Abstract
A large number of miRNAs have recently been discovered in plants and animals. Development of reverse genetic approaches that act to inhibit microRNA function would facilitate the study of this new class of noncoding RNA. Here we show that 2′-O-methyl oligoribonucleotides, but not 2′-deoxyoligonucleotides specifically inactivate the RNAi activity associated with miRNA–protein complexes in human cell extracts as well as in cultured human cells.
Keywords: RNA interference, miRNA, miRNP, posttranscriptional gene regulation, translational regulation, mRNA degradation
INTRODUCTION
RNA silencing is a fundamental mechanism of gene regulation that uses double-stranded RNA (dsRNA) derived 21– to 28 nucleotide (nt) small RNAs to guide mRNA degradation, control mRNA translation, or chromatin modification (for recent reviews, see Carrington and Ambros 2003; Dykxhoorn et al. 2003; Grewal and Moazed 2003; Pickford and Cogoni 2003). In the past 2 years, several hundred novel genes were identified in plants (Llave et al. 2002a; Mette et al. 2002; Park et al. 2002; Reinhart et al. 2002; Palatnik et al. 2003) and animals (Lee et al. 1993; Reinhart et al. 2000; Lagos-Quintana et al. 2001, 2002, 2003; Lau et al. 2001; Lee and Ambros 2001; Mourelatos et al. 2002; Ambros et al. 2003; Aravin et al. 2003; Brennecke et al. 2003; Dostie et al. 2003; Grad et al. 2003; Houbaviy et al. 2003; Lai et al. 2003; Lim et al. 2003a,b) that encode transcripts that contain short dsRNA hairpins. Defined 22-nt RNAs, referred to as microRNAs (miRNAs), are excised by dsRNA-specific endonucleases from the hairpin precursors (Grishok et al. 2001; Hutvágner et al. 2001; Ketting et al. 2001; Lee et al. 2002, 2003). The miRNAs are incorporated into ribonucleoprotein particles (miRNPs; Caudy et al. 2002; Hutvágner and Zamore 2002; Lee et al. 2002, 2003; Mourelatos et al. 2002).
Plant miRNAs target mRNAs containing sequence segments with high complementarity for degradation (Llave et al. 2002b; Rhoades et al. 2002; Emery et al. 2003; Palatnik et al. 2003; Tang et al. 2003) or suppress translation of partially complementary mRNAs (Aukerman and Sakai 2003; Chen 2003). Animal miRNAs appear to act predominantly as translational repressors (Lee et al. 1993; Wightman et al. 1993; Moss et al. 1997; Olsen and Ambros 1999; Reinhart et al. 2000; Slack et al. 2000; Seggerson et al. 2002; Abrahante et al. 2003; Brennecke et al. 2003; Lin et al. 2003). However, these miRNAs can also guide RNA degradation if presented an artificial substrate with a high degree of complementarity (Hutvágner & Zamore 2002; Brennecke and Cohen 2003; Brennecke et al. 2003; Zeng et al. 2003). This indicates that animal miRNPs act like small interfering RNA (siRNA)-induced silencing complexes (RISCs). Cell-based reporter assays further support the idea that RISC and miRNPs have a similar composition and that their activity is determined by the degree of complementarity to the target mRNA (Doench et al. 2003; Saxena et al. 2003; Zeng et al. 2003). Considering that non-Watson-Crick base pairs are tolerated at certain positions with respect to RISC-mediated target degradation (Jackson et al. 2003; Saxena et al. 2003), it remains a possibility that some animal miRNAs may target mRNA degradation to a certain extent.
Investigation of miRNA expression combined with genetic and molecular studies in Caenorhabditis elegans and plants have identified cell lineage-specific miRNA expression patterns, which are required to control timing of development and tissue specification (for reviews, see Slack and Ruvkun 1997; Pasquinelli and Ruvkun 2002; Ambros 2003; Carrington and Ambros 2003; Hake 2003). Recently, two miRNAs were identified that control cell death by regulating proapoptotic genes in Drosophila melanogaster (Brennecke et al. 2003; Xu et al. 2003). Deregulated expression of certain miRNAs has been linked to human proliferate diseases such as B-cell chronic lymphocytic leukemia (Calin et al. 2002; Lagos-Quintana et al. 2003) and colorectal neoplasia (Michael et al. 2003). Thus far, miRNA profiling was based on cloning of small RNAs, a technique that is limited by the availability of tissue (Pfeffer et al. 2003). The development of array technology for the systematic analysis of miRNA expression profiles may help to overcome these limitations (Krichevsky et al. 2003).
Understanding the biological function of miRNAs requires knowledge of their mRNA targets. Bioinformatic approaches have been used to predict mRNA targets, among which transcription factors and proapoptotic genes were prominent candidates (Rhoades et al. 2002; Enright et al. 2003; Stark et al. 2003). Processes such as Notch signaling, cell proliferation, morphogenesis, and axon guidance appear to be controlled by miRNA genes.
To validate the predicted miRNA targets, new and rapid methods for sequence-specific inactivation of miRNPs are needed. We adapted an antisense-based strategy to block miRNA function in cell extracts as well as cultured mammalian cells. The use of nuclease-resistant 2′-O-methyl oligoribonucleotides allowed us to sequence-specifically inactivate miRNPs in HeLa cell extracts. Transfection of 2′-O-methyl oligoribonucleotides, but not of antisense deoxyoligonucleotides into cultured mammalian cells efficiently blocked the RISC-associated miRNP activity. This demonstrates that 2′-O-methyl oligoribonucleotides are useful inhibitors of miRNAs that will be valuable reagents for the study of miRNA function.
RESULTS AND DISCUSSION
To assess the ability of modified oligonucleotides to specifically interfere with miRNA function, we used our previously described mammalian biochemical system developed for assaying RISC activity (Martinez et al. 2002). Zamore and colleagues (Hutvágner and Zamore 2002) showed that crude cytoplasmic cell lysates and eIF2C2 immunoprecipitates prepared from these lysates contain let-7 RNPs that specifically cleave let-7-complementary target RNAs. We previously reported that in HeLa cells, numerous miRNAs are expressed including several let-7 miRNA variants (Lagos-Quintana et al. 2001). To assess if other HeLa cell miRNAs are also engaged in RISC-like miRNPs we examined the cleavage of a 32P-cap-labeled substrate RNA with a complementary site to the highly expressed miR-21 (Lagos-Quintana et al. 2001; Mourelatos et al. 2002). Sequence-specific target RNA degradation was readily observed and appeared to be approximately two- to fivefold more effective than cleavage of a similar let-7 target RNA (Fig. 1A ▶, lane 1; data not shown). We therefore decided to interfere with miR-21 guided target RNA cleavage.
FIGURE 1.
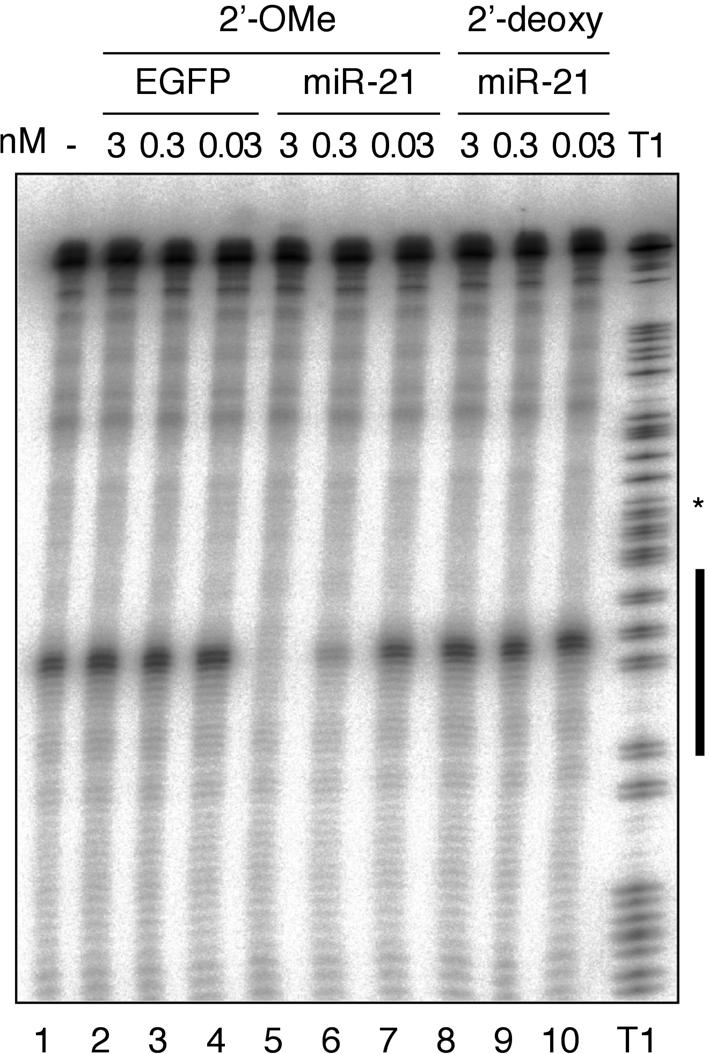
Antisense 2′-O-methyl oligoribonucleotide specifically inhibit miR-21 guided cleavage activity in HeLa cell S100 cytoplasmic extracts. The black bar to the left of the RNase T1 ladder represents the region of the target RNA complementary to miR-21. Oligonucleotides complementary to miR-21 were preincubated in S100 extracts prior to the addition of 32P-cap-labeled cleavage substrate. Cleavage bands and T1 hydrolysis bands appear as doublets after a 1-nt slipping of the T7 RNA polymerase near the middle of the transcript indicated by the asterisk.
2′-O-Methyl modification is frequently used to protect oligoribonucleotides from degradation in cell extracts or cultured cells (reviewed in Lamond and Sproat 1993; Verma and Eckstein 1998). 2′-O-Methyl oligoribonucleotides are also well known for their rapid and very stable hybridization to single-stranded RNA (Cummins et al. 1995; Majlessi et al. 1998). We chemically synthesized a 24-nt 2′-O-methyl oligoribonucleotide that contained a 3′ C7 aminolinker and was complementary to the longest form of the miR-21. The aminolinker was introduced to enable postsynthetic conjugation of nonnucleotidic residues such as biotin.
Increasing concentrations of anti-miR-21 2′-O-methyl oligoribonucleotide and a control 2′-O-methyl oligoribonucleotide cognate to an EGFP sequence were added to the S100 extract 20 min prior to the addition of 32P-cap-labeled substrate. We determined the concentration of miR-21 in the S100 extract by quantitative Northern blotting to be 50 pM (Lim et al. 2003b). The control EGFP oligonucleotide did not interfere with miR-21 cleavage even at the highest applied concentration (Fig. 1A ▶, lanes 2,3). In contrast, the activity of miR-21 was completely blocked at a concentration of only 3 nM (Fig. 1A ▶, lane 5), and a concentration of 0.3 nM showed a substantial 60–70% reduction of cleavage activity (Fig. 1 ▶, lane 6). At a concentration of 0.03 nM, the cleavage activity of miR-21 was not affected when compared to the lysate alone (Fig. 1 ▶, lanes 1,7). It was previously reported that antisense 2′-deoxyoligonucleotides were able to sequence-specifically inactivate miRNPs in microinjected D. melanogaster embryos (Boutla et al. 2003). However, at concentrations identical to those of 2′-O-methyl oligoribonucleotides, we could not detect blockage of miR-21 induced cleavage (Fig. 1A ▶, lanes 8–10). The 2′-deoxynucleotides used in this study were protected against 3′-exonucleases by the addition of three 2′-O-methyl ribonucleotide residues.
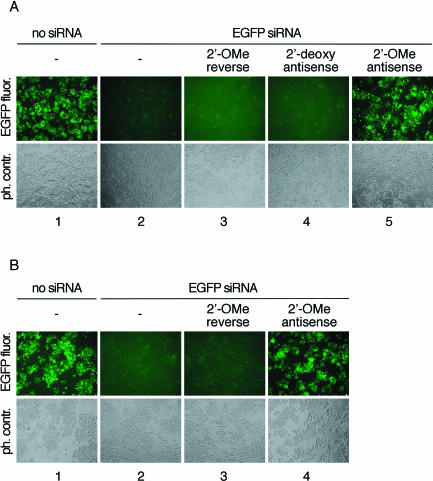
To assess the ability of 2′-O-methyl oligoribonucleotides for interfering with RISC in living cells, we first examined if siRNA duplex initiated RISC was blocked by 2′-O-methyl oligoribonucleotides complementary to the guide siRNA strand. Control or inhibitor oligonucleotides were cotransfected in equimolar amounts with preannealed siRNA duplex targeting EGFP into HeLa S3 cells stably expressing EGFP (Fig. 2A ▶). The 2′-O-methyl oligoribonucleotide complementary to the guide siRNA strand specifically blocked duplex siRNA function (Fig. 2A ▶, lane 5) whereas a 2′-O-methyl oligoribonucleotide of the reversed sequence did not interfere with silencing or compromise the viability of the treated cells (Fig. 2A ▶, lane 3). Cotransfection of the reverse 2′-O-methyl oligoribonucleotide with nonspecific, luciferase-targeting siRNA had also no effect on GFP expression (data not shown). A 2′-deoxyoligonucleotide complementary to the guide EGFP siRNA was unable to block EGFP RISC activity (Fig. 2A ▶, lane 4). Inhibition of EGFP RISC and recovery of EGFP fluorescence was also observed when the inhibitory 2′-O-methyl oligoribonucleotide was transfected 24 h after the initial introduction of the EGFP-targeting siRNA duplex (Fig. 2B ▶).
FIGURE 2.
Antisense 2′-O-methyl oligoribonucleotides block EGFP-targeting RISC in EGFP stably expressing HeLa cells. (A) EGFP expressing HeLa cells were cotransfected with EGFP siRNA duplex and the indicated single-stranded oligonucleotides. EGFP fluorescence was recorded 48 h past transfection. The antisense 2′-OMe oligoribonucleotide is complementary to the guide siRNA strand. The reversed sequence (not the reverse complement) of antisense 2′-OMe oligoribonucleotide was used as control. The guide siRNA is complementary to a segment of the EGFP mRNA. The top row shows fluorescence microscopy images recording the EGFP fluorescence; the bottom row shows phase contrast images. (B) EGFP expressing HeLa cells were first transfected with EGFP siRNA duplex followed by transfection of and the indicated single-stranded 2′-OMe oligoribonucleotides after 24 h.
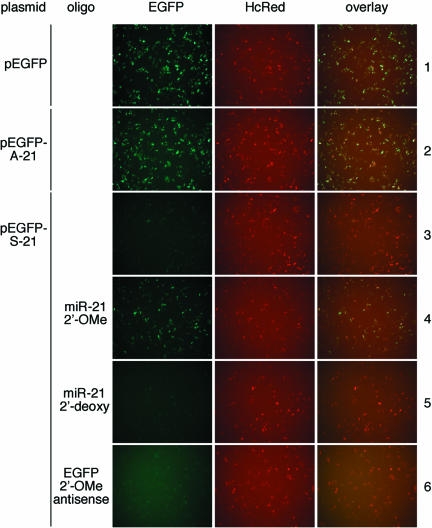
To monitor the activity of miR-21 in HeLa cells, we constructed reporter plasmids that express EGFP mRNA that contains in its 3′ UTR a 22-nt sequence complementary to miR-21 (pEGFP-S-21) or in sense orientation to miR-21 (p-EGFP-A-21). Endogenous miRNAs have previously been shown to act like siRNAs by cleaving reporter mRNAs carrying sequences perfectly complementary to miRNA (Zeng et al. 2002, 2003; Brennecke et al. 2003; Stark et al. 2003). To monitor transfection efficiency and specific interference with the EGFP indicator plasmids, the far-red fluorescent protein encoding plasmid pHcRed-C1 was cotransfected. Expression of EGFP was observed in HeLa cells transfected with pEGFP and pEGFP-A-21 (Fig 3 ▶, rows 1,2), but not from those transfected with pEGFP-S-21 (Fig. 3 ▶, row 3). However, expression of EGFP from pEGFP-S-21 was restored upon cotransfection with anti miR-21 2′-O-methyl oligoribonucleotide (Fig. 3 ▶, row 4). Consistent with our previous observation, the 2′-deoxy anti miR-21 oligonucleotide showed no effect (Fig. 3 ▶, row 5). Similarly, cotransfection of the EGFP 2′-O-methyl oligoribonucleotide in sense orientation with respect to the EGFP mRNA (or antisense to EGFP guide siRNA) had no effect (Fig. 3 ▶, row 6).
FIGURE 3.
Antisense 2′-O-methyl oligoribonucleotides interfere with endogenous miR-21 RNP cleavage in HeLa cells. HeLa cells were transfected with pHcRed and pEGFP or its derivatives, with or without inhibitory or control oligonucleotides. EGFP and HcRed protein fluorescence were excited and recorded individually by fluorescence microscopy 24 h after transfection. Coexpression of cotransfected reporter plasmids was documented by superimposing of the fluorescence images in the right panel.
In summary, we have demonstrated that RISC and miRNP complexes can be effectively and sequence-specifically inhibited with 2′-O-methyl oligoribonucleotides antisense to the guide strand positioned in the RNA silencing complex. This technique will be essential to assess the function of miRNA genes expressed in cultured cells. It is also conceivable that 2′-O-methyl oligoribonucleotides or other antisense reagents could be valuable for therapeutic intervention to control miRNA activity in disease states.
MATERIALS AND METHODS
Oligonucleotide synthesis
siRNAs and miR-21 were synthesized using 5′-silyl, 2′-ACE phosphoramidites (Dharmacon) on 0.2-μmole synthesis columns using a modified ABI 394 synthesizer (Scaringe 2001a,b). The phosphate methyl group was removed by flushing the column with 2 mL of 0.2 M 2-carbamoyl-2-cyanoethylene-1,1-dithiolate trihydrate in DMF/water (98:2 v/v) for 30 min at room temperature. The reagent was removed and the column rinsed with 10 mL water followed by 10 mL acetonitrile. The oligonucleotide was cleaved and eluted from the solid support by flushing with 1.6 mL of 40% aqueous methylamine over 2 min, collected in a screw-cap vial, and incubated for 10 min at 55°C. Subsequently, the base-treated oligonucleotide was dried down in an Eppendorf concentrator to remove methylamine and water. The residue was dissolved in sterile 2′-deprotection buffer (400 μL of 100 mM acetate-TEMED, pH 3.8, for a 0.2 μmole scale synthesis) and incubated for 30 min at 60°C to remove the 2′ ACE group. The oligoribonucleotide was precipitated from the acetate-TEMED solution by adding 24 μL 5 M NaCl and 1.2 mL of absolute ethanol. The pellets were dissolved in 500 μL of water and siRNA duplexes were formed as previously described (Elbashir et al. 2002).
2′-O-Methyl oligoribonucleotides were synthesized using 5′-DMT, 2′-O-methyl phosphoramidites (Proligo) on 1-μmole synthesis columns loaded with 3′-aminomodifier (TFA) C7 Icaa control pore glass support (Chemgenes). The aminolinker was added to also use the oligonucleotides for conjugation to amino group reactive reagents, such as biotin succinimidyl esters. The synthesis products were deprotected for 16 h at 55°C in 30% aqueous ammonia and then precipitated by the addition of 12 mL absolute 1-butanol. The full-length product was then gel-purified using a denaturing 20% polyacrylamide gel. 2′-Deoxyoligonucleotides were prepared using 0.2-μmole scale synthesis and standard DNA synthesis reagents (Proligo).
The sequences of the 2′-O-methyl oligoribonucleotides were 5′-GUCAACAUCAGUCUGAUAAGCUAL (L, 3′ aminolinker) for 2′-OMe miR-21, 5′-AAGGCAAGCUGACCCUGAAGUL for EGFP 2′-OMe antisense, 5′-UGAAGUCCCAGUCGAACGGAAL for EGFP 2′-OMe reverse; the sequence of chimeric 2′-OMe/DNA oligonucleotides was 5′-GTCAACATCAGTCTGATAAGCTAGC GL for 2′-deoxy miR-21 (underlined, 2′-OMe residues) and 5′-AAGGCAAGCTGACCCTGAAGTGCGL for EGFP 2′-deoxy antisense; the sequences of the EGFP siRNAs were GGCAAGCUGAC CCUGAAGUUT for sense and 5′-pACUUCAGGGUCAGCUU GCCUT for antisense (p, 5′-monophosphate; T, 2′-deoxythymidine).
The miR-21 cleavage substrate was prepared by PCR-based extension of the partially complementary synthetic DNA oligonucleotides 5′-GAACAATTGCTTTTACAGATGCACATATCGA GGTGAACATCACGTACGTCAACATCAGTCTGATAAGCTATC GGTTGGCAGAAGCTAT and 5′-GGCATAAAGAATTGAAGA GAGTTTTCACTGCATACGACGATTCTGTGATTTGTATTCAG CCCATATCGTTTCATAGCTTCTGCCAACCGA. The extended dsDNA was then used as template for a new PCR with primers 5′-TAATACGACTCACTATAGAACAATTGCTTTTACAG and 5′ATTTAGGTGACACTATAGGCATAAAGAATTGAAGA to introduce the T7 and SP6 promoter sequences for in vitro transcription. The PCR product was ligated into pCR2.1-TOPO (Invitrogen). Plasmids isolated from sequence-verified clones were used as templates for PCR to produce sufficient template for run-off in vitro transcription reactions using phage RNA polymerases (Elbashir et al. 2001). 32P-Cap-labeling was performed as reported (Martinez et al. 2002).
Plasmids
Plasmids pEGFP-S-21 and pEGFP-A-21 were generated by T4 DNA ligation of preannealed oligodeoxynucleotides 5′-GGCCT CAACATCAGTCTGATAAGCTAGGTACCT and 5′-GGCCAGGT ACCTAGCTTATCAGACTGATGTTGA into NotI digested pEGFPN-1 (Clontech). The plasmid pHcRed-C1 was from Clontech.
HeLa extracts and miR-21 quantification
HeLa cell extracts were prepared as described (Dignam et al. 1983); 5 × 109 cells from HeLa suspension cultures were collected by centrifugation and washed with PBS (pH 7.4). The cell pellet (approx. 15 mL) was resuspended in two times of its volume with 10 mM KCl/1.5 mM MgCl2/0.5 mM dithiothreitol/10mM HEPES-KOH (pH 7.9) and homogenized by douncing. The nuclei were then removed by centrifugation of the cell lysate at 1000 × g for 10 min. The supernatant was spun in an ultracentrifuge for 1 h at 105,000 × g to obtain the cytoplasmic S100 extract. The concentration of KCl of the S100 extract was subsequently raised to 100 mM by the addition of 1 M KCl. The extract was then supplemented with 10% glycerol and frozen in liquid nitrogen.
Two hundred eighty micrograms of total RNA were isolated from 1 mL of S100 extract using the acidic guanidinium thiocyanate–phenol–chloroform extraction method (Chomczynski and Sacchi 1987). A calibration curve for miR-21 Northern signals was produced by loading increasing amounts (10 to 30,000 pg) of synthetically made miR-21 (Lim et al. 2003b). Northern blot analysis was performed as described using 30 μg of total RNA per well (Lagos-Quintana et al. 2001).
In vitro miRNA cleavage and inhibition assay
2′-O-Methyl oligoribonucleotides or 2′-deoxyoligonucleotides were preincubated with HeLa S100 at 30°C for 20 min prior to the addition of the cap-labeled miR-21 target RNA. The concentration of the reaction components were 5 nM target RNA, 1 mM ATP, 0.2 mM GTP, 10 U/mL RNasin (Promega) and 50% HeLa S100 extract in a final reaction volume of 25 μL. The reaction time was 1.5 h at 30°C. The reaction was stopped by addition of 200 μL of 300 mM NaCl/25 mM EDTA/20% w/v SDS/200 mM Tris HCl (pH 7.5). Subsequently, proteinase K was added to a final concentration of 0.6 mg/mL and the sample was incubated for 15 min at 65°C. After phenol/chloroform extraction, the RNA was ethanol-precipitated and separated on a 6% denaturing polyacrylamide gel. Radioactivity was detected by phosphorimaging.
Cell culture and transfection
HeLa S3 and HeLa S3/GFP were grown in 5% CO2 at 37°C in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum (FBS), 100 unit/mL penicillin, and 100 μg/mL streptomycin. One day before transfection, 105 cells were plated in 500 μL DMEM containing 10% FBS per well of a 24-well plate. Plasmid and plasmid/oligonucleotide transfection was carried out with Lipofectamine2000 (Invitrogen). pEGFP (0.2 μg) or its derivatives were cotransfected with 0.3 μg pHcRed with or without 10 pmole of 2′-O-methyl oligoribonucleotide or 10 pmole of 2′-deoxyoligonucleotide per well. For siRNA transfection, 20 pmole of siRNA duplex and 60 pmole of 2′-O-methyl oligoribonucleotide or 60 pmole of 2′-deoxyoligonucleotide were used per well. Fluorescent cell images were recorded on a Zeiss Axiovert 200 inverted fluorescence microscope (Plan-Apochromat 10×/0.45) equipped with Chroma Technology Corp. filter sets 41001 (EGFP) and 41002c (HcRed) and AxioVision 3.1 software.
Acknowledgments
We thank P.Y. (Jeff) Chen and Jutta Meyer for expert technical assistance, R. Lührmann (MPIBPC, Göttingen, Germany) for HeLa cell cytoplasmic extracts, and J. Martinez for discussion. G.M. was supported by fellowship ATLF-219-2003 of the European Molecular Biology Organization. This work was also supported by NIH Grant R01 GM068476-01.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.rnajournal.org/cgi/doi/10.1261/rna.5235104.
REFERENCES
- Abrahante, J.E., Daul, A.L., Li, M., Volk, M.L., Tennessen, J.M., Miller, E.A., and Rougvie, A.E. 2003. The Caenorhabditis elegans hunchback-like gene lin-57/hbl-1 controls developmental time and is regulated by microRNAs. Dev. Cell 4: 625–637. [DOI] [PubMed] [Google Scholar]
- Ambros, V. 2003. MicroRNA pathways in flies and worms: Growth, death, fat, stress, and timing. Cell 113: 673–676. [DOI] [PubMed] [Google Scholar]
- Ambros, V., Lee, R.C., Lavanway, A., Williams, P.T., and Jewell, D. 2003. MicroRNAs and other tiny endogenous RNAs in C. elegans. Curr. Biol. 13: 807–818. [DOI] [PubMed] [Google Scholar]
- Aravin, A.A., Lagos-Quintana, M., Yalcin, A., Zavolan, M., Marks, D., Snyder, B., Gaasterland, T., Meyer, J., and Tuschl, T. 2003. The small RNA profile during Drosophila melanogaster development. Dev. Cell 5: 337–350. [DOI] [PubMed] [Google Scholar]
- Aukerman, M.J. and Sakai, H. 2003. Regulation of flowering time and floral organ identity by a microRNA and its APETALA2-like target genes. Plant Cell 15: 2730–2741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boutla, A., Delidakis, C., and Tabler, M. 2003. Developmental defects by antisense-mediated inactivation of microRNAs 2 and 13 in Drosophila and the identification of putative target genes. Nucleic Acids Res. 31: 4973–4980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennecke, J. and Cohen, S.M. 2003. Towards a complete description of the microRNA complement of animal genomes. Genome Biol. 4: 228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennecke, J., Hipfner, D.R., Stark, A., Russell, R.B., and Cohen, S.M. 2003. bantam encodes a developmentally regulated microRNA that controls cell proliferation and regulates the proapoptotic gene hid in Drosophila. Cell 113: 25–36. [DOI] [PubMed] [Google Scholar]
- Calin, G.A., Dumitru, C.D., Shimizu, M., Bichi, R., Zupo, S., Noch, E., Aldler, H., Rattan, S., Keating, M., Rai, K., et al. 2002. Frequent deletions and down-regulation of microRNA genes miR-15 and miR-16 at 13q14 in chronic lymphocytic leukemia. Proc. Natl. Acad. Sci. 99: 15524–15529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrington, J.C. and Ambros, V. 2003. Role of microRNAs in plant and animal development. Science 301: 336–338. [DOI] [PubMed] [Google Scholar]
- Caudy, A.A., Myers, M., Hannon, G.J., and Hammond, S.M. 2002. Fragile X-related protein and VIG associate with the RNA interference machinery. Genes & Dev. 16: 2491–2496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, X. 2003. A microRNA as a translational repressor of APETALA2 in Arabidopsis flower development. Science 11: 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chomczynski, P. and Sacchi, N. 1987. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal. Biochem. 162: 156–159. [DOI] [PubMed] [Google Scholar]
- Cummins, L.L., Owens, S.R., Risen, L.M., Lesnik, E.A., Freier, S.M., McGee, D., Guinosso, C.J., and Cook, P.D. 1995. Characterization of fully 2′-modified oligoribonucleotide hetero- and homoduplex hybridization and nuclease sensitivity. Nucleic Acids Res. 23: 2019–2024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dignam, J.D., Lebovitz, R.M., and Roeder, R.G. 1983. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 11: 1475–1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doench, J.G., Petersen, C.P., and Sharp, P.A. 2003. siRNAs can function as miRNAs. Genes & Dev. 17: 438–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dostie, J., Mourelatos, Z., Yang, M., Sharma, A., and Dreyfuss, G. 2003. Numerous microRNPs in neuronal cells containing novel microRNAs. RNA 9: 180–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dykxhoorn, D.M., Novina, C.D., and Sharp, P.A. 2003. Killing the messenger: Short RNAs that silence gene expression. Nat. Rev. Mol. Cell Biol. 4: 457–467. [DOI] [PubMed] [Google Scholar]
- Elbashir, S.M., Martinez, J., Patkaniowska, A., Lendeckel, W., and Tuschl, T. 2001. Functional anatomy of siRNAs for mediating efficient RNAi in Drosophila melanogaster embryo lysate. EMBO J. 20: 6877–6888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbashir, S.M., Harborth, J., Weber, K., and Tuschl, T. 2002. Analysis of gene function in somatic mammalian cells using small interfering RNAs. Methods 26: 199–213. [DOI] [PubMed] [Google Scholar]
- Emery, J.F., Floyd, S.K., Alvarez, J., Eshed, Y., Hawker, N.P., Izhaki, A., Baum, S.F., and Bowman, J.L. 2003. Radial patterning of Arabidopsis shoots by class III HD-ZIP and KANADI genes. Curr. Biol. 13: 1768–1774. [DOI] [PubMed] [Google Scholar]
- Enright, A.J., John, B., Gaul, U., Tuschl, T., Sander, C., and Marks, D. 2003. MicroRNA targets in Drosophila melanogaster. Genome Biol. 5: R1.1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grad, Y., Aach, J., Hayes, G.D., Reinhart, B.J., Church, G.M., Ruvkun, G., and Kim, J. 2003. Computational and experimental identification of C. elegans microRNAs. Mol. Cell 11: 1253–1263. [DOI] [PubMed] [Google Scholar]
- Grewal, S.I. and Moazed, D. 2003. Heterochromatin and epigenetic control of gene expression. Science 301: 798–802. [DOI] [PubMed] [Google Scholar]
- Grishok, A., Pasquinelli, A.E., Conte, D., Li, N., Parrish, S., Ha, I., Baillie, D.L., Fire, A., Ruvkun, G., and Mello, C.C. 2001. Genes and mechanisms related to RNA interference regulate expression of the small temporal RNAs that control C. elegans developmental timing. Cell 106: 23–34. [DOI] [PubMed] [Google Scholar]
- Hake, S. 2003. MicroRNAs: A role in plant development. Curr. Biol. 13: R851–R852. [DOI] [PubMed] [Google Scholar]
- Houbaviy, H.B., Murray, M.F., and Sharp, P.A. 2003. Embryonic stem cell-specific MicroRNAs. Dev. Cell 5: 351–358. [DOI] [PubMed] [Google Scholar]
- Hutvágner, G. and Zamore, P.D. 2002. A microRNA in a multiple-turnover RNAi enzyme complex. Science 297: 2056–2060. [DOI] [PubMed] [Google Scholar]
- Hutvágner, G., McLachlan, J., Bálint, É., Tuschl, T., and Zamore, P.D. 2001. A cellular function for the RNA interference enzyme Dicer in small temporal RNA maturation. Science 93: 834–838. [DOI] [PubMed] [Google Scholar]
- Jackson, A.L., Bartz, S.R., Schelter, J., Kobayashi, S.V., Burchard, J., Mao, M., Li, B., Cavet, G., and Linsley, P.S. 2003. Expression profiling reveals off-target gene regulation by RNAi. Nat. Biotechnol. 21: 635–637. [DOI] [PubMed] [Google Scholar]
- Ketting, R.F., Fischer, S.E., Bernstein, E., Sijen, T., Hannon, G.J., and Plasterk, R.H. 2001. Dicer functions in RNA interference and in synthesis of small RNA involved in developmental timing in C. elegans. Genes & Dev. 15: 2654–2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krichevsky, A.M., King, K.S., Donahue, C.P., Khrapko, K., and Kosik, K.S. 2003. A microRNA array reveals extensive regulation of microRNAs during brain development. RNA 9: 1274–1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagos-Quintana, M., Rauhut, R., Lendeckel, W., and Tuschl, T. 2001. Identification of novel genes coding for small expressed RNAs. Science 294: 853–858. [DOI] [PubMed] [Google Scholar]
- Lagos-Quintana, M., Rauhut, R., Yalcin, A., Meyer, J., Lendeckel, W., and Tuschl, T. 2002. Identification of tissue-specific microRNAs from mouse. Curr. Biol. 12: 735–739. [DOI] [PubMed] [Google Scholar]
- Lagos-Quintana, M., Rauhut, R., Meyer, J., Borkhardt, A., and Tuschl, T. 2003. New microRNAs from mouse and human. RNA 9: 175–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai, E.C., Tomancak, P., Williams, R.W., and Rubin, G.M. 2003. Computational identification of Drosophila microRNA genes. Genome Biol. 4: R42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamond, A.I. and Sproat, B.S. 1993. Antisense oligonucleotides made of 2′-O-alkylRNA: Their properties and applications in RNA biochemistry. FEBS Lett. 325: 123–127. [DOI] [PubMed] [Google Scholar]
- Lau, N.C., Lim, L.P., Weinstein, E.G., and Bartel, D.P. 2001. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science 294: 858–862. [DOI] [PubMed] [Google Scholar]
- Lee, R.C. and Ambros, V. 2001. An extensive class of small RNAs in Caenorhabditis elegans. Science 294: 862–864. [DOI] [PubMed] [Google Scholar]
- Lee, R.C., Feinbaum, R.L., and Ambros, V. 1993. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 75: 843–854. [DOI] [PubMed] [Google Scholar]
- Lee, Y., Jeon, K., Lee, J.T., Kim, S., and Kim, V.N. 2002. MicroRNA maturation: Stepwise processing and subcellular localization. EMBO J. 21: 4663–4670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, Y., Ahn, C., Han, J., Choi, H., Kim, J., Yim, J., Lee, J., Provost, P., Radmark, O., Kim, S., et al. 2003. The nuclear RNase III Drosha initiates microRNA processing. Nature 425: 415–419. [DOI] [PubMed] [Google Scholar]
- Lim, L.P., Glasner, M.E., Yekta, S., Burge, C.B., and Bartel, D.P. 2003a. Vertebrate microRNA genes. Science 299: 1540. [DOI] [PubMed] [Google Scholar]
- Lim, L.P., Lau, N.C., Weinstein, E.G., Abdelhakim, A., Yekta, S., Rhoades, M.W., Burge, C.B., and Bartel, D.P. 2003b. The microRNAs of Caenorhabditis elegans. Genes & Dev. 17: 991–1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, S.Y., Johnson, S.M., Abraham, M., Vella, M.C., Pasquinelli, A., Gamberi, C., Gottlieb, E., and Slack, F.J. 2003. The C. elegans hunchback homolog, hbl-1, controls temporal patterning and is a probable microRNA target. Dev. Cell 4: 639–650. [DOI] [PubMed] [Google Scholar]
- Llave, C., Kasschau, K.D., Rector, M.A., and Carrington, J.C. 2002a. Endogenous and silencing-associated small RNAs in plants. Plant Cell 14: 1605–1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llave, C., Xie, Z., Kasschau, K.D., and Carrington, J.C. 2002b. Cleavage of Scarecrow-like mRNA targets directed by a class of Arabidopsis miRNA. Science 297: 2053–2056. [DOI] [PubMed] [Google Scholar]
- Majlessi, M., Nelson, N.C., and Becker, M.M. 1998. Advantages of 2′-O-methyl oligoribonucleotide probes for detecting RNA targets. Nucleic Acids Res. 26: 2224–2229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez, J., Patkaniowska, A., Urlaub, H., Lührmann, R., and Tuschl, T. 2002. Single-stranded antisense siRNAs guide target RNA cleavage in RNAi. Cell 110: 563–574. [DOI] [PubMed] [Google Scholar]
- Mette, M.F., van der Winden, J., Matzke, M., and Matzke, A.J. 2002. Short RNAs can identify new candidate transposable element families in Arabidopsis. Plant Physiol. 130: 6–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michael, M.Z., O’Connor, S.M., van Holst Pellekaan, N.G., Young, G.P., and James, R.J. 2003. Reduced accumulation of specific microRNAs in colorectal neoplasia. Mol. Cancer Res. 1: 882–891. [PubMed] [Google Scholar]
- Moss, E.G., Lee, R.C., and Ambros, V. 1997. The cold shock domain protein LIN-28 controls developmental timing in C. elegans and is regulated by the lin-4 RNA. Cell 88: 637–646. [DOI] [PubMed] [Google Scholar]
- Mourelatos, Z., Dostie, J., Paushkin, S., Sharma, A., Charroux, B., Abel, L., Rappsilber, J., Mann, M., and Dreyfuss, G. 2002. miRNPs: A novel class of ribonucleoproteins containing numerous microRNAs. Genes & Dev. 16: 720–728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen, P.H. and Ambros, V. 1999. The lin-4 regulatory RNA controls developmental timing in Caenorhabditis elegans by blocking LIN-14 protein synthesis after the initiation of translation. Dev. Biol. 216: 671–680. [DOI] [PubMed] [Google Scholar]
- Palatnik, J.F., Allen, E., Wu, X., Schommer, C., Schwab, R., Carrington, J.C., and Weigel, D. 2003. Control of leaf morphogenesis by microRNAs. Nature 425: 257–263. [DOI] [PubMed] [Google Scholar]
- Park, W., Li, J., Song, R., Messing, J., and Chen, X. 2002. CARPEL FACTORY, a Dicer homolog, and HEN1, a novel protein, act in microRNA metabolism in Arabidopsis thaliana. Curr. Biol. 12: 1484–1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasquinelli, A.E. and Ruvkun, G. 2002. Control and developmental timing by microRNAs and their targets. Annu. Rev. Cell Dev. Biol. 18: 495–513. [DOI] [PubMed] [Google Scholar]
- Pfeffer, S., Lagos-Quintana, M., and Tuschl, T. 2003. Cloning of small RNA molecules. In: Current protocols in molecular biology (eds. F.M. Ausubel et al.), pp. 26.24.21–26.24.16. John Wiley and Sons, New York. [DOI] [PubMed]
- Pickford, A.S. and Cogoni, C. 2003. RNA-mediated gene silencing. Cell. Mol. Life Sci. 60: 871–882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhart, B.J., Slack, F.J., Basson, M., Pasquinelli, A.E., Bettinger, J.C., Rougvie, A.E., Horvitz, H.R., and Ruvkun, G. 2000. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature 403: 901–906. [DOI] [PubMed] [Google Scholar]
- Reinhart, B.J., Weinstein, E.G., Rhoades, M.W., Bartel, B., and Bartel, D.P. 2002. MicroRNAs in plants. Genes & Dev. 16: 1616–1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhoades, M., Reinhart, B., Lim, L., Burge, C., Bartel, B., and Bartel, D. 2002. Prediction of plant microRNA targets. Cell 110: 513–520. [DOI] [PubMed] [Google Scholar]
- Saxena, S., Jonsson, Z.O., and Dutta, A. 2003. Small RNAs with imperfect match to endogenous mRNA repress translation: implications for off-target activity of siRNA in mammalian cells. J. Biol. Chem. 278: 44312–44319. [DOI] [PubMed] [Google Scholar]
- Scaringe, S.A. 2001a. Advanced 5′-silyl-2′-orthoester approach to RNA oligonucleotide synthesis. Methods Enzymol. 317: 3–18. [DOI] [PubMed] [Google Scholar]
- ———. 2001b. RNA oligonucleotide synthesis via 5′-silyl-2′-orthoester chemistry. Methods 23: 206–217. [DOI] [PubMed] [Google Scholar]
- Seggerson, K., Tang, L., and Moss, E.G. 2002. Two genetic circuits repress the Caenorhabditis elegans heterochronic gene lin-28 after translation initiation. Dev. Biol. 243: 215–225. [DOI] [PubMed] [Google Scholar]
- Slack, F. and Ruvkun, G. 1997. Temporal pattern formation by heterochronic genes. Annu. Rev. Genet. 31: 611–634. [DOI] [PubMed] [Google Scholar]
- Slack, F.J., Basson, M., Liu, Z., Ambros, V., Horvitz, H.R., and Ruvkun, G. 2000. The lin-41 RBCC gene acts in the C. elegans heterochronic pathway between the let-7 regulatory RNA and the LIN-29 transcription factor. Mol. Cell 5: 659–669. [DOI] [PubMed] [Google Scholar]
- Stark, A., Brennecke, J., Russell, R.B., and Cohen, S.M. 2003. Identification of Drosophila microRNA targets. PLoS Biol. 1: 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang, G., Reinhart, B.J., Bartel, D.P., and Zamore, P.D. 2003. A biochemical framework for RNA silencing in plants. Genes & Dev. 17: 49–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma, S. and Eckstein, F. 1998. Modified oligonucleotides: Synthesis and strategy for users. Annu. Rev. Biochem. 67: 99–134. [DOI] [PubMed] [Google Scholar]
- Wightman, B., Ha, I., and Ruvkun, G. 1993. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell 75: 855–862. [DOI] [PubMed] [Google Scholar]
- Xu, P., Vernooy, S.Y., Guo, M., and Hay, B.A. 2003. The Drosophila microRNA mir-14 suppresses cell death and is required for normal fat metabolism. Curr. Biol. 13: 790–795. [DOI] [PubMed] [Google Scholar]
- Zeng, Y., Wagner, E.J., and Cullen, B.R. 2002. Both natural and designed microRNAs can inhibit the expression of cognate mRNAs when expressed in human cells. Mol. Cell 9: 1327–1333. [DOI] [PubMed] [Google Scholar]
- Zeng, Y., Yi, R., and Cullen, B.R. 2003. MicroRNAs and small interfering RNAs can inhibit mRNA expression by similar mechanisms. Proc. Natl. Acad. Sci. 100: 9779–9784. [DOI] [PMC free article] [PubMed] [Google Scholar]