess whether there is a physical association between miRNP proteins and miRNAs in the polyribosomal fractions, we performed immunoprecipitation experiments from polyribosome-containing fractions (prepared from Weri cells treated with cycloheximide) and from the lighter fractions at the top of the gradient that contain 15S miRNPs. The polyribosomal fractions were pelleted over a sucrose cushion (see Materials and Methods). The purity of the preparations was assessed by Western blots against PABP, which is normally enriched in polyribosomal fractions. As shown in Figure 2A ▶, there is substantial enrichment of PABP in the polyribosomal-containing preparation. We performed immunoprecipitations with 8C7 (an anti-eIF2C2 monoclonal antibody) or nonimmune mouse IgG (negative control) from polyribosomal or light fractions, and the immunoprecipitates were analyzed by Western blots with anti-Gemin3 and anti-Gemin4 antibodies. As shown in Figure 2B ▶, Gemin3 and Gemin4 coimmunoprecipitate with eIF2C2 from polyribosomal and light fractions. The amount of Gemin3 and Gemin4 proteins is considerably higher in the light fraction (which contains the 15S miRNP) and this is consistent with the sedimentation profile of eIF2C2, Gemin3, and Gemin4 as shown in Figure 1A ▶. We note however that the amount of Gemin3 and Gemin4 is likely to be underrepresented in the experiment of Figure 2B ▶ because the amount shown represents ~10% of the 8C7 immunoprecipitate from the polyribosomal pellet compared to ~50% of the 8C7 immunoprecipitate from the light fraction. The Gemin3 and Gemin4 proteins do not immunoprecipitate with nonimmune mouse immunoglobulin (Fig. 2B ▶). Numerous miRNAs are found in the 15S miRNPs (Mourelatos et al. 2002). To determine whether miRNAs or ribosomes associate with eIF2C2 in polyribosomal fractions, RNA was isolated from the polyribosomal pellet and from 8C7 or nonimmune mouse IgG immunoprecipitates (performed from the polyribosomal pellet) and probed with antisense oligonucleotide probes against the let-7b miRNA and the 28S ribosomal RNA. As shown in Figure 2C ▶, both let-7b and 28S rRNA associate with eIF2C2 in the polyribosomal fraction. These findings indicate that eIF2C2 associates with Gemin3, Gemin4, and miRNAs not only as part of the 15S miRNP but also in polyribosomal fractions.
FIGURE 2.
Demonstration of a physical association between miRNP proteins and a miRNA (let-7b) in polyribosome-containing fractions. Weri cell lysates were separated in heavy, polyribosome-containing (P) and non-polyribosome-containing, light fractions (L). (A) Western blot (WB) against PABP. (B) Immunoprecipitations were performed from P and L fractions with an antibody against eIF2C2 (8C7) or nonimmune mouse IgG as a negative control. Approximately 10% of the 8C7 and IgG immunoprecipitates from the polyribosomal pellet and ~50% of the 8C7 immunoprecipitate from the light fraction were loaded on the gel and analyzed by Western blots with anti-Gemin3 (12H12) and anti-Gemin4 (17D10) antibodies. Asterisk shows nondissociated IgG molecules. (C) Immunoprecipitations were performed from the polyribosomal fraction with 8C7, or nonimmune mouse IgG; RNA was isolated and analyzed by Northern blots using oligonucleotide probes against let-7b or 28S rRNA. (T) ~10% of the total lysate from polyribosomal pellet.
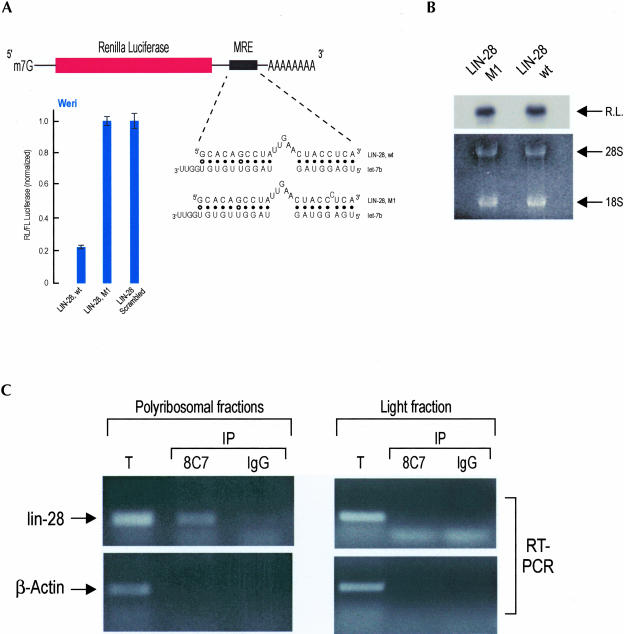
In C. elegans, the lin-4 miRNA cosediments with polyribosomes that contain lin-14 and lin-28 mRNAs, two of its targets, implying that there may be a physical association between a miRNA and its mRNA target in polyribosomes (Olsen and Ambros 1999; Seggerson et al. 2002). As yet, no demonstration for such an association has been presented experimentally for any miRNA. Prior studies have demonstrated that MRE sequences are necessary and sufficient to confer miRNA-dependent translational repression in MRE-bearing target mRNAs (Moss et al. 1997; Reinhart et al. 2000; Zeng et al. 2002; Doench et al. 2003). In plants, the computational identification of miRNA targets was facilitated by the extensive complementarity between plant miRNAs and their mRNA targets (Llave et al. 2002a,b; Reinhart et al. 2002; Rhoades et al. 2002). However, the partial complementarity between animal miRNA and their mRNA targets and the lack of knowledge of the rules that dictate animal miRNA:MRE recognition have precluded their identification. We have devised a computational–experimental approach that describes important rules governing animal miRNA:MRE recognition and allows prediction of animal miRNA targets (this algorithm will be described elsewhere). Based on this approach we have identified a putative MRE for let-7b miRNA, in the 3′ UTR of the human lin-28 mRNA (Fig. 3A ▶). This MRE is also found in the 3′ UTR of mouse lin-28 (the sequence of let-7b miRNA is 100% conserved between human and mouse). This finding is particularly interesting because in C. elegans, the translation of lin-28 mRNA is repressed by the lin-4 miRNA, and the lin-28, lin-4, and let-7 genes function in the same developmental pathway (Ambros 2000). A direct role for the let-7 miRNA in the regulation of C. elegans lin-28 mRNA has not been shown; however, lin-28 is also regulated by a lin-4-independent pathway (Seggerson et al. 2002). Because there are four let-7 paralogs in C. elegans (Lim et al. 2003), it is possible that one of them regulates lin-28. Because MREs are necessary and sufficient to confer miRNA-dependent translational repression, placement of the predicted lin-28 MRE for let-7b in the 3′ UTR of a reporter construct, followed by transfections in cells expressing let-7b, should lead to a decrease of the reporter protein levels. We cloned the predicted lin-28 MRE in the 3′ UTR of a renilla luciferase (RL) reporter construct. As negative controls, we cloned in the 3′ UTR of RL a scrambled sequence of the lin-28 MRE or a single point mutation of lin-28 MRE that is predicted to disrupt base-pairing with let-7b (Fig. 3A ▶). We cotransfected the RL-MRE-bearing constructs along with a plasmid encoding firefly luciferase (FL) in Weri cells (which normally express let-7b and other let-7 paralogs; Fig. 2C ▶; Dostie et al. 2003; P. Nelson and Z. Mourelatos, unpubl. data). Forty-eight hours after transfections, we quantitated the levels of normalized RL/FL using luminometric assays. As shown in Figure 3A ▶, we consistently observe an approximately fivefold reduction in the protein levels of RL bearing the lin-28 MRE versus RL bearing the scrambled MRE or the lin-28-M1 point mutant. To determine whether the reduction of the renilla luciferase activity was secondary to let-7b-mediated translational repression versus mRNA destruction, we performed Northern blots on total RNA isolated from Weri cells that had been transfected with LIN-28-wt or LIN-28-M1 constructs. As shown in Figure 3B ▶, the mRNA levels between these two constructs were the same, ruling out the possibility that the observed reduction of the renilla luciferase activity in the construct bearing the wild-type LIN-28 MRE (LIN-28-wt) was secondary to destabilization of its mRNA. Similar results were obtained when we transfected other cell lines that express let-7b with the above constructs (HeLa cells and the mouse motor neuronal cell line, MN-1) and Northern blots showed that the mRNA levels of all transfected constructs remained the same (P. Nelson and Z. Mourelatos, unpubl. data). These findings indicate that the lin-28 MRE and, by extension, the lin-28 mRNA are likely to be recognized and regulated by let-7b in Weri cells. The Moss laboratory has shown recently that the expression of LIN-28 protein is developmentally regulated in Drosophila, mouse, and Xenopus and in various human and mouse cell lines (Moss and Tang 2003). LIN-28 protein is present during a portion of development and absent from terminally differentiated cells, a pattern that is similar to the expression pattern of LIN-28 protein in C. elegans (Moss and Tang 2003). The Moss laboratory has also identified the same MRE for let-7b in the 3′ UTR of the human and mouse lin-28 mRNA (Moss and Tang 2003). Collectively, these findings suggest that human lin-28 mRNA constitutes a target for let-7b.
FIGURE 3.
A miRNA recognition element (MRE) for let-7b, naturally found in the 3′ UTR of human and mouse lin-28 mRNAs, is sufficient to confer let-7b-mediated translational repression to a luciferase reporter construct. (A) Potential base-pairing between LIN-28 wild-type or mutant (LIN-28, M1) MREs with let-7b is shown. Weri cells were cotransfected with indicated renilla luciferase (RL) constructs along with firefly luciferase (FL). Results shown are average values (with standard deviations) of normalized RL/FL activities obtained from three experiments. (B) Weri cells were transfected with indicated constructs, total RNA was isolated, and RL mRNA was visualized with Northern blotting; the ethidium bromide-stained gel is shown underneath. (C) Endogenous, human lin-28 mRNA associates with miRNPs in polyribosome-containing fractions only. Immunoprecipitations were performed either from polyribosomal or light fractions with anti-eIF2C2 (8C7) or nonimmune mouse IgG. RNA was isolated and amplified with RT-PCR using primers specific for lin-28 or β-actin mRNAs. (T) ~10% of the total lysate from polyribosomal or light fractions.
To determine whether the lin-28 miRNA associates physically with miRNPs in polyribosomes, we performed immunoprecipitations with 8C7 or IgG (negative control) from the polyribosomal or light fractions. RNA was isolated from the immunoprecipitates or from the total polyribosomal and light fractions and was reverse transcribed and amplified by polymerase chain reaction (RT-PCR) using primers specific for the human lin-28 or β-actin mRNAs. As shown in Figure 3C ▶, lin-28, but not β-actin, mRNA is present in the 8C7 immunoprecipitates from the polyribosomal fractions only; both lin-28 and β-actin mRNAs are absent from the negative control IgG immunoprecipitates. These findings demonstrate a physical association between eIF2C2 and lin-28 mRNA in polyribosome-containing fractions. These results also indicate that either miRNPs do not associate with mRNAs in non-polyribosome-containing fractions, or if they do, our RT-PCR method is not sufficiently sensitive to detect them. We favor the former possibility because the majority of miRNPs are found in nonpolyribosome containing fractions and we would have expected them to be enriched for target mRNAs, if mRNAs were associated with miRNPs in the lighter, non-polyribosome-containing fractions.
Our findings demonstrate that miRNAs are present in the form of miRNPs in polyribosome-containing fractions and they likely form a stable association with their mRNA targets. Our experiments do not discriminate whether the interaction between a miRNA (let-7b) and its putative mRNA target (lin-28) is direct or bridged by other factors. Treatment of immunopurified miRNPs with ribonucleases (RNAse A or micrococcal nuclease) destroys the miRNAs (but not the association between the miRNP proteins eIF2C2, Gemin3, and Gemin4; P. Nelson and Z. Mourelatos, unpubl. data) and it is not possible to address whether the observed lin-28-miRNP association is miRNA mediated. Experiments to address this issue will likely require the development of efficient miRNA:MRE cross-linking techniques and in vitro systems that recapitulate miRNA-dependent translational regulation, both of which are lacking as of now. The contribution of let-7b to the regulation of endogenous lin-28 remains to be determined. For example, the observed down-regulation of lin-28 protein in terminally differentiated mammalian cells (Moss and Tang 2003) may also be secondary to transcriptional down-regulation or increased protein turnover. It is also possible that other miRNAs regulate human lin-28. A putative binding site for miR-125a (a human homolog of the C. elegans lin-4 miRNA) has been described (Moss and Tang 2003) and it would be of great interest to investigate further this putative interaction. Further proof that let-7b regulates endogenous lin-28 in vivo would require the demonstration that in a let7-deficient cell line, the protein levels of lin-28 are not repressed. Ultimate proof that let-7b exerts its regulation on lin-28 through the predicted MRE, as proposed by the Moss laboratory and in this study, would require the replacement (“knock-in”) of the human lin-28 gene with one lacking this predicted MRE.
Immunopurified, let-7a-containing human miRNPs can cleave an RNA that bears a fully complementary site to let-7a in vitro, demonstrating that the elusive RNAi endonuclease is part of the miRNP complex (Hutvagner and Zamore 2002). Here we show that miRNPs are found in polyribosomes and are likely to direct translational repression of mRNA targets. The identity of the RNAi endonuclease and the mechanism of translational repression by miRNAs/miRNPs remain unknown. We favor the hypothesis that Argonaute proteins (such as the eIF2C2 protein of human miRNPs) bound to miRNAs (forming ribonucleoproteins that we term miRgonautes) are the most critical molecules that mediate miRNA function. We speculate that certain miRgonautes function as endonucleases when the complementarity between miRNAs and their mRNA targets are extensive. Our hypothesis is strengthened by the findings that Argonaute proteins are the only protein components that miRNPs (Mourelatos et al. 2002) and Drosophila (Hammond et al. 2001) and human (Martinez et al. 2002) RISCs have in common. In this scenario, extensive complementarity between a miRNA and its mRNA target might activate the nuclease domain of Argonaute proteins. If the complementarity is partial, miRgonautes may repress the translation of their mRNA targets either alone or by recruiting other protein factors to regulate components of the translational machinery. The (DEAD-box helicase) Gemin3 and Gemin4 proteins may be accessory molecules that assist miRgonautes in target RNA recognition and translational repression. Gemin3 and Gemin4 may also assist in the maturation of miRNAs from pre-miRNAs and in the assembly of functional miRgonautes. A number of helicases have been implicated in RNAi (for review, see Hannon 2002) and it is possible that they may assist at multiple steps during the maturation and function of miRNAs; Gemin3 may thus be part of this family of RNAi-related helicases.
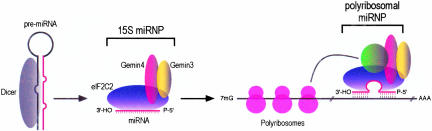
It was demonstrated recently that a fraction of siRNAs, both of sense and antisense polarity, are found in translating polyribosomes in Trypanosoma (Djikeng et al. 2003). This finding indicates that siRNAs and siRNA-associated proteins may associate with the translational machinery independent of their association with their RNA targets. Based on this finding, it was proposed that siRNAs and possibly RISCs associate with translating ribosomes, perhaps as a surveillance system that targets mRNAs for siRNA-mediated destruction. However, mRNA translation is not required for target mRNA recognition and endonucleolytic cleavage by siRNAs or miRNAs in vitro (Tuschl et al. 1999; Hammond et al. 2000; Zamore et al. 2000; Hutvagner and Zamore 2002; Martinez et al. 2002). It is possible that a fraction of miRNPs may also associate with the translational machinery in a target RNA-independent manner. The demonstration, however, that lin-28 mRNA (but not β-actin mRNA) is found associated with let-7b-containing miRNPs only in polyribosomal fractions strongly suggests that there is preferential and perhaps more stable association of miRNAs with their mRNA targets in polyribosomes. It is also possible that additional, as yet unidentified factors associate with polyribosomal miRNPs and function in translational regulation of mRNAs targeted by miRNAs. A model describing how miRNAs might associate with their mRNA targets is proposed in Figure 4 ▶. In summary, this study suggests that the biochemical isolation and cloning of miRNA targets from polyribosome-associated miRNPs may be possible. Furthermore, the Argonaute protein eIF2C2 and the Gemin3 and Gemin4 proteins are likely to play important roles in the recognition and translational repression of miRNA-associated mRNAs.
FIGURE 4.
Proposed model for miRNA:target mRNA association. We hypothesize that miRNAs and the miRNP proteins associate with their mRNA targets in polyribosomes. Additional, as yet unidentified factors (shown in green), may also associate with the polyribosomal miRNP. We favor a direct miRNA:mRNA association guided by miRNP proteins; however, direct interactions between miRNP proteins and the translational machinery (depicted with an arched line) are also likely to occur.
MATERIALS AND METHODS
Cell fractionation and sucrose gradient centrifugation
Weri cells (108) were washed in lysis buffer (LyB: 20 mM Tris-HCl at pH 7.4, 200 mM NaCl, 14 mM MgCl2, 100 μg/mL cycloheximide, 0.1 U/μL RNAsin; Promega), Complete protease inhibitors, EDTA-free (Roche), or a buffer (LyBE) that was identical to LyB except EDTA (30 mM) was added, MgCl2 was decreased to 0.2 mM, and cycloheximide was excluded. Cells were lysed on ice for 5 min in LyB or LyBE buffers containing 0.1% Triton-X 100, and clarified by centrifugation at 14,000g for 10 min at 4°C. All ultracentrifugations were carried with a SW41 rotor at 4°C. For fractionation, clarified lysates were loaded on 15%–45% sucrose gradients and resolved by centrifugation at 35,000 rpm for 130 min. Fractions were collected with a Biocomp collector. For the experiments shown in Figures 2 ▶ and 3C ▶, polysomal pellet preparations (P) were prepared as follows: fractions #12–18 were collected from a 15%–45% sucrose gradient, resuspended in LyB, loaded >2 mL of a 29% sucrose-LyB cushion and centrifuged for 2 h at 35,000 rpm; the polyribosome-containing pellet was washed twice with LyB and resuspended in LyB containing 0.01% Triton-X 100. For nonpolyribosomal, light (L) fraction preparations, the top fraction (fraction #2 in Fig. 1 ▶) of the 15%–45% sucrose gradient, which contains the majority of free miRNPs, was collected. RNA was isolated with Trizol LS reagent (Invitrogen).
Immunoprecipitations, Western blots, and Northern blots
Monoclonal antibodies were used against the following human proteins: the Argonaute protein eIF2C2, (8C7); Gemin2 (2E17), Gemin3 (12H12), Gemin4 (17D10), SMN (2B1), PABP (10E10). The α-Dicer antibody was from affinity-purified rabbit polyclonal antiserum (Doi et al. 2003). Equal volume aliquots from fractions were used for all experiments. For Northern blots of miRNAs, RNA was fractionated on 15% urea-PAGE, blotted on Hybond+ (Amersham) membranes and hybridized with 5′-end [γ-32P]-ATP-labeled oligonucleotides probes, antisense to let-7b or miR-124a. For the Northern blot of Figure 3B ▶, RNA was fractionated on a 1% formaldehyde-agarose gel and probed with a 5′-end [γ-32P]-ATP-labeled oligonucleotide probe, antisense to the 28S ribosomal RNA (AAAACGATCAGAGTAGTGGTATTTCACCG; sequence is from Gu et al. 2001).
Transfections and dual luciferase assays
MRE sequences were subcloned into the NotI-XbaI site of the pRL-TK plasmid (Promega) and resultant constructs (1 μg) were cotransfected along with PGL-3 reporter plasmid (1 μg) into Weri (2 × 104) cells using Lipofectamine 2000 (Invitrogen). Luciferase activities were determined 48 h after transfection using the Dual-Luciferase Reporter Assay System (Promega).
RT-PCR
RNA from polyribosomal fractions and immunoprecipitates was prepared as described above. Equal amounts from each sample were used for reverse transcription using SuperScript II (Invitrogen). Primers include: β-actin, as in Li et al. (2003); lin-28, forward: ACTCCAACCATGCTCTGTCC, reverse: CTTGGCTCCAT GAATCTGGT. PCR cycle program for all primer pairs was: 94°C for 4 min; 32 cycles of 94°C for 40 sec; 55°C for 50 sec; and 72°C for 1 min. From each 50-μL PCR reaction, 25 μL were resolved on a 1% agarose gel.
Acknowledgments
We are grateful to Dr. G. Dreyfuss for 8C7, 12H12, 17D10, 2B1, 2E17, and 10E10 antibodies; Dr. K. Saigo for anti-Dicer antibody; and Dr. M. Kiriakidou for constructs and insightful guidance. This research was supported by NIH grants to P.N. (AG00255) and Z.M. (NS02199) and by a University of Pennsylvania Genomics Institute award to A.H. and Z.M.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.rnajournal.org/cgi/doi/10.1261/rna.5181104.
REFERENCES
- Ambros, V. 2000. Control of developmental timing in Caenorhabditis elegans. Curr. Opin. Genet. Dev. 10: 428–433. [DOI] [PubMed] [Google Scholar]
- Bartel, B. and Bartel, D.P. 2003. MicroRNAs: At the root of plant development? Plant Physiol. 132: 709–717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein, E., Caudy, A.A., Hammond, S.M., and Hannon, G.J. 2001. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature 409: 363–366. [DOI] [PubMed] [Google Scholar]
- Carmell, M.A., Xuan, Z., Zhang, M.Q., and Hannon, G.J. 2002. The Argonaute family: Tentacles that reach into RNAi, developmental control, stem cell maintenance, and tumorigenesis. Genes & Dev. 16: 2733–2742. [DOI] [PubMed] [Google Scholar]
- Caudy, A.A., Myers, M., Hannon, G.J., and Hammond, S.M. 2002. Fragile X-related protein and VIG associate with the RNA interference machinery. Genes & Dev. 16: 2491–2496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caudy, A.A., Ketting, R.F., Hammond, S.M., Denli, A.M., Bathoorn, A.M., Tops, B.B., Silva, J.M., Myers, M.M., Hannon, G.J., and Plasterk, R.H. 2003. A micrococcal nuclease homologue in RNAi effector complexes. Nature 425: 411–414. [DOI] [PubMed] [Google Scholar]
- Djikeng, A., Shi, H., Tschudi, C., Shen, S., and Ullu, E. 2003. An siRNA ribonucleoprotein is found associated with polyribosomes in Trypanosoma brucei. RNA 9: 802–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doench, J.G., Petersen, C.P., and Sharp, P.A. 2003. siRNAs can function as miRNAs. Genes & Dev. 17: 438–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doi, N., Zenno, S., Ueda, R., Ohki-Hamazaki, H., Ui-Tei, K., and Saigo, K. 2003. Short-interfering-RNA-mediated gene silencing in mammalian cells requires Dicer and eIF2C translation initiation factors. Curr. Biol. 13: 41–46. [DOI] [PubMed] [Google Scholar]
- Dostie, J., Mourelatos, Z., Yang, M., Sharma, A., and Dreyfuss, G. 2003. Numerous microRNPs in neuronal cells containing novel microRNAs. RNA 9: 180–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbashir, S.M., Lendeckel, W., and Tuschl, T. 2001. RNA interference is mediated by 21- and 22-nucleotide RNAs. Genes & Dev. 15: 188–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu, Y.C., Talts, J.F., Gullberg, D., Timpl, R., and Ekblom, M. 2001. Glucocorticoids down-regulate the extracellular matrix proteins fibronectin, fibulin-1 and fibulin-2 in bone marrow stroma. Eur. J. Haematol. 67: 176–184. [DOI] [PubMed] [Google Scholar]
- Hamilton, A.J. and Baulcombe, D.C. 1999. A species of small antisense RNA in posttranscriptional gene silencing in plants. Science 286: 950–952. [DOI] [PubMed] [Google Scholar]
- Hammond, S.M., Bernstein, E., Beach, D., and Hannon, G.J. 2000. An RNA-directed nuclease mediates post-transcriptional gene silencing in Drosophila cells. Nature 404: 293–296. [DOI] [PubMed] [Google Scholar]
- Hammond, S.M., Boettcher, S., Caudy, A.A., Kobayashi, R., and Hannon, G.J. 2001. Argonaute2, a link between genetic and biochemical analyses of RNAi. Science 293: 1146–1150. [DOI] [PubMed] [Google Scholar]
- Hannon, G.J. 2002. RNA interference. Nature 418: 244–251. [DOI] [PubMed] [Google Scholar]
- Hutvagner, G. and Zamore, P.D. 2002. A microRNA in a multiple-turnover RNAi enzyme complex. Science 297: 2056–2060. [DOI] [PubMed] [Google Scholar]
- Hutvagner, G., McLachlan, J., Pasquinelli, A.E., Balint, E., Tuschl, T., and Zamore, P.D. 2001. A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7 small temporal RNA. Science 293: 834–838. [DOI] [PubMed] [Google Scholar]
- Ishizuka, A., Siomi, M.C., and Siomi, H. 2002. A Drosophila fragile X protein interacts with components of RNAi and ribosomal proteins. Genes & Dev. 16: 2497–2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasschau, K.D., Xie, Z., Allen, E., Llave, C., Chapman, E.J., Krizan, K.A., and Carrington, J.C. 2003. P1/HC-Pro, a viral suppressor of RNA silencing, interferes with Arabidopsis development and miRNA function. Dev. Cell 4: 205–217. [DOI] [PubMed] [Google Scholar]
- Ketting, R.F., Fischer, S.E., Bernstein, E., Sijen, T., Hannon, G.J., and Plasterk, R.H. 2001. Dicer functions in RNA interference and in synthesis of small RNA involved in developmental timing in C. elegans. Genes & Dev. 15: 2654–2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight, S.W. and Bass, B.L. 2001. A role for the RNase III enzyme DCR-1 in RNA interference and germ line development in Caenorhabditis elegans. Science 293: 2269–2271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagos-Quintana, M., Rauhut, R., Lendeckel, W., and Tuschl, T. 2001. Identification of novel genes coding for small expressed RNAs. Science 294: 853–858. [DOI] [PubMed] [Google Scholar]
- Lagos-Quintana, M., Rauhut, R., Yalcin, A., Meyer, J., Lendeckel, W., and Tuschl, T. 2002. Identification of tissue-specific microRNAs from mouse. Curr. Biol. 12: 735–739. [DOI] [PubMed] [Google Scholar]
- Lau, N.C., Lim, L.P., Weinstein, E.G., and Bartel, D.P. 2001. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science 294: 858–862. [DOI] [PubMed] [Google Scholar]
- Lee, R.C. and Ambros, V. 2001. An extensive class of small RNAs in Caenorhabditis elegans. Science 294: 862–864. [DOI] [PubMed] [Google Scholar]
- Lee, R.C., Feinbaum, R.L., and Ambros, V. 1993. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 75: 843–854. [DOI] [PubMed] [Google Scholar]
- Li, A., Zhu, X., Brown, B., and Craft, C.M. 2003. Gene expression networks underlying retinoic acid-induced differentiation of human retinoblastoma cells. Invest. Ophthalmol. Vis. Sci. 44: 996–1007. [DOI] [PubMed] [Google Scholar]
- Lim, L.P., Lau, N.C., Weinstein, E.G., Abdelhakim, A., Yekta, S., Rhoades, M.W., Burge, C.B., and Bartel, D.P. 2003. The microRNAs of Caenorhabditis elegans. Genes & Dev. 17: 991–1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llave, C., Kasschau, K.D., Rector, M.A., and Carrington, J.C. 2002a. Endogenous and silencing-associated small RNAs in plants. Plant Cell 14: 1605–1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llave, C., Xie, Z., Kasschau, K.D., and Carrington, J.C. 2002b. Cleavage of Scarecrow-like mRNA targets directed by a class of Arabidopsis miRNA. Science 297: 2053–2056. [DOI] [PubMed] [Google Scholar]
- Martinez, J., Patkaniowska, A., Urlaub, H., Luhrmann, R., and Tuschl, T. 2002. Single-stranded antisense siRNAs guide target RNA cleavage in RNAi. Cell 110: 563–574. [DOI] [PubMed] [Google Scholar]
- Moss, E.G. and Tang, L. 2003. Conservation of the heterochronic regulator Lin-28, its developmental expression and microRNA complementary sites. Dev. Biol. 258: 432–442. [DOI] [PubMed] [Google Scholar]
- Moss, E.G., Lee, R.C., and Ambros, V. 1997. The cold shock domain protein LIN-28 controls developmental timing in C. elegans and is regulated by the lin-4 RNA. Cell 88: 637–646. [DOI] [PubMed] [Google Scholar]
- Mourelatos, Z., Dostie, J., Paushkin, S., Sharma, A., Charroux, B., Abel, L., Rappsilber, J., Mann, M., and Dreyfuss, G. 2002. miRNPs: A novel class of ribonucleoproteins containing numerous microRNAs. Genes & Dev. 16: 720–728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson, P., Kiriakidou, M., Sharma, A., Maniataki, E., and Mourelatos, Z. 2003. The microRNA world: Small is mighty. Trends Biochem. Sci. 28: 534–540. [DOI] [PubMed] [Google Scholar]
- Olsen, P.H. and Ambros, V. 1999. The lin-4 regulatory RNA controls developmental timing in Caenorhabditis elegans by blocking LIN-14 protein synthesis after the initiation of translation. Dev. Biol. 216: 671–680. [DOI] [PubMed] [Google Scholar]
- Paushkin, S., Gubitz, A.K., Massenet, S., and Dreyfuss, G. 2002. The SMN complex, an assemblyosome of ribonucleoproteins. Curr. Opin. Cell Biol. 14: 305–312. [DOI] [PubMed] [Google Scholar]
- Reinhart, B.J., Slack, F.J., Basson, M., Pasquinelli, A.E., Bettinger, J.C., Rougvie, A.E., Horvitz, H.R., and Ruvkun, G. 2000. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature 403: 901–906. [DOI] [PubMed] [Google Scholar]
- Reinhart, B.J., Weinstein, E.G., Rhoades, M.W., Bartel, B., and Bartel, D.P. 2002. MicroRNAs in plants. Genes & Dev. 16: 1616–1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhoades, M.W., Reinhart, B.J., Lim, L.P., Burge, C.B., Bartel, B., and Bartel, D.P. 2002. Prediction of plant microRNA targets. Cell 110: 513–520. [DOI] [PubMed] [Google Scholar]
- Seggerson, K., Tang, L., and Moss, E.G. 2002. Two genetic circuits repress the Caenorhabditis elegans heterochronic gene lin-28 after translation initiation. Dev. Biol. 243: 215–225. [DOI] [PubMed] [Google Scholar]
- Tuschl, T., Zamore, P.D., Lehmann, R., Bartel, D.P., and Sharp, P.A. 1999. Targeted mRNA degradation by double-stranded RNA in vitro. Genes & Dev. 13: 3191–3197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wightman, B., Ha, I., and Ruvkun, G. 1993. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell 75: 855–862. [DOI] [PubMed] [Google Scholar]
- Zamore, P.D., Tuschl, T., Sharp, P.A., and Bartel, D.P. 2000. RNAi: Double-stranded RNA directs the ATP-dependent cleavage of mRNA at 21 to 23 nucleotide intervals. Cell 101: 25–33. [DOI] [PubMed] [Google Scholar]
- Zeng, Y., Wagner, E.J., and Cullen, B.R. 2002. Both natural and designed micro RNAs can inhibit the expression of cognate mRNAs when expressed in human cells. Mol. Cell 9: 1327–1333. [DOI] [PubMed] [Google Scholar]