Abstract
The genomic RNA of the gypsy retroelement from Drosophila melanogaster exhibits features similar to other retroviral RNAs because its 5′ untranslated (5′ UTR) region is unusually long (846 nucleotides) and potentially highly structured. Our initial aim was to search for an internal ribosome entry site (IRES) element in the 5′ UTR of the gypsy genomic RNA by using various monocistronic and bicistronic RNAs in the rabbit reticulocyte lysate (RRL) system and in cultured cells. Results reported here show that two functionally distinct and independent RNA domains control the production of gypsy encoded proteins. The first domain corresponds to the 5′ UTR of the env subgenomic RNA and exhibits features of an efficient IRES (IRESE) both in the reticulocyte lysate and in cells. The second RNA domain that encompasses the gypsy insulator can function as an IRES in the rabbit reticulocyte lysate but strongly represses translation in cultured cells. Taken together, these results suggest that expression of the gypsy encoded proteins from the genomic and subgenomic RNAs can be regulated at the level of translation.
Keywords: cap-independent, IRES, eIF4G, translation, gypsy, retrovirus
INTRODUCTION
For the majority of eukaryotic mRNAs, translation occurs by the ribosome scanning mechanism in which the 40 S subunit of the ribosome (carrying Met-tRNA and other initiation factors) binds to the cap structure at the 5′ end of the mRNA and moves along the 5′ leader, unwinding the RNA secondary structures, until an initiator AUG is encountered (Kozak 1989). For efficient recognition, this AUG should be in an appropriate context (A/G)CCAUGG, the purine at position −3 being particularly important together with a G at position +4 (Kozak 1991). An alternative mechanism of translation has first been described for picornaviruses. It is now well established that the initiation of protein synthesis on picornavirus RNAs, which are uncapped and present a long 5′ untranslated region (5′ UTR), occurs by a cap-independent mechanism that is directed by an internal ribosome entry site (IRES) located within the 5′ UTR of the genomic RNA (Jackson et al. 1994; Jackson and Kaminski 1995; Belsham and Sonenberg 2000). The mechanism of internal initiation has been extended to other viruses and a growing number of cellular genes from yeast, Drosophila, and mammals (for a recent review, see Vagner et al. 2001). Recently, IRESs have been identified within the genomic 5′ UTR of retroviruses such as Moloney murine leukemia virus (M-MLV; Vagner et al. 1995), Friend murine leukemia virus (F-MLV; Berlioz and Darlix 1995; Deffaud and Darlix 2000), the rat VL30 region of Harvey murine sarcoma virus (Berlioz et al. 1995), avian reticuloendotheliosis virus type A (Lopez-Lastra et al. 1997), human T-cell leukemia virus (Attal et al. 1996), simian immunodeficiency virus (SIV; Ohlmann et al. 2000) and human immunodeficiency virus type 1 (HIV-1; Buck et al. 2001; Brasey et al. 2003). The mouse VL30 retrotransposon was also found to contain an IRES (Lopez-Lastra et al. 1999). In retroviruses and retrotransposons, the 5′ untranslated region (UTR) is formed of stable secondary structures that are required for several steps of virus replication such as genomic RNA dimerization, packaging and reverse transcription (Berkowitz et al. 1996; Corbin and Darlix 1996; Paillart et al. 1996; Telesnitsky 1997). These stable secondary structures are thought to strongly interfere with ribosome scanning, and IRES-dependent translation might provide a way for the ribosomes to gain access to the gag initiation codon.
The genetic structure of gypsy displays striking similarities with that of vertebrate onco-retroviruses. The provirus is 7.5 kpb in length with two LTRs flanking three open reading frames that are homologous to the retroviral gag, pol, and env (Bayev et al. 1984; Marlor et al. 1986). The Gag and Pol polyproteins are expressed from the genome-length RNA and correspond to the structural proteins and enzymes of the particle, whereas Env is expressed from the subgenomic transcript (Pelisson et al. 1994). The gypsy Env protein was shown to be functional because it can pseudotype M-MLV that can infect Drosophila culture cells (Teysset et al. 1998). In vivo, the gypsy retroelement moves unpredictably and at low frequency, but some Drosophila strains have been described in which gypsy transposes at high frequency (Kim et al. 1990; Kim and Belyaeva 1991; Prud’homme et al. 1995). Genetic analyses identified a X-linked flamenco (flam) gene as a major regulator of gypsy retrotransposition (Prud’homme et al. 1995). At the molecular level, proviral mobilization in the progeny of flam permissive females correlates with a dramatic derepression of gypsy expression in the ovaries of these females, and in accordance, the full-length genomic RNAs has been detected in follicle cells (Pelisson et al. 1994; Song et al. 1994; Chalvet et al. 1999). The Pol proteins are expressed in both follicle and nurse cells (from stage 10A to stages 13 to 14), whereas the Env proteins are mainly detected in the follicle cells, starting at stage 9 of oogenesis to stages 13 to 14 (Pelisson et al. 1994; Song et al. 1997). However, these proteins do not accumulate at detectable levels in the oocyte, but viral-like particles can be observed by immunoelectron microscopy in the perivitelline space starting at stage 10 (Song et al. 1997). Therefore, gypsy retrotransposition and spread result from the transfer of gypsy-encoding genetic material from follicle cells toward the germline cells of the mother and its subsequent insertion into the chromosomes of the progeny. In vivo, the gypsy proviral DNA is also transcribed in a tissue-specific manner at the embryonic, larval, and adult stages (Smith and Corces 1995). However, little is known on possible translational controls of gypsy genomic and subgenomic RNAs. The 5′ UTR of the genomic RNA is long and has the potential to form stable secondary structures, possibly impairing translation initiation by ribosome scanning. By investigating the translation of mono- and dicistronic RNAs, we show that the 5′ UTR of the gypsy RNA can be divided in two distinct separate domains. The first sequence (env) can efficiently drive translation in a cap-independent manner both in the RRL and in cultured cells and was therefore named IRESE. The second domain (named D1) can function as an IRES in the rabbit reticulocyte but not in cultured cells. In fact, our results show that D1 represses translation in several cell types, including 293T and Drosophila SL2 cells.
RESULTS
Features of the 5′ UTR of the gypsy genomic RNA
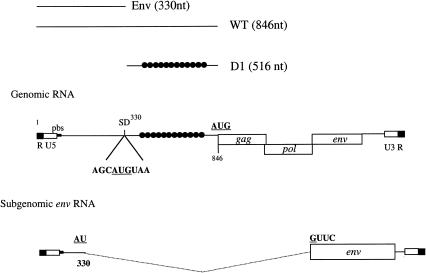
The 5′ UTR of the genomic RNA of gypsy is unusually long (846 nt) with potential stable secondary structures and contains an internal AUG codon (position 330) immediately followed by a stop codon (UAA; Fig. 1 ▶). This Gag initiation codon overlaps with the splice donor site for the env subgenomic mRNA (Fig. 1 ▶). Thus, upon splicing, the UAA codon is removed, and the AUG330 becomes the initiation codon for the env reading frame (Fig. 1 ▶).
FIGURE 1.
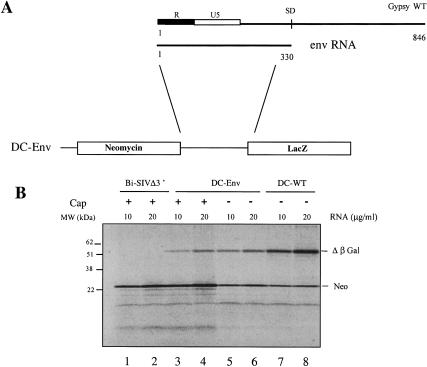
Schematic diagram of the gypsy genomic and subgenomic RNAs. The 5′ UTR and the gag, pol, and env open reading frames are depicted on the figure. Position of the cap site (+1), splice donor (SD), and AUGs are indicated. The position of the two RNA domains described in this work (Env and D1) are shown on the figure, and the black dots represent the position of the gypsy insulator. Numbering is with respect to the +1 site of transcription on the genomic RNA. pbs indicates primer binding site.
The gypsy 5′ UTR drives translation of mono- and bicistronic RNAs in the RRL
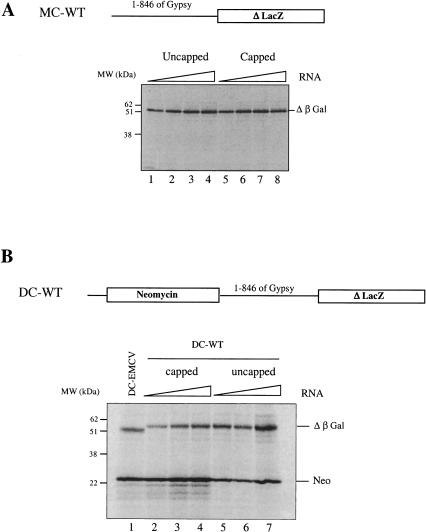
The complete 5′ UTR (1–846) of gypsy was inserted upstream of a lacZ reporter gene, in the pMC-WT construct, and capped and uncapped RNAs generated in vitro (see Materials and Methods) were translated in the reticulocyte lysate (RRL) system. Results (Fig. 2A ▶) show that there was little difference, if any, in the amount of β-galactosidase (β-Gal) synthesized from capped (lanes 1–4) versus uncapped (lanes 5–8) RNAs over a wide range of RNA concentrations. Thus, the presence of a cap structure at the 5′ end of the mRNA did not enhance expression driven by the gypsy 5′ UTR in the RRL system, suggesting that translation initiation may occur via a cap-independent mechanism. This prompted us to look whether the 5′ UTR of gypsy could drive protein synthesis in the context of a bicistronic construct. The 846 nt of the gypsy 5′ UTR up to the authentic AUG of gag were inserted into the NheI site of a dicistronic vector previously described (Ohlmann et al. 2000) downstream of the neomycin and upstream of the lacZ genes creating the pDC-WT construct (Fig. 2B ▶). Capped and uncapped bicistronic RNAs were translated in the RRL at various RNA concentrations together with 10 μg/mL of capped bicistronic RNA derived from pEMCV-D260–837 (hereafter, designed pDC-EMCV) containing the IRES of encephalomyocarditis virus (EMCV). Data reported in Figure 2B ▶ show that, as expected, the amount of neomycin synthesized (first gene) was significantly higher when the RNA was capped (cf. lanes 2–4 and 5–7). Interestingly, detection of radiolabeled β-Gal protein indicated that the gypsy 5′ UTR can direct translation of the 3′ cistron in a bicistronic context. These results indicate that the complete gypsy 5′ UTR is able to efficiently drive gene expression in the absence of a 5′ cap structure and in the context of a bicistronic RNA.
FIGURE 2.
The 5′ UTR of gypsy promotes translation in the context of mono- and dicistronic RNAs. (A) The complete 5′ UTR (846 nt) of gypsy was inserted upstream of the lacZ reporter gene (pMC-WT construct). Several concentrations of uncapped (lane 1, 5μg/mL; 2, 10 μg/mL; 3, 20 μg/mL; 4, 40 μg/mL) and capped RNAs (lane 5, 5μg/mL; 6, 10 μg/mL; 7, 20 μg/mL; 8, 40 μg/mL) were translated for 60 min in the RRL (10 μL). (B) The 5′ UTR was inserted between the two cistrons of a bicistronic construct coding for neomycin and β-Gal proteins (pDC-WT). Several concentrations of capped (lane 2, 10 μg/mL; 3, 20 μg/mL; 4, 40 μg/mL) and uncapped (lane 5, 10 μg/mL; 6, 20 μg/mL; 7, 40 μg/mL) bicistronic RNAs were translated as described above. A control assay with 10 μg/mL of capped bicistronic EMCV-D260-837 (DC-EMCV) was set in parallel (lane 1). At the end of a 60-min incubation, 1 μL sample of each assay was analyzed on a 15% SDS-PAGE and subjected to autoradiography. Positions of molecular weight markers (kD), neomycin (Neo), and β-Gal translation products are indicated. Note that β-Gal synthesized from the EMCV construct migrates slightly faster on SDS-PAGE (lane 1), which is due to linearization of the DNA at a different site (see Materials and Methods). This difference in size does not alter the methionine content of the protein.
Translation driven by the gypsy 5′ UTR is not inhibited by protease-mediated cleavage of eIF4G
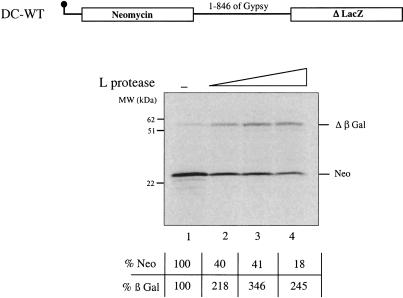
Next, we used the L protease from the foot-and-mouth-disease virus (FMDV) in order to cleave the initiation factor eIF4G in the reticulocyte lysate. Upon FMDV, polio, and rhino, virus infection, eIF4G is cleaved early in the viral life cycle by picornavirus-encoded proteases (proteases L and 2A, respectively), causing an inhibition of cap-dependent protein synthesis (for a recent review, see Gale et al. 2000). In the RRL, addition of in vitro translated L protease results in complete processing of the initiation factor eIF4G (data not shown) and creates conditions in which cap-dependent translation is inhibited, whereas IRES-mediated protein synthesis is unaffected or even stimulated (Ziegler et al. 1995; Ohlmann et al. 1996; Borman et al. 1997a). Increasing amounts of FMDV-L protease were added to the rabbit reticulocyte lysate programmed with 5′ capped DC-WT RNA (Fig. 3 ▶). As expected, addition of the L protease impaired 5′ cap-dependent translation (Neo) in a dose-dependent manner (cf. lanes 1 and 2–4). At the highest dose of L protease used, expression of the 5′ cistron was <20% of the control assay (cf. lanes 1 and 4). In contrast, β-Gal synthesis driven by the gypsy 5′ UTR was clearly stimulated by addition of the L protease with a maximum increase of ~3.5-fold over the control level (lane 3). This indicates that the gypsy 5′ UTR contains sequences that can efficiently drive internal initiation of translation in the RRL system.
FIGURE 3.
Translation driven by the gypsy 5′ UTR is not inhibited by addition of the FMDV L protease. Capped DC-WT RNAs (20 μg/mL) were translated in the RRL (10 μL) in the absence (lane 1) or presence (lanes 2, 0.2 μL; 3, 0.4 μL; 4, 0.8 μL) of in vitro translated L protease. 1 μL sample of each assay was analyzed on a 15% SDS-PAGE, subjected to autoradiography, and quantified by using a STORM 850 phoshoimager. Positions of the molecular weight markers (kD), neomycin (Neo), and β-Gal translation products are indicated. Results of the quantitation are expressed as percentage of the control (no L protease added) for both neomycin (first cistron), and β-Gal (second cistron) as indicated.
The RNA sequence from position 530–790 has IRES activity in the RRL
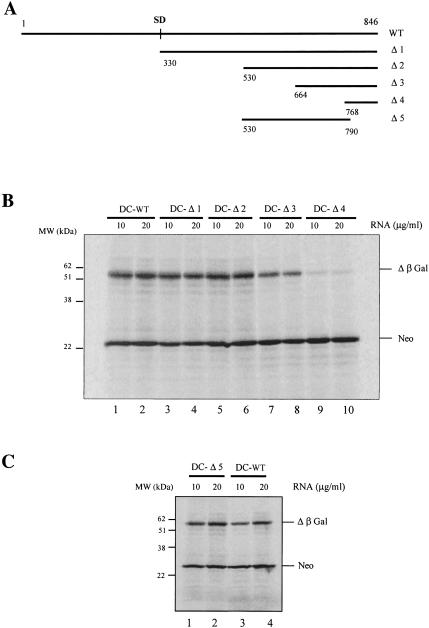
To delineate the minimal sequence required to promote internal initiation of translation, 5′ and 3′ truncated versions of the gypsy 5′ UTR (Fig. 4A ▶, Δ1 to Δ5) were generated by PCR and cloned in the intercistronic spacer of pNeo-LacZ in the NheI site to generate pDC-Δ1 to pDC-Δ5. It should be noted that all RNA transcripts with 5′ and 3′ deletions contain the initiation codon (and its surrounding context) corresponding to the authentic AUG of gag. Uncapped transcripts containing 5′ deletions were translated in the RRL at two RNA concentrations (Fig. 4B ▶). Interestingly, removal of the 5′ first 530 nt (Δ2) of gypsy did not affect expression of β-Gal (lanes 1–6), and only a moderate decrease in the yield of β-Gal was observed upon deletion of nucleotides 530–664 (Δ3; lanes 7,8). However, further deletions in this region up to position 768 (Δ4) had a drastic negative effect on translation (lanes 9,10). To delineate more precisely the region involved in internal initiation, a 3′ deletion removing the last 62 nt (Δ5) was generated. Data show that translation of uncapped mRNAs harboring this deletion was not affected (Fig. 4C ▶, cf. lanes 1,2 and 3,4).
FIGURE 4.
Boundaries of a gyspy genetic element harboring an IRES activity. (A) Schematic representation of deletions introduced in the 5′ UTR. The numbering is with respect to the 5′ cap site. Segments corresponding to 5′ (B) and 3′ (C) deletions of the 5′ UTR were inserted in between the two cistrons of the bicistronic RNA described in Figure 2B ▶. The resulting uncapped bicistronic RNAs were translated in the RRL (10 μL) at the concentrations indicated. Control assay with uncapped bicistronic RNA containing the entire 5′ UTR was set in parallel (B, lanes 1,2; C, lanes 3,4). At the end of a 60-min incubation, 1μL sample of each assay was analyzed on a 15% SDS-PAGE and subjected to autoradiography. Positions of the molecular weight markers (kD), neomycin (Neo), and β-Gal translation products are indicated.
Taken together, these results indicate that the gypsy 5′ UTR harbors an IRES contained within the region between nucleotides 530–790. Moreover, the inability of the 3′ sequence (nucleotides 768–846) to confer internal initiation (Fig. 4B ▶), together with the fact that the last 62 nt are dispensable for ribosome entry (Fig. 4C ▶), suggests that the actual entry site is located at least 50 nt upstream from the authentic AUG gag codon and that ribosomes must be transferred, probably by scanning, from the entry site to the AUG initiator at position 846.
The gypsy env RNA contains an IRES
The data presented above suggest that the segment of gypsy 5′ UTR spanning from the cap (+1 of transcription) to the major splice donor site (nucleotide 330) is most probably not involved in the delivery of a 40 S subunit to the AUG of gag because the 5′ truncation (Δ1) does not affect the yield of β-Gal synthesized by using the bicistronic construct (Fig. 4B ▶, cf. DC-Δ1 and DC-WT). However, it cannot be ruled out that ribosomes possibly enter the mRNA at AUG330 in an initiation-competent manner and are then arrested at the stop codon, which lies immediately downstream (UAA333). This is especially interesting because upon splicing, this AUG is used for Env translation (Fig. 1 ▶). To examine this possibility, we have constructed a monocistronic RNA with the gypsy 5′ UTR up to the AUG initiatior codon of env RNA in frame with the LacZ coding sequence. Translation of this monocistronic RNA in the RRL revealed that capping of the transcript did not increase protein synthesis (data not shown) as it was found for the full-length genomic RNA (Fig. 2A ▶). This prompted us to look for an IRES element within this region. The strategy used was essentially similar to that described before, and the env 5′ UTR was inserted in between the two cistrons of the pNeo-LacZ bicistronic vector (Fig. 5A ▶). The resulting capped and uncapped RNAs were translated in parallel with uncapped DC-WT in the RRL system, and results show that the env 5′ UTR has the ability to promote expression of β-Gal in the context of a bicistronic RNA (Fig. 5B ▶, lanes 3–6). To assess the efficiency of the env 5′ UTR to drive translation in a bicistronic construct, the Bi-SIVΔ3′ RNA, which contains an inactive truncated version of the SIV IRES (Ohlmann et al. 2000), was translated in parallel. Taken together, the results show that the env 5′ UTR can function as an efficient IRES in the rabbit reticulocyte lysate.
FIGURE 5.
The gypsy domain upstream of the splice donor site drives translation of bicistronic RNAs. (A) Scheme of the DC-Env construct. (B) Capped (lanes 3,4), uncapped (lanes 5,6) DC-Env, uncapped (lanes 7,8) DC-WT RNA, and capped (lanes 1,2) Bi-SIVΔ3′ RNAs were translated in the RRL (10 μL) at various RNA concentration as indicated; 1μL of each assay was analyzed on a 15% SDS-PAGE subjected to autoradiography. Positions of the molecular weight markers (kD), neomycin (Neo), and β-Gal translation products are indicated.
The IRES from the gypsy env RNA is not inhibited by the L protease
The in vitro translated FMDV-L protease was added to a RRL programmed with capped bicistronic env RNAs. Addition of the enzyme to the assay inhibited cap-dependent translation of the 5′ cistron (neo) in a dose-dependent manner, with maximum inhibition being obtained at the highest dose of L protease (Fig. 6 ▶, lanes 1–4). However, effects of the enzyme on the translation of the 3′ cistron clearly differed because a four- to fivefold stimulation of β-Gal expression was observed. This indicates that the region spanning the cap site to the AUG at position 330 also contains sequences that can promote internal translation initiation in the RRL system. Taken together, these data obtained in the RRL system indicate that two non-overlapping IRES elements are present within the gypsy 5′ UTR.
FIGURE 6.
Translation of the DC-Env RNA is not affected by the FMDV-L protease. Capped DC-Env RNA (20 μg/mL) was translated in the RRL (10 μL) in the absence (lane 1) or presence (lane 2, 0.2μL; lane 3, 0.4 μL; lane 4, 0.8 μL) of in vitro translated FMDV-L protease; 1μL sample of each assay was analyzed on a 15% SDS-PAGE, subjected to autoradiography, and quantified by using a STORM 850 phosphoimager. Positions of the molecular weight markers (kD), neomycin (Neo), and β-Gal translation products are indicated. Results of the quantitation are expressed as percentage of the control (no L protease added) for both neomycin (first cistron) and β-Gal (second cistron) as indicated.
The IRESE but not the 5′ UTR of gypsy directs translation of a monocistronic RNA in 293T cells
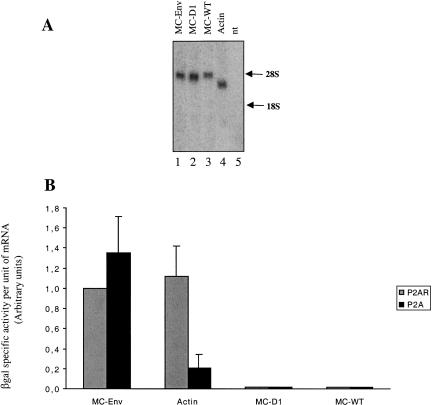
To substantiate the data obtained in the RRL system, DNA transfection experiments were performed in cultured cells. For this, human 293T cells were selected in order to use the poliovirus 2A protease as a way to cleave the initiation factor eIF4G. As previously documented, this proteolytic event inhibits cap-dependent but not IRES-driven translation (Fouillot et al. 1993; Berlioz and Darlix 1995; Berlioz et al. 1995). The monocistronic constructs containing the full-length 5′ UTR, the Δ1 deletion (named D1 thereafter), and the env 5′ UTR (pMC-WT, pMC-D1, and pMC-Env) were each transfected into 293T cells either with a plasmid encoding poliovirus protease 2A (pMLP-P2A) or with a plasmid containing the protease 2A coding sequences in a reverse orientation (negative control, pMLP-P2AR). The pβ-actin-lacZ plasmid that contains the 5′ UTR of β-actin gene directing the synthesis of the lacZ reporter gene was used as a control to monitor cap-dependent translation. Twenty-four hours after transfection, levels of mRNA were examined and quantified either by Northern blot (Fig. 7A ▶) or slot blot with a lacZ probe, and the intensity of the signal was quantified by scanning densitometry. The relative translational efficiency of each RNA was determined from the ratio between β-Gal specific activity over the concentration of lacZ mRNA (β-Gal/per unit of lacZ mRNA; Fig. 7B ▶). In the absence of P2A, the enzymatic activity of both MC-Env and β-actin-lacZ reached similar levels. Upon P2A expression, a drastic decrease of β-Gal expression from the β-actin-lacZ RNA (over fivefold decrease) was observed, whereas translation driven by the env 5′ UTR was moderately stimulated. Thus, it can be concluded that the env 5′ UTR contains a functional IRES ex vivo.
FIGURE 7.
Expression of gypsy-derived monocistronic constructs in 293T cells in presence of the poliovirus 2A protease. Constructs MC-Env, MC-D1, MC-WT, and β-actin-lacZ (Actin) were each cotransfected into 293T cells together with (P2A, black bars) or without (P2AR, grey bars) a plasmid encoding the 2A protease from poliovirus (see Materials and Methods). Cell extracts were harvested 24 h after transfection, and LacZ mRNA and β-Gal activities were measured as described in Materials and Methods. (A) Northern blot analysis of mRNA; 5 μg of total RNAs from transfected cells were separated on a 1.5% formaldehyde-agarose gels, transferred to nylon membrane, and hybridized with a 32P-lacZ probe (ClaI-ClaI fragment). The positions of 28S (4718 nt) and 18S (1874 nt) RNAs are indicated on the right. Nt indicates nontransfected cells. (B) Efficiency of translation is given as units of β-Gal specific activity per arbitrary units of lacZ mRNA. Experiments were repeated three times, and results are expressed as mean ± SE. To calibrate the data from different experiments, the value generated by the MC-Env construct cotransfected with pMLP-P2AR was set to one.
However, virtually no production of β-Gal from translation of both MC-D1 and MC-WT could be detected in 293T cells (Fig. 7B ▶), and this occurred whether the 293T cells were cotransfected with the control plasmid pMLP-P2AR or pMLP-P2A. Histochemical staining confirmed that only a few cells transfected with MC-D1 or MC-WT expressed β-Gal (data not shown). Northern blotting analysis revealed the presence of MC-Δ1 and MC-WT RNA transcripts of the expected size (Fig. 7A ▶), indicating that the lack of β-Gal expression was not due to a defect in RNA synthesis or mRNA instability. Essentially similar results were obtained with several other cell lines such as HeLa cells and hamster ovary cells (CHO), as revealed by lacZ histochemical staining (data not shown). Taken together, these data confirm the IRES activity of the 1–330 RNA segment (hereafter, named IRESE) and also show that, ex vivo, the 3′ region of the gypsy (D1) 5′ UTR harbors an RNA sequence that inhibits translation.
The complete 5′ UTR and the D1 RNA fragment of gypsy inhibit translation in SL2 Drosophila cells
The observation that the D1 RNA segment has the ability to inhibit translation in 293T cells prompted us to extend this investigation to SL2 Drosophila cells. For this, monocistronic constructs containing the full-length 5′ UTR, the D1 deletion, and the env 5′ UTR (pMC-WT, pMC-D1, and pMC-Env, respectively) were each transfected into SL2 cells (see Materials and Methods). Then, β-Gal activity was quantified, and the values obtained were expressed with respect to the concentration of lacZ transcripts as described above. To this end, SL2 cell extracts and total RNAs were collected 48 h after transfection, and the relative translational efficiency for each individual RNA tested was expressed as a ratio of β-Gal specific activity per unit of lacZ mRNA (Fig. 8 ▶). As previously observed in mammalian cell cultures, translation driven by both D1 or the complete 5′ UTR was very low compared with expression promoted by the env sequence (Fig. 8 ▶). Nevertheless, the contrast between pMC-WT, pMC-D1, and pMC-Env was not as sharp as that previously observed in mammalian cells (Fig. 7 ▶). Thus, these results indicate that the genomic 5′ UTR is functional in SL2 Drosophila cells, although this activity remains very low.
FIGURE 8.
Expression of gypsy-derived monocistronic RNAs in Drosophila SL2 cells. Construct pMC-Env, pMC-D1, and pMC-WT were each transfected into SL2 cells. Cell extracts were harvested 48 h after transfection, and the level of LacZ mRNA and β-Gal activity was assessed as described in Materials and Methods. Efficiency of translation is given as units of β-Gal specific activity per arbitrary units of lacZ mRNA. Experiments were repeated at least three times, and results are expressed as mean ± SE. To calibrate the data from different experiments, the value generated by the MC-Env construct was set to one.
The gypsy IRES Env drives translation in the context of a bicistronic RNA in cell culture
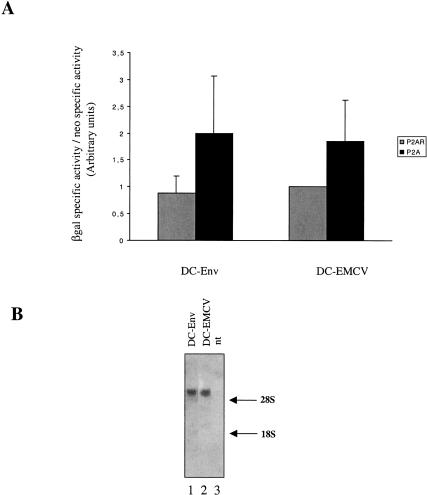
Finally, the ability of the env sequence to promote internal initiation was assayed in the context of a dicistronic construct in 293T cell culture. To this end, plasmids pDC-Env and pDC-EMCV were each transfected into 293T cells together with pMLP-P2A or the control pMLP-P2AR. Twenty-four hours after transfection, cell extracts were collected, and neomycin and β-Gal activities were determined (see Materials and Methods). Both DC-Env and DC-EMCV exhibited enhancement of the β-Gal/neo ratio in the presence of protease P2A, indicating that the two cistrons were not equally affected by expression of the poliovirus protease and that the RNA segment 1–330 was able to direct cap-independent translation initiation in a bicistronic context (Fig. 9A ▶). Northern blot analysis showed that both the DC-Env and DC-EMCV RNAs migrated at the expected size (5.6 kb), therefore ruling out the possibility that the 3′ cistron was expressed from a monocistronic RNA generated from a cryptic promoter or cryptic splice site (Fig. 9B ▶). Thus, it can be concluded that the 1–330 RNA element functions as an IRES in 293T cells.
FIGURE 9.
The gypsy Env IRES is active in the context of a bicistronic RNA. DNA constructs DC-Env and DC-EMCV were cotransfected into 293T cells together with pMLP-P2A or pMLP-P2AR DNA. Cells extracts were harvested 24 h after transfection. Neo and β-Gal activities were measured as described in Materials and Methods. (A) The histogram represents the ratio of β-Gal and neomycin specific activities measured in presence of P2A (black bars) or in presence of P2AR (grey bars). To calibrate the data from different experiments, the value generated by the DC-EMCV construct cotransfected with pMLP-P2AR was set to one. Experiments were repeated three times, and results are expressed as mean ± SE. (B) Integrity of the RNA transcripts synthesized from the dicistronic constructs. Total RNAs were isolated from cells transfected with DC-Env, DC-EMCV, or from nontransfected cells (nt); 10 μg of total RNAs were subjected to Northern blot analysis by using a 32PlacZ probe (ClaI-ClaI fragment). The positions of 28S and 18S RNA are indicated on the right.
DISCUSSION
The Drosophila melanogaster gypsy retroelement has a genetic organization similar to that of vertebrate oncoretroviruses because it contains two LTRs flanking the three canonical open reading frames gag, pol, and env. Gypsy can retrotranspose in the progeny of some strains (vertical transmission), and under defined experimental conditions, it can be horizontally transmitted between individuals (Kim et al. 1994; Song et al. 1994). Transposition of gypsy might be deleterious for the host genome, as gypsy carries a strong DNA insulator (Gdula et al. 1996) that has the ability to disrupt activity of the genes into which gypsy is integrated, by blocking the interactions of distal enhancers with the target promoter. Thus, the inability of gypsy to extensively invade the genome is conditioned by its capacity to be repressed, and yet, this downregulation of gene expression has been shown to mainly take place at the transcriptional level.
The aim of this study was to investigate whether gypsy expression might also be regulated at the translational level. Thus, the RNA sequence located between the cap site and the major splice donor (1–330) that corresponds to the 5′ UTR of the env sugenomic RNA was shown to display IRES activity in vitro. This was demonstrated by showing that this sequence can drive efficient translation in the context of a bicistronic construct even under conditions when the initiation factor eIF4G was cleaved by the L protease from FMDV (Figs. 5 ▶, 6 ▶). Because the env RNA fragment also exhibited IRES activity in 293T cells (Figs. 7 ▶, 9 ▶) it was called IRESE.
By using the rabbit reticulocyte system, we have also shown that the complete 5′ UTR (from the cap site to the authentic AUG of gag) can promote internal initiation of protein synthesis (Figs. 2 ▶, 3 ▶). By deletion analyses, an RNA sequence exhibiting features of an IRES in vitro was mapped between nucleotides 530 and 790 (Fig. 4 ▶). However, in cultured 293T cells, neither the complete 5′ UTR nor an RNA segment spanning from the splice donor to the AUG codon (named D1) could direct translation, whereas expression driven by the env and actin RNAs was very efficient (Fig. 7 ▶). Similar results were obtained in other mammalian cell lines such as HeLa and CHO cells (data not shown). This lack of protein synthesis could not be attributed to inefficient transcription or premature degradation, as Northern blot analysis revealed no alteration of the mRNA transcripts (Fig. 7 ▶). Transfection of the same monocistronic RNAs in SL2 Drosophila cultured cells showed that expression driven by the complete 5′ UTR or the D1 deletion was very low compared with expression of the env RNA, although the level of inhibition was not as drastic as that observed in 293T cells (Fig. 8 ▶).
Therefore, it is possible that the D1 RNA segment folds into very stable secondary and tertiary structures that would impede ribosome scanning and would result in virtually no translation. However, data obtained in the RRL indicate that the D1 sequence has the ability to drive translation in both mono- and bicistronic contexts (Figs. 2 ▶, 3 ▶), suggesting that the reticulocyte lysate contains factors that can relieve translation inhibition. Thus, we favor the model in which the D1 RNA segment does fold into an IRES, which is very efficient in the RRL (Figs. 2 ▶–4 ▶ ▶) but silenced or impaired in most cell types (Figs. 7 ▶, 8 ▶). A similar mechanism has been previously described for the IRES of the hepatitis A virus (HAV). Replication of HAV in cell culture is very low (Day et al. 1992; Funkhouser et al. 1999), and translation driven by the HAV IRES is almost undetectable in cultured cells of different origins (Borman et al. 1997b). However, the HAV IRES is functional in the RRL system with an activity similar to enterovirus IRESes (Borman et al. 1995). It has been shown that both viral and cellular IRES elements are regulated by ITAFs (IRES trans-acting factors) in a cell-specific manner (Belsham and Sonenberg 2000; Martinez-Salas et al. 2001; Pestova et al. 2001). Thus, translational activity of D1 most probably relies on the presence of one or several specific proteins that are highly expressed in the RRL but absent or in limiting amount in cell cultures. In the context of our data, the factors required for translation of the genomic RNA would be absent from mammalian cell lines (293T, HeLa, and CHO), present in limiting amount in SL2 Drosophila cells, and present in sufficient amount in the reticulocyte lysate.
Our data also show that expression of the Env proteins from the subgenomic RNA is driven by an IRES element that is very efficient in vitro and in cell cultures of dif-ferent origins (Figs. 5 ▶, 7 ▶, 8 ▶, 9 ▶). Interestingly, in the context of the genomic RNA, ribosomes are delivered by IRESE to the entry site at AUG330, and translation is immediately terminated at the next stop codon at position 333 (UGA333; Fig. 1 ▶). Therefore, such a genetic organization ensures that protein synthesis driven by IRESE can only take place in the context of the subgenomic RNA after removal of UGA333 by splicing.
In cultured cells, our data suggest that tight translational control is mainly exerted by the D1 3′ RNA sequence and seriously affects Gag and Gag-Pol protein production in several cell types. Interestingly, the RNA sequence that constitutes D1(nt 330–846) corresponds to the DNA region that exhibits the insulator activity of gypsy. This insulator blocks the activity of transcriptional enhancer when inserted between enhancer and promoters (Gdula et al. 1996), and is suspected to play a role in the organization of the chromatin fiber (Labrador and Corces 2002). The gypsy insulator is 430 bp long and consists of a cluster of 12 binding sites for the Su(Hw) zinc-finger protein (Geyer et al. 1988; Spana et al. 1988). Our results show that the RNA sequence corresponding to this region can also inhibit protein expression when positionned upstream of a reporter gene (Figs. 7 ▶, 8 ▶), although the mechanism by which this occurs remains to be elucidated. The fact that gypsy transposition occurs suggests that this translational repression must be alleviated under certain physiological conditions that remained to be found.
Numerous studies have demonstrated that Gypsy transposition was controlled in a tissue-specific manner and restricted to certain stages of development. However, these stringent controls were shown to be exerted at the level of RNA synthesis. The findings reported here indicate that expression of all Gypsy encoded proteins (Gag, Pol, and Env) are synthesized by use of two adjacent, non-overlapping, and independent IRESes. The first IRES is used for synthesis of the Env proteins and only becomes active upon splicing of the genomic RNA, whereas the second IRES drives synthesis of the Gag and GagPol proteins and its activity appears to be tightly regulated by unknown cellular trans-acting factors. Therefore, we propose that these peculiar features of the Gypsy retroelement provide further possibilities for the regulation of its expression at the translational level. Thus, a better understanding of these mechanisms would certainly help to know more about the regulation of Gypsy retrotransposition.
MATERIALS AND METHODS
Plasmid construction
Standard procedures were used for restriction nuclease digestion and plasmid DNA construction and purification. All numberings are with respect to the genomic RNA cap site (position +1). The gypsy RNA 5′ UTR sequences were amplified by PCR from a complete gypsy clone (accession no. M12927; kind gift of A. Bucheton, IGH, Montpellier, France), digested with NheI (PCR added restriction site) and cloned into the unique NheI site of the monocistronic construct pMLV-CB93 first digested with NheI (Berlioz and Darlix 1995). To construct dicistronic vectors, each fragment was inserted between the neo and lacZ genes of the pMLV-CB63, previously digested with NheI (pNeo-LacZ; Berlioz and Darlix 1995). The structures of the resulting DNA constructs (pMC and pDC, respectively) were verified by restriction enzyme digestion and sequencing. In the mono- and bicistronic constructs, the neo and lacZ sequences are under the control of the T7 RNA polymerase promoter for in vitro experiments, and the cytomegalovirus early promoter for expression in eucaryotic cells. pMC-WT and pDC-WT contain the gypsy 5′ UTR sequence, from position 1 to 846. pMC/DC-Δ1 (position 330–846), pMC/DC-Δ2 (position 530–846), pMC/DC-Δ3 (position 664–846), pMC/DC-Δ4 (position 768–846), pMC/DC-Δ5 (position 530–790), and pMC/DC-Δ6 (position 1–329) are derivatives of the pMC/DC-WT constructs. pDC-EMCV is the pEMCV-D260–837 previously described (Ohlmann et al. 2000). The pβ-actin-lacZ plasmid (gift of P. Savatier, Lyon, France) contains the lacZ gene under the control of the rat β-actin promoter. Plasmids pMLP-P2A and pMLP-P2AR (kind gift of N. Fouillot, Orsay, France) contain the poliovirus protease 2A coding sequences derived from poliovirus type 1 (Mahoney strain). The plasmid pMLP-P2AR contains the insert in the reverse orientation to serve as a control.
In vitro transcription
For in vitro transcription, DNA was linearized with AflIII truncating the lacZ gene (position 1,314 within pMC-WT and 2,327 within pDC-WT) except for the pEMCV-D260-837, which was linearized at the Ssp1 site at position 2252. Transcription reactions were carried out with the bacteriophage T7 RNA polymerase as previously described (Ohlmann et al. 1995). For synthesis of capped transcripts, GTP concentration was reduced to 0.48 mM and m7GpppG cap analog (New England BioLabs) added at a concentration of 1.92 mM. At the end of the incubation period, the transcripts were purified on a microspin S-400 microcolumn (Amersham Pharmacia Biotech) and precipitated with LiCl (7.5 M). The integrity of the RNAs was checked by electrophoresis on 1% agarose gels, and their concentration was measured by spectrophotometry.
In vitro translation
Capped and uncapped RNAs were translated in nuclease-treated rabbit reticulocyte lysate (Promega) in the presence of KCl (75 mM), MgCl2 (0.5 mM), 2-aminopurine (15 mM), and 20 μM each amino acid (except methionine). The mixture was incubated for 1 h at 30°C in the presence of 0.6 mCi/mL of [35S]methionine. Translation products were then separated on SDS-PAGE; the gel was dried and subjected to autoradiography for 12 h by using Biomax films (Kodak).
The in vitro synthesized L protease was prepared in the reticulocyte lysate from translation of the pMM1 clone as previously described (Ohlmann et al. 1995).
Cell culture and DNA transfection
293T cells were cultured in Dulbecco’s modified Eagle’s medium (GIBCO-Invitrogen) at 37°C with 10% newborn calf serum in a 5% CO2 atmosphere. SL-2 cells are Drosophila embryonic cells having an epithelial phenotype (Schneider 1972). SL2 cells were cultured in Schneider Drosophila medium (Invitrogen) with 10% fetal calf serum and incubated at 25°C, without CO2. Twenty-four hours prior to transfection, 293T cells were seeded at 8 × 105 cells per six-well plates. DNA transfection was performed at 70%–80% confluency by the calcium phosphate method (Chen and Okayama 1988), using 1.5 μg of plasmid DNA and 4.5 μg of either pMLP-P2A or pMLP-P2AR plasmid. SL-2 cells were transfected in suspension by the calcium phosphate procedure, by mixing 5 × 106 cells with 5 μg of plasmid DNA during 30 min. Subsequently, cells were plated into six-well plates.
Cell extracts
Cell extracts were collected 24 h (293T) or 48 h (SL2) after DNA transfection. Cells were washed twice with 1× phosphate buffer saline (PBS), either trypsinized (293T) or collected by pipetting (SL2), separated in two aliquots and pelleted by centrifugation at 600g. The first aliquot was resuspended in NP40 buffer (0.5% NP-40, 140 mM NaCl, 30 mM Tris-HCl at pH 7.5). Nuclei were removed by a 10-min centrifugation at 14,000g. Cell extracts were used as substrate for subsequent enzymatic assays. Total RNAs were extracted from the second aliquot and analyzed by Northern blot or slot blot by using a lacZ probe (see below).
Enzymatic activities
Protein concentration was determined using the Biorad protein assay reagent. The neomycin phosphotransferase activity (Neo) in cell extract was measured by [γ–32P]ATP phosphate transfer to kanamycin (Ramesh and Osborne 1991). β-Gal activity was determined spectrophotometrically. To check for linearity of the assays and to determine the relative activities of the neomycin and β-Gal in each cell extract, an internal standard curve was generated by serial dilutions of the cell extract giving the strongest Neo or β-Gal activity.
Northern and slot blot analysis
Extraction of cellular RNAs from transfected cells was performed by using the Trizol reagent (Invitrogen) according to the manufacturer’s instructions. Northen blot and slot blot analysis were performed as previously described (Ronfort et al. 1995). A 32P-labeled probe complementary to the lacZ gene (ClaI-ClaI fragment), was generated by random priming using the Random Primer DNA labeling system (Invitrogen). Signals on blots were quantified with a phosphoimager (Molecular Dynamics).
Acknowledgments
This work was supported by grants from ANRS and ARC. We thank Dr. Didier Décimo for helpful discussion, Dr. Pascal Leblanc and Dr. Déborah Prévôt for critical reading of the manuscript, Christelle Daudé for excellent technical assistance, and N. Fouillot and P. Savatier for plasmids pMLP-P2A, pMLP-P2AR and pβactin-lacZ, respectively. Thanks are also due to Alain Bucheton for providing us the gypsy clone.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.rnajournal.org/cgi/doi/10.1261/rna.5185604.
REFERENCES
- Attal, J., Theron, M.C., Taboit, F., Cajero-Juarez, M., Kann, G., Bolifraud, P., and Houdebine, L.M. 1996. The RU5 (‘R’) region from human leukaemia viruses (HTLV-1) contains an internal ribosome entry site (IRES)-like sequence. FEBS Lett. 392: 220–224. [DOI] [PubMed] [Google Scholar]
- Bayev Jr., A.A., Lyubomirskaya, N.V., Dzhumagaliev, E.B., Ananiev, E.V., Amiantova, I.G., and Ilyin, Y.V. 1984. Structural organization of transposable element mdg4 from Drosophila melanogaster and a nucleotide sequence of its long terminal repeats. Nucleic Acids Res. 12: 3707–3723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belsham, G.J. and Sonenberg, N. 2000. Picornavirus RNA translation: Roles for cellular proteins. Trends Microbiol. 8: 330–335. [DOI] [PubMed] [Google Scholar]
- Berkowitz, R., Fisher, J., and Goff, S.P. 1996. RNA packaging. Curr. Top. Microbiol. Immunol. 214: 177–218. [DOI] [PubMed] [Google Scholar]
- Berlioz, C. and Darlix, J.L. 1995. An internal ribosomal entry mechanism promotes translation of murine leukemia virus gag polyprotein precursors. J. Virol. 69: 2214–2222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berlioz, C., Torrent, C., and Darlix, J.L. 1995. An internal ribosomal entry signal in the rat VL30 region of the Harvey murine sarcoma virus leader and its use in dicistronic retroviral vectors. J. Virol. 69: 6400–6407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borman, A.M., Bailly, J.L., and Kean, K.M. 1995. Picornavirus internal ribosome entry segments: Comparison of translation efficiency and the requirements for optimal internal initiation of translation in vitro. Nucleic Acids Res. 25: 925–932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borman, A.M., Kirchweger, R., Ziegler, E., Rhoads, R.E., Skern, T., and Kean, K.M. 1997a. elF4G and its proteolytic cleavage products: Effect on initiation of protein synthesis from capped, uncapped, and IRES-containing mRNAs. RNA 3: 186–196. [PMC free article] [PubMed] [Google Scholar]
- Borman, A.M., Le Mercier, P., Girard, M., and Kean, K.M. 1997b. Comparison of picornaviral IRES-driven internal initiation of translation in cultured cells of different origins. Nucleic Acids Res. 25: 925–932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brasey, A., Lopez-Lastra, M., Ohlmann, T., Beerens, N., Berkhout, B., Darlix, J.L., and Sonenberg, N. 2003. The leader of human immunodeficiency virus type 1 genomic RNA harbors an internal ribosome entry segment that is active during the G2/M phase of the cell cycle. J. Virol. 77: 3939–3949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buck, C.B., Shen, X., Egan, M.A., Pierson, T.C., Walker, C.M., and Siliciano, R.F. 2001. The human immunodeficiency virus type 1 gag gene encodes an internal ribosome entry site. J. Virol. 75: 181–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalvet, F., Teysset, L., Terzian, C., Prud’homme, N., Santamaria, P., Bucheton, A., and Pelisson, A. 1999. Proviral amplification of the Gypsy endogenous retrovirus of Drosophila melanogaster involves env-independent invasion of the female germline. EMBO J. 18: 2659–2669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, C.A. and Okayama, H. 1988. Calcium phosphate-mediated gene transfer: A highly efficient transfection system for stably transforming cells with plasmid DNA. Biotechniques 6: 632–638. [PubMed] [Google Scholar]
- Corbin, A. and Darlix, J.L. 1996. Functions of the 5′ leader of murine leukemia virus genomic RNA in virion structure, viral replication and pathogenesis, and MLV-derived vectors. Biochimie 78: 632–638. [DOI] [PubMed] [Google Scholar]
- Day, S.P., Murphy, P., Brown, E.A., and Lemon, S.M. 1992. Mutations within the 5′ nontranslated region of hepatitis A virus RNA which enhance replication in BS-C-1 cells. J. Virol. 66: 6533–6540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deffaud, C. and Darlix, J.L. 2000. Characterization of an internal ribosomal entry segment in the 5′ leader of murine leukemia virus env RNA. J. Virol. 74: 846–850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fouillot, N., Tlouzeau, S., Rossignol, J.M., and Jean-Jean, O. 1993. Translation of the hepatitis B virus P gene by ribosomal scanning as an alternative to internal initiation. J. Virol. 67: 4886–4895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funkhouser, A.W., Schultz, D.E., Lemon, S.M., Purcell, R.H., and Emerson, S.U. 1999. Hepatitis A virus translation is rate-limiting for virus replication in MRC-5 cells. Virology 254: 268–278. [DOI] [PubMed] [Google Scholar]
- Gale Jr., M., Tan, S.L., and Katze, M.G. 2000. Translational control of viral gene expression in eukaryotes. Microbiol. Mol. Biol. Rev. 64: 239–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gdula, D.A., Gerasimova, T.I., and Corces, V.G. 1996. Genetic and molecular analysis of the gypsy chromatin insulator of Drosophila. Proc. Natl. Acad. Sci. 93: 9378–9383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geyer, P.K., Green, M.M., and Corces, V.G. 1988. Mutant gene phenotypes mediated by a Drosophila melanogaster retrotransposon require sequences homologous to mammalian enhancers. Proc. Natl. Acad. Sci. 85: 8593–8597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson, R.J. and Kaminski, A. 1995. Internal initiation of translation in eukaryotes: The picornavirus paradigm and beyond. RNA 1: 985–1000. [PMC free article] [PubMed] [Google Scholar]
- Jackson, R.J., Hunt, S.L., Gibbs, C.L., and Kaminski, A. 1994. Internal initiation of translation of picornavirus RNAs. Mol. Biol. Rep. 19: 147–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, A.I. and Belyaeva, E.S. 1991. Transposition of mobile elements gypsy (mdg4) and hobo in germ-line and somatic cells of a genetically unstable mutator strain of Drosophila melanogaster. Mol. Gen. Genet. 229: 437–444. [DOI] [PubMed] [Google Scholar]
- Kim, A.I., Belyaeva, E.S., and Aslanian, M.M. 1990. Autonomous transposition of gypsy mobile elements and genetic instability in Drosophila melanogaster. Mol. Gen. Genet. 224: 303–308. [DOI] [PubMed] [Google Scholar]
- Kim, A., Terzian, C., Santamaria, P., Pelisson, A., Purd’homme, N., and Bucheton, A. 1994. Retroviruses in invertebrates: The gypsy retrotransposon is apparently an infectious retrovirus of Drosophila melanogaster. Proc. Natl. Acad. Sci. 91: 1285–1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak, M. 1989. The scanning model for translation: an update. J. Cell. Biol. 108: 229–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ———. 1991. Structural features in eukaryotic mRNAs that modulate the initiation of translation. J. Biol. Chem. 266: 19867–19870. [PubMed] [Google Scholar]
- Labrador, M. and Corces, V.G. 2002. Setting the boundaries of chromatin domains and nuclear organization. Cell 111: 151–154. [DOI] [PubMed] [Google Scholar]
- Lopez-Lastra, M., Gabus, C., and Darlix, J.L. 1997. Characterization of an internal ribosomal entry segment within the 5′ leader of avian reticuloendotheliosis virus type A RNA and development of novel MLV-REV-based retroviral vectors. Hum. Gene Ther. 8: 1855–1865. [DOI] [PubMed] [Google Scholar]
- Lopez-Lastra, M., Ulrici, S., Gabus, C., and Darlix, J.L. 1999. Identification of an internal ribosome entry segment in the 5′ region of the mouse VL30 retrotransposon and its use in the development of retroviral vectors. J. Virol. 73: 8393–8402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marlor, R.L., Parkhurst, S.M., and Corces, V.G. 1986. The Drosophila melanogaster gypsy transposable element encodes putative gene products homologous to retroviral proteins. Mol. Cell. Biol. 6: 1129–1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Salas, E., Ramos, R., Lafuente, E., and Lopez de Quinto, S. 2001. Functional interactions in internal translation initiation directed by viral and cellular IRES elements. J. Gen. Virol. 82: 973–984. [DOI] [PubMed] [Google Scholar]
- Ohlmann, T., Rau, M., Morley, S.J., and Pain, V.M. 1995. Proteolytic cleavage of initiation factor eIF-4 γ in the reticulocyte lysate inhibits translation of capped mRNAs but enhances that of uncapped mRNAs. Nucleic Acids Res. 23: 334–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohlmann, T., Rau, M., Pain, V.M., and Morley, S.J. 1996. The C-terminal domain of eukaryotic protein synthesis initiation factor (eIF) 4G is sufficient to support cap-independent translation in the absence of eIF4E. EMBO J. 15: 1371–1382. [PMC free article] [PubMed] [Google Scholar]
- Ohlmann, T., Lopez-Lastra, M., and Darlix, J.L. 2000. An internal ribosome entry segment promotes translation of the simian immunodeficiency virus genomic RNA. J. Biol. Chem. 275: 11899–11906. [DOI] [PubMed] [Google Scholar]
- Paillart, J.C., Marquet, R., Skripkin, E., Ehresmann, C., and Ehresmann, B. 1996. Dimerization of retroviral genomic RNAs: Structural and functional implications. Biochimie 78: 639–653. [DOI] [PubMed] [Google Scholar]
- Pelisson, A., Song, S.U., Prud’homme, N., Smith, P.A., Bucheton, A., and Corces, V.G. 1994. Gypsy transposition correlates with the production of a retroviral envelope-like protein under the tissue-specific control of the Drosophila flamenco gene. EMBO J. 13: 4401–4411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pestova, T.V., Kolupaeva, V.G., Lomakin, I.B., Pilipenko, E.V., Shatsky, I.N., Agol, V.I., and Hellen, C.U. 2001. Molecular mechanisms of translation initiation in eukaryotes. Proc. Natl. Acad. Sci. 98: 7029–7036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prud’homme, N., Gans, M., Masson, M., Terzian, C., and Bucheton, A. 1995. Flamenco, a gene controlling the gypsy retrovirus of Drosophila melanogaster. Genetics 139: 697–711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramesh, N. and Osborne, W.R. 1991. Assay of neomycin phosphotransferase activity in cell extracts. Anal. Biochem. 193: 316–318. [DOI] [PubMed] [Google Scholar]
- Ronfort, C., Girod, A., Cosset, F.L., Legras, C., Nigon, V.M., Chebloune, Y., and Verdier, G. 1995. Defective retroviral endogenous RNA is efficiently transmitted by infectious particles produced on an avian retroviral vector packaging cell line. Virology 207: 271–275. [DOI] [PubMed] [Google Scholar]
- Schneider, I. 1972. Cell lines derived from late embryonic stages of Drosophila melanogaster. J. Embryol. Exp. Morphol. 27: 353–365. [PubMed] [Google Scholar]
- Smith, P.A. and Corces, V.G. 1995. The suppressor of Hairy-wing protein regulates the tissue-specific expression of the Drosophila gypsy retrotransposon. Genetics 139: 215–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song, S.U., Gerasimova, T., Kurkulos, M., Boeke, J.D., and Corces, V.G. 1994. An env-like protein encoded by a Drosophila retroelement: Evidence that gypsy is an infectious retrovirus. Genes & Dev. 8: 2046–2057. [DOI] [PubMed] [Google Scholar]
- Song, S.U., Kurkulos, M., Boeke, J.D., and Corces, V.G. 1997. Infection of the germ line by retroviral particles produced in the follicle cells: A possible mechanism for the mobilization of the gypsy retroelement of Drosophila. Development 124: 2789–2798. [DOI] [PubMed] [Google Scholar]
- Spana, C., Harrison, D.A., and Corces, V.G. 1988. The Drosophila melanogaster suppressor of Hairy-wing protein binds to specific sequences of the gypsy retrotransposon. Genes & Dev. 2 1414–1423. [DOI] [PubMed] [Google Scholar]
- Telesnitsky. 1997. Reverse transcriptase and the generation of retroviral DNA. In Retroviruses (ed. J.M. Coffin), pp. 161–204. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY. [PubMed]
- Teysset, L., Burns, J.C., Shike, H., Sullivan, B.L., Bucheton, A., and Terzian, C. 1998. A Moloney murine leukemia virus-based retroviral vector pseudotyped by the insect retroviral gypsy envelope can infect Drosophila cells. J. Virol. 72: 853–856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vagner, S., Waysbort, A., Marenda, M., Gensac, M.C., Amalric, F., and Prats, A.C. 1995. Alternative translation initiation of the Moloney murine leukemia virus mRNA controlled by internal ribosome entry involving the p57/PTB splicing factor. J. Biol. Chem. 270: 20376–20383. [DOI] [PubMed] [Google Scholar]
- Vagner, S., Galy, B., and Pyronnet, S. 2001. Irresistible IRES: Attracting the translation machinery to internal ribosome entry sites. EMBO Rep. 2: 893–898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziegler, E., Borman, A.M., Kirchweger, R., Skern, T., and Kean, K.M. 1995. Foot-and-mouth disease virus Lb proteinase can stimulate rhinovirus and enterovirus IRES-driven translation and cleave several proteins of cellular and viral origin. J. Virol. 69: 3465–3474. [DOI] [PMC free article] [PubMed] [Google Scholar]