Abstract
cDNA expression libraries displayed on lambda phage have been successfully employed to identify partners involved in antibody–antigen, protein– protein and DNA–protein interactions and represent a novel approach to functional genomics. However, as in all other cDNA expression libraries based on fusion to a carrier polypeptide, a major issue of this system is the absence of control over the translation frame of the cDNA. As a consequence, a large number of clones will contain lambda D/cDNA fusions, resulting in the foreign sequence being translated on alternative reading frames. Thus, many phage will not display natural proteins, but could be selected, as they mimic the binding properties of the real ligand, and will hence interfere with the selection outcome. Here we describe a novel lambda vector for display of exogenous peptides at the C-terminus of the capsid D protein. In this vector, translation of fusion peptides in the correct reading frame allows efficient in vivo biotinylation of the chimeric phage during amplification. Using this vector system we constructed three libraries from human hepatoma cells, mouse hepatocytic MMH cells and from human brain. Clones containing open reading frames (ORFs) were rapidly selected by streptavidin affinity chromatography, leading to biological repertoires highly enriched in natural polypeptides. We compared the selection outcome of two independent experiments performed using an anti-GAP-43 monoclonal antibody on the human brain cDNA library before and after ORF enrichment. A significant increase in the efficiency of identification of natural target peptides with very little background of false-positive clones was observed in the latter case.
INTRODUCTION
Building a comprehensive map of protein–protein or nucleic acid–protein interactions is a major target in post-genomic research, and in the last few years a number of different strategies have been adopted for the rapid identification of potential interaction partners (1–3).
Affinity selectable biological repertoires have been successfully exploited to identify ligands for several ligates (4,5). Filamentous phage M13/fd has been proven to be the vector of choice to generate small peptide libraries or display specialized repertoires where variability is confined to a few amino acids in the context of a fixed scaffold (i.e. antibody libraries). In contrast, M13 display of cDNA libraries has met with only limited success, presumably due to some peculiar biological features of this phage, such as the requirement for the fusion products to be secreted prior to phage assembly (6). This requirement may introduce a bias during phage production because of inefficient recombinant protein translocation (7), which in turn would lead to under representation, or even the absence, of many polypeptides in the library. Thus, the M13 phage is not the ideal presentation vehicle for complex repertoires from natural sources, such as cDNA libraries.
As an alternative to filamentous phage a few laboratories, including our own, have chosen lytic phage as display vectors for exogenous proteins. With these vectors, encapsidation of the fusion protein is an intracellular event, thus making assembly of chimeric phage a less demanding process. Both T7 and lambda have been reported as suitable systems to expose large polypeptides of different nature (8–15).
We previously described the construction and affinity selection of cDNA expression libraries from viruses and bacteria, as well as complex repertoires from mammalian tissues or whole organisms, which were displayed on the lambda surface as fusion to the C-terminus of the D protein (16,17). Even though these libraries can be successfully surveyed with mono or polyclonal antibodies and even with proteins as baits (15), a large number of clones representing peptide mimics that specifically bind to the selector molecules were often isolated, and in some cases overcame selection of the natural ligand (17). These clones are generated by fusions of gene fragments to the D protein in a different frame from the correct one, but eventually give rise to short additions to the C-terminus of the D protein.
To solve this problem, we have generated a new lambda display vector where the sequence for a 13 amino acid long peptide representing the target for the biotinylating BirA enzyme is engineered downstream of the cloning site. In this way, only cDNA fragments in-frame with both D and tag sequences will lead to the corresponding chimeric phage being biotinylated in Escherichia coli strains containing the BirA activity. We generated three libraries from mouse hepatic cells, human hepatoma and human brain, and demonstrated efficient separation of open reading frame (ORF)-encoding phage from non-ORF clones by affinity selection on streptavidin (SA) beads. ORF-enriched libraries were shown to perform much better than the untagged libraries in selection experiments.
MATERIALS AND METHODS
λD-bio vector construction
The DNA sequence coding for the 13 amino acid peptide LNDIFEAQKIEWH was inserted at the 3′ of the lambda D gene in plasmid λ171 (17), by ligation of the following annealed oligos: OL197, 5′-CTAGTTTTTAATTGCGGCCGCGTGGTTCAGGCCTGAACGACATCTCGAAGCTCAGAAAATCGAATGGCACTAATCGGCCGC-3′; OL198, 5′-GGCCGATTAGTGCCATTCGATTTTCTGAGCTTCGAAGATGTCGTTAGGCCTGAACCACGCGGCCGCAATTAAAAA-3′ to the SpeI and NotI digested vector. The resulting vector (p171bio1) was digested with RsrII and SpeI restriction enzymes, and then used for inserting an amber codon upstream of the SpeI restriction site, by ligation with the following annealed oligos: OL3, 5′-GACCGCGTTTGCCGGAACGGCAATCAGCATCGTTTAGA-3′; OL4, 5′-CTAGTCTAAACGATGCTGATTGCCGTTCCGGCAAACGCG-3′.
The lambda D gene was then amplified using a primer annealing on the 5′ of the D gene, and containing the restriction site NcoI (OL5, 5′-CACATCCCATGGGCACGAGCAAAGAAACCTTTACCCAT-3′) and a primer annealing on the DNA sequence coding for the 13 amino acid peptide, downstream of the D gene, and containing the restriction sites XbaI and EcoRI (OL6, 5′-GCGGAATTCTAGATCATCTTAGTGCCATTCGATTTTCTGAGC-3′). The PCR product was cut with NcoI and EcoRI and cloned into a p171 backbone, digested with NcoI and EcoRI. The lambda D gene in the resulting vector (p171bio3) encodes for a protein that differs from the wild-type only for an additional Gly residue after the initial Met.
For the construction of a new lambda vector (λD-bio), the p171bio3 plasmid region containing lacIq and lambda D genes was amplified using a primer annealing on lacIq promoter and containing the restriction sites PacI and SfiI (OL7, 5′-CCTTAATTAAGTCTACGGCCCTTAAGGCCATCGAATGGCGCAAAACCTTTC-3′) and the primer OL6. The amplification product was cut with PacI and XbaI and cloned into λ171loxP– backbone vector (14), generating the λID-bio vector.
The ligated lambda vector DNA was phenol/chloroform extracted, ethanol precipitated and packaged in vitro using the Ready to Go Lambda Packaging Kit (Amersham Pharmacia Biotech) according to the provided instructions.
Finally, a PCR fragment containing the β-lactamase gene and ColE1 ori was cloned into the λID-bio vector, to generate the λD-bio vector used for the construction of the cDNA libraries. The PCR fragment was obtained by amplification of λ171 plasmid with OL8 (5′-TGCTTAATTAATGCAGCCCGGGCTCAAATTAAGCAGAAGGCCATCCT-3′) containing the PacI restriction site and OL9 (5′-CTGAAGGCC TTAAGGGCCATTTAAATAGGCTCCGCCCCCCTGACG-3′) containing the restriction site SfiI, and cloned into the λD-bio vector, digested with PacI and SfiI restriction enzymes.
Construction of the libraries
cDNA inserts of the brain library were obtained by tagged random-primed elongation and PCR amplification from a commercially available cDNA library, as previously described (17). For the construction of human hepatoma HepG2 library and mouse MMH E14 hepatocytic cells library (18,19), total RNA was extracted from the cells according to standard protocol. Then, PolyA+ RNA was purified by a commercial kit (Clontech) and used for generating double-stranded, full-length cDNAs using the SMART PCR cDNA Synthesis Kit (Clontech). Library inserts were produced by tagged random priming elongation and PCR amplification, using the HepG2 and MMH E14 cDNAs as templates. The PCR products were then subjected to three subsequent purification steps, to eliminate small inserts. They were first purified through Wizard columns (Promega), then through S400 MicroSpin columns (Pharmacia Biotech) according to the manufacturer’s instructions.
Fragments in the size range of 300–1200 bp were finally eluted from a 1.5% low melting point agarose gel. The size-selected PCR products were then digested with SpeI and NotI restriction enzymes (200 U each enzyme for 1.5 µg of DNA) and purified through QIAquick PCR purification columns (QIAGEN) according to the manufacturer’s instructions.
To evaluate the performance of the size selection procedure, equal amounts of DNA (5 ng) before and after each purification step were analyzed by PCR using the NotI primer (5′-GGAGGCTCGAGCGGCCGCAAC-3′) and the SpeI–FseI primer (5′-GCACTAGTGGCCGGCCAAC-3′). The PCR conditions were as follows: 30 s at 94°C, 30 s at 52°C, 1 min at 72°C for 20 cycles, followed by an elongation of 7 min at 72°C.
The λD-bio vector DNA was digested with SpeI and NotI restriction enzymes and purified by isopropanol precipitation. 130 ligation reactions were set up, each one containing 2 µg of vector and 44 ng of inserts, in the presence of 4000 U of ligase (New England Biolabs), and incubated at 16°C overnight. The ligation mixture was phenol, phenol/chloroform extracted, ethanol precipitated and in vitro packaged (Amersham Pharmacia Biotech). The packaging mixture was then used for infecting BB4 cells (OD600 = 2.00), plated into 100 square (23 × 23 cm) plates. Phage elution was achieved by scraping the agarose from the plates and shaking it night and day at 4°C in SM buffer (100 mM NaCl, 8.1 mM MgSO4, 50 mM Tris–HCl pH 7.5, 0.01% gelatin). Ten separate phage preparations were obtained, each one deriving from the scraping of 10 plates. Phage were PEG precipitated, and complete EDTA free protease inhibitor cocktail (Boehringer) was added, according to the provided instructions.
Streptavidin selection
SA selection of the library was performed using the CELLection Biotin Binder Kit (Dynal). Free biotin, eventually present in the library preparation, was eliminated by an overnight dialysis in SM buffer. A total of 2 × 1010 plaque forming units (p.f.u.) from each of the 10 library stocks were separately incubated in SM buffer/0.015% Tween-20 with 200 µl of Dynabeads (∼8 × 107 beads), formerly washed with SM buffer/0.1% Tween-20 (four washing steps, 5 min each). The incubation was performed at room temperature on a wheel for 90 min. The washing steps were performed in SM buffer/0.1% Tween-20 (four washings on a wheel, 15 min each), then in SM buffer (two washings of 15 min each). The bound phage were detached from the beads by incubation (30 min on a wheel at room temperature) with 1 ml of SM buffer containing 8 µl of Releasing Buffer (provided by the kit). The released phages were collected and amplified by infecting BB4 cells and plating in 50 square (23 × 23 cm) plates. The eluted phage were concentrated by PEG precipitation and titrated.
Affinity selection with mAb growth-associated protein (GAP)-43
Affinity selection with mAb GAP-43 (Sigma G-9264) was performed in microtiter plates (Nunc) as described (14). A total of 1 × 1010 p.f.u. were used for each selection. Bound phage were recovered by infection of BB4 cells (OD600 = 2.00) and plated in two square 23 × 23 cm plates. The eluted phage were concentrated by PEG precipitation, and subjected to a second round of selection.
ELISA
ELISA with affinity selected phage was performed as follows. Multiwell plates (Nunc) were coated overnight at 4°C with 100 µl of an anti-lambda phage rabbit purified IgG fraction, 1:500 dilution of a 0.17 mg/ml stock in coating buffer (50 mM NaHCO3 pH 9.6). After blocking (1× PBS, 5% milk, 0.05% Tween-20) for 2 h at 37°C, 109 p.f.u./well were incubated for 2 h at room temperature. The plates, washed in 1× PBS/0.05% Tween-20, were then incubated overnight at 4°C with anti-GAP-43 mAb diluted 1:1000 in blocking buffer. After washing, the plates were incubated for 2 h at room temperature with an anti-mouse alkaline phosphatase-conjugated antibody (diluted 1:2000, Sigma A7434). The reaction was detected by adding 100 µl/well of a 1 mg/ml solution of p-nitrophenyl phosphate in substrate buffer (10% diethanolamine, 0.5 mM MgCl2, 0.05% NaN3, adjusted to pH 9.8 with HCl). The results were recorded as the difference between the absorbance at 405 and 620 nm as determined with an automated ELISA reader (Labsystems Multiskan, Finland).
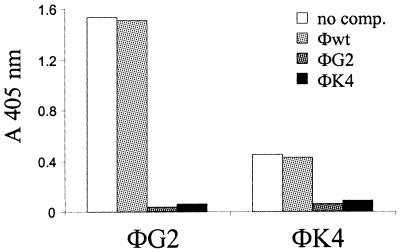
Cross-competition experiments were performed with lambda clones ΦG2, expressing GAP-43 (amino acids 131–231) and ΦK4, expressing (h-myomegalin) KIAA0477 (amino acids 863–911).
An ELISA multiwell plate (Nunc) was coated overnight at 4°C with 2.5 × 108 and 1 × 109 p.f.u. of lambda ΦG2 and ΦK4 clones, respectively, and blocked for 2 h at 37°C. Anti-GAP-43 mAb was pre-incubated overnight at 4°C with no phage, lambda wild-type vector, ΦG2 or ΦK4 lambda clones (1 × 109 p.f.u.) in blocking buffer. The anti-GAP-43 final concentration was 1:64 000 and 1:8000 for ΦG2 and ΦK4 coated phages, respectively. After washing, pre-incubated mAb anti-GAP-43 was added to the plate and incubated overnight at 4°C. After washing, the plate was incubated for 2 h at room temperature with anti-mouse alkaline phosphatase-conjugated antibody (diluted 1:2000).
Rabbit immunization
Multiple antigenic peptide (MAP) was prepared as described (20). The branched lysine core was formed by seven lysine residues giving rise to an octameric peptide structure. Each arm of the MAP had the sequence: GSGLNDIFEAQKIEWHGG.
New Zealand white female rabbits were immunized with the MAP injected at a final concentration of 0.1 mg/ml.
Immunoscreening
Phage plaques from infected BB4 cells were transferred onto a dry nitrocellulose filter (Schleicher & Schuell, Keene, NH) for 4 h at room temperature.
Immunoscreening with the anti-MAP polyclonal antibody was performed as follows: the filters were blocked for 2 h at room temperature in 1× PBS, 5% milk, 0.05% NP-40. The anti-MAP polyclonal antibody was pre-incubated for 4 h at 4°C with 4% bacterial extract in blocking buffer at a final dilution of 1:3000. After washing (with 1× PBS, 0.05% NP-40), pre-incubated antibody was added to the filters and incubated overnight at 4°C. The washed filters were then incubated for 2 h at room temperature with anti-rabbit IgG alkaline phosphatase-conjugated antibody (Sigma) diluted 1:25 000 in blocking buffer. Filters were washed and developed using as substrates p-nitro blue tetrazolium chloride (NBT) and 5-bromo-4-chloro-3-indolyl phosphate, p-toluidine salt (BCIP) (Sigma).
Immunoscreening with the anti-GAP-43 mAb was performed as described (17).
Immunoscreening with alkaline phosphatase-conjugated SA was performed as follows: the nitrocellulose filters were pre-incubated overnight at 4°C with a rabbit anti-lambda polyclonal antibody (purified IgG fraction, 1.7 µg/ml) in coating buffer and blocked (1× PBS, 3% BSA, 0.05% Tween-20) for 3 h at room temperature. Phage plaques were then transferred overnight at 4°C onto the dried pre-incubated filter. The washed filter was incubated 3 h at room temperature with alkaline phosphate-conjugated SA (1:1000) in blocking buffer (Kirkegaard & Perry Laboratories) and developed as described above.
DNA sequencing
Lambda plaques were isolated and eluted in 50 µl of SM buffer and 2 µl of chloroform. A 1 µl sample was then utilized for PCR reaction with primer 5′-CTGACGTTCTACAATCCGGC-3′ and primer 5′-TCAGCAGCTACAGTCAGAATT-3′. The PCR conditions were: 30 cycles, 94°C for 1 min, 60°C for 1 min, 72°C for 1 min and the elongation was 5 min at 72°C. The resulting amplification products were sequenced by the Ampli-Taq cycle sequencing method and run on an automated sequencer (ABI373A; Perkin Elmer).
RESULTS
Construction and characterization of the λD-bio vector
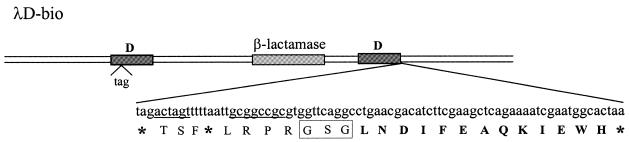
A schematic diagram of the λD-bio expression vector is shown in Figure 1. The key device of this vector is based on a 13 amino acid long sequence inserted downstream of the cDNA cloning restriction sites and spaced by a flexible linker (see Materials and Methods for construction details). This peptide sequence has been formerly described to be efficiently biotinylated in vivo by biotin holoenzyme synthetase (BirA), endogenously present in E.coli (21), henceforth referred to throughout this work as biotag. The biotag is not expressed by the empty vector, due to the presence of a TAA stop codon between the SpeI and NotI restriction sites. Similarly, insertion of cDNA fragments containing stop codons should not generate a substrate for biotinylation. In contrast, in-frame fusions of cDNAs which do not contain stop codons will allow phage exposing biotinylated D recombinant proteins to be generated in the presence of BirA. Selection of biotinylated phage by affinity chromatography on a SA matrix should provide a sub-population of phage where ORF cDNAs are enriched.
Figure 1.
Schematic representation of the λD-bio vector. The DNA sequence introduced at the 3′ end of the D gene and the corresponding amino acid sequence are shown. Amino acid residues corresponding to the biotinylation target peptide are in bold. Unique restriction sites are underlined. The box encloses the linker. * indicates termination codons.
The λD-bio display vector is based on a two-gene system where a second copy of the D gene carrying an amber mutation is present. Phage growth in an amber suppressor strain ensures the concomitant expression of both recombinant and wild-type D proteins, the latter being necessary for correct assembly, especially in those cases where the D/cDNA fusions result in large proteins. Because amber suppressors often have high efficiency (30–70%) (22), to avoid cDNA inserts carrying an amber codon giving rise to biotinylated D fusion products, an additional amber codon was inserted immediately upstream of the SpeI cloning site.
To set up the conditions for in vivo biotinylation of chimeric phage we used a clone expressing a 106 amino acid long fragment of the hepatitis C virus NS3 protein (λNS31370–1475) (16). λNS31370–1475 lysates were produced in E.coli BB4 cells in the presence or absence of a compatible plasmid expressing BirA. Equal amounts of phage from the two lysates were then affinity purified, using an excess of SA immobilized on magnetic beads via a DNA linker. After washing unbound particles, bound phage were recovered by adding a releasing buffer containing DNase, and the ratio of bound versus unbound phage was determined by titration on competent bacteria. High and comparable efficiency of selection was observed with both λNS31370–1475 preparations (∼70% of the total number of input phage), while 2000 less infectious particles were recovered by affinity selection over SA beads of phage produced in the same conditions using the λD-bio empty vector (data not shown). We concluded that the endogenous BirA activity present in E.coli BB4 cells is sufficient to modify the majority of phage particles so as to ensure efficient selection by SA chromatography. These data are consistent with the results of a comparative analysis of the same lysates for their reactivity to SA by ELISA (data not shown). Similar results were obtained by using phage clones carrying inserts of different length.
Generation of ORF-enriched λD-bio display cDNA libraries
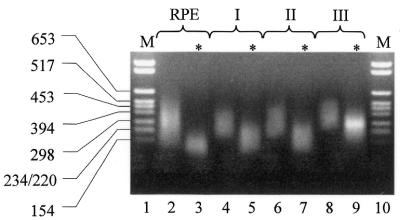
cDNA libraries from three different sources were generated. In the first case, cDNA inserts were obtained from a commercially available library from human brain, while the other two repertoires were generated by reverse transcription of poly-A-enriched mRNAs from human hepatoma HepG2 cells and from mouse MMH E14 hepatic cells (18,19). In all cases cDNA products were subjected to tagged random primed elongation and PCR amplification as previously described (23). The resulting pool of inserts was subjected to a three-step size selection procedure, with a final cut-off of 300 bp. Since small-size DNA inserts are preferentially amplified during a PCR reaction, we monitored the efficacy of each step by PCR. As shown in Figure 2, a significant reduction in the number of small fragments was detected after the third purification step. The pools of size-selected inserts were then utilized to construct libraries in the λD-bio vector.
Figure 2.
Size selection of brain cDNA library inserts. cDNA inserts obtained from tagged-random priming (RPE, lane 2), followed by Wizard column purification (lane 4), S400 MicroSpin column purification (lane 6) and agarose gel fractionation (lane 8) were analyzed by agarose gel electrophoresis. Equal amounts of DNA from each size-selection step were subjected to PCR and corresponding amplification products were analyzed (*, lanes 3, 5, 7, 9). The relevant size (bp) of the molecular weight markers (M, lanes 1, 10) are indicated on the left of the figure.
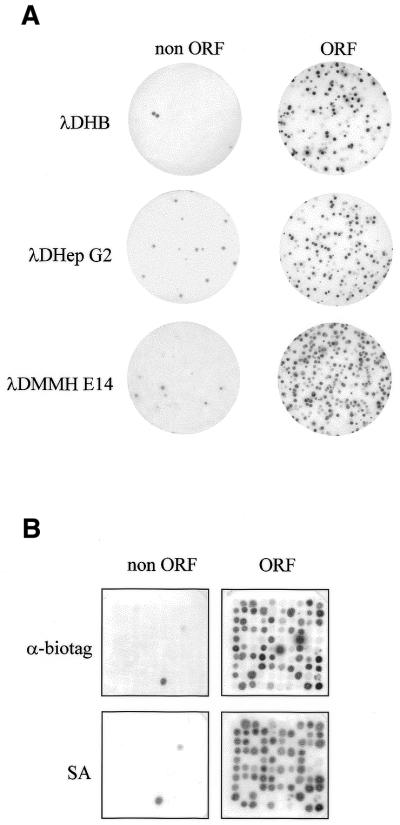
Following ligation, in vitro packaging and infection of bacteria, 3 × 107, 5 × 107 and 2.5 × 108 plaques were obtained, respectively, for the HepG2 (λDG2), MMH E14 (λDMMH) and human brain (λDHB) libraries. Amplifying inserts of several randomly chosen clones confirmed that the majority of inserts (∼90%) were above 250 bp, and only a small percentage of them were found to contain ORFs by sequence analysis (data not shown). For each library, the sub-population of biotinylated recombinant phage was affinity purified, using SA-coated beads as described above. To ensure the entire repertoire was represented, a phage input 1000-fold higher than the estimated complexity of the libraries was subjected to SA purification. The enrichment of phage expressing full ORF peptides was monitored by immunoscreening, probing phage plaques with an antibody raised in rabbits against the peptide substrate for biotinylation (see Materials and Methods). Of the clones, 80, 95 and 70% were found positive in the SA-enriched λDG2-bio, λDMMH-bio and λDHB-bio sub-libraries, respectively, while only ∼1.9–9.5% of phage clones were stained by the anti-biotag antibodies in the starting input libraries (Fig. 3A). Phage recovery after binding to SA was very efficient as judged by titration of anti-biotag-positive virus liberated from the beads upon DNase digestion of the SA-bead linker: between 25 and 50% of the biotinylated input phage was eluted from the beads (data not shown). At present we do not know why there is no quantitative rescue of biotinylated phage. DNase used to release the phage does not affect phage titer and we can also exclude an inefficient binding to the SA beads, since only approximately 10 times less modified phage are found in the population of unbound virus (data not shown).
Figure 3.
Enrichment of biotin-tagged D-protein phage after SA selection. (A) Phage deriving from the brain cDNA library (λDHB), the HepG2 library (λDHepG2) and the MMH E14 library (λDMMH E14) were probed with a polyclonal antibody recognizing the biotinylation target peptide, before (non ORF) and after (ORF) SA selection. (B) Phage deriving from the original λDHB library (non ORF) and the corresponding SA-enriched phage (ORF) were replica plated and probed with a polyclonal antibody recognizing the biotinylation target peptide (α-biotag) or with SA.
To show that ORF-encoding clones were indeed biotinylated, 100 randomly chosen clones from the λDHB library before and after SA selection were replica-plated and screened in parallel with the anti-biotag antibodies and with alkaline phosphatase-conjugated SA. The data obtained with the two assays were highly consistent, indicating that all the expression products from in-frame cDNA inserts were effectively biotinylated during amplification in BB4 cells and were purified from the bulk library by SA selection (Fig. 3B).
Affinity selection of the λDHB-bio library with a monoclonal antibody (mAb) recognizing the neural-specific protein GAP-43
We previously reported the selection of an untagged human brain cDNA library displayed on lambda using a mouse mAb specific for the GAP-43 which is expressed exclusively in the peripheral and central nervous systems (24–26). This antibody (mAb GAP-43) exhibits wide interspecies cross-reactivity, and can be used for affinity selection of the human brain library (24). In these experiments, three rounds of affinity selection yielded some phage clones encoding a 40 amino acid long GAP-43 peptide fragment, but also several other cross-reacting phage, including GAP-43 mimotopes (17).
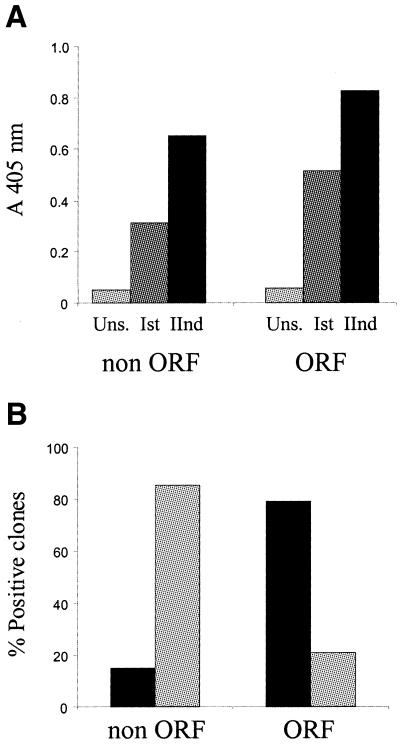
Since the goal of the λD-bio system was to improve the efficiency of selecting clones displaying authentic polypeptides, and concomitantly reduce the number of background clones encoding for peptide mimics (mimotopes) of natural ligands, we used mAb GAP-43 selection as a model system to evaluate the performance of the ORF-enriched display library. To this end, parallel selections of the λDHB and the ORF-enriched λDHB-bio repertories were carried out. Following two rounds of affinity selection with mAb GAP-43, a comparable enrichment of specific clones was observed, as monitored by a similar increase of reactivity in ELISA with phage pools from each selection step (Fig. 4A). Immunological screening of phage pools from the second round of selection confirmed the ELISA data, showing a similar frequency of positive clones in both enriched libraries (19 and 17% for the λDHB and the λDHB-bio libraries, respectively; data not shown).
Figure 4.
(A) ELISA reactivity of phage affinity selected from the original λDHB (non ORF) and ORF-enriched (ORF) libraries with mAb GAP-43. Pools of phage obtained after each round of selection–amplification (Ist, IInd) were tested, and compared with the reactivity of the unselected libraries (Uns.). Average values (A405nm) from two independent experiments were determined. (B) Frequency of proteins (black bars) and mimotopes (gray bars) among mAb GAP-43-positive clones selected from the λDHB library (non ORF) and ORF-enriched (ORF) libraries. Proteins refer to cDNA inserts with the correct reading frame and encoding domain of naturally occurring proteins. Mimotopes refer to cDNA inserts in a non-correct orientation, or with no homology with known proteins.
From each of the two pools of enriched phage, 34 clones specifically reacting with mAb GAP-43 were randomly chosen and analyzed by sequencing to classify their inserts into cDNA that encode for authentic polypeptides, or mimotopes deriving from out-of-frame protein D/cDNA fusions. The results of this analysis highlighted a strong difference in the selection outcome from the two repertoires: the frequency of mimotopes among the λDHB selected clones was 85%, while the majority of phage isolated from the ORF-enriched λDHB-bio library (79%) encoded for peptide fragments of known proteins in the correct frame (Fig. 4B).
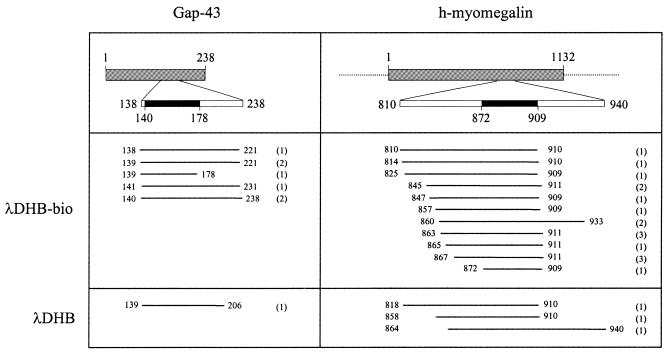
Among phage selected from the ORF-enriched library, seven displayed GAP-43 peptides (Fig. 5). The length of the isolated GAP-43 fragments varied between 40 and 99 residues, and all of them included amino acid sequences from positions 141 to 178, previously reported to contain the epitope recognized by mAb GAP-43 (17). The majority of the remaining isolates from the ORF-enriched λDHB-bio library displayed peptide regions of the KIAA0477 gene, which is highly homologous (77.8%) to the rat myomegalin (27,28), hence referred to throughout this work as human myomegalin (h-myomegalin). Also in this case, several independent clones were selected, encoding protein fragments comprised of between 38 and 101 amino acids. Finally, one phage encoded for the neurofilament triplet M protein (29) and two contained human sequences encoding for an unknown ORF (accession no. CAB70910). Specific reactivity of all isolated phage was confirmed by ELISA (Fig. 6 and data not shown), suggesting that mAb GAP-43 can cross-react with epitopes from h-myomegalin and the neurofilament triplet M protein. This hypothesis is also supported by the observation that all h-myomegalin clones share a common amino acid sequence (from amino acids 872 to 909; Fig. 5) most probably representing the epitope recognized by mAb GAP-43. To prove that GAP-43 and h-myomegalin phage contain epitopes cross-reacting for the same paratope on mAb GAP-43, we performed cross-inhibition assays between clones ΦG2 and ΦK4, which represent the GAP-43 and the h-myomegalin families, respectively. Each of the two clones reciprocally abolished recognition by mAb GAP-43 when used as competitors, while no inhibition was observed using wild-type phage (Fig. 6). These results allowed us to conclude that GAP-43 and h-myomegalin are antigenically cross-reactive. Similar conclusions could be drawn from competition experiments between phage displaying GAP-43 and the neurofilament triplet M protein or the unknown ORF (data not shown).
Figure 5.
Characterization of phage clones selected with mAb GAP-43 from the λDHB and ORF-enriched (λDHB-bio) libraries. GAP-43 and h-myomegalin are represented as gray boxes. Numbers refer to the amino acid position. Black boxes represent the minimal binding domain of mAb GAP-43, identified by the selected phage. White boxes represent the largest regions covered by the selected phage. Numbers in parentheses indicate the number of times each sequence was independently isolated.
Figure 6.
Competition between mAb GAP-43 selected clones. Inhibition of mAb GAP-43 binding was measured by ELISA. Coated phage are indicated at the bottom. Competitor phage are indicated at the top. Wild-type phage (wt) was used as control. Average values (A405nm) from two independent experiments are reported.
Among phage selected from the λDHB library, only one clone displayed a GAP-43 peptide, three phage encoded for the human homolog of myomegalin and one contained a peptide from the neurofilament triplet M protein (Fig. 5). All of the other isolates (29 out of 34) encoded short peptides, due to the presence of stop codons in the cDNA inserts. Among them, 10 phage displayed the same peptide, indicating that they were actively selected from the library.
DISCUSSION
Isolation of false-positive clones represents a major drawback of many, if not all, cDNA expression systems, as highlighted by the number of different strategies that have been developed to solve this problem (30–34). We and others have developed phage systems for efficient display of cDNA expression libraries from complex mRNA repertoires (15,17,35). One of the major advantages of these affinity selectable libraries over more conventional repertoires is that they can be rapidly surveyed by subsequent cycles of selection and amplification to identify the natural targets of monoclonal and polyclonal antibodies, proteins or DNA (14,15,17). However, this process can concomitantly lead to the isolation of phage displaying short peptides which derive from out-of-frame insertion of cDNA fragments. The frequency of isolation of these clones is often very high due to a growth advantage over phage expressing longer peptides during the amplification steps, thus significantly affecting the outcome of selection. To overcome this limitation we constructed a novel lambda vector where clones whose inserts contain an ORF encoding for a natural protein ligand can be readily separated from the bulk of the library by virtue of an affinity selectable peptide tag located at the 3′ end of the protein D/cDNA fusion sequence.
In constructing the libraries we have utilized a tagged random primed elongation and amplification procedure, which proved very efficient in generating randomly distributed cDNA fragments that can be cloned without using linkers (16). Thus, clones containing out-of-frame inserts represented the majority of the library population. In fact, taking into account only the frame and orientation, on average only one out of 18 clones should display an authentic polypeptide in-frame with both lambda D protein and the C-terminal biotag. Assuming that on average the non-coding part of mRNAs accounts for 35% of the molecule (36), the theoretical number of productive clones should be 3.6% of the phage rescued after in vitro packaging and infection. As a matter of fact, all three libraries showed a frequency of phage expressing the tag peptide similar to that predicted (2, 1.9 and 9.5% for the human brain, MMH E14 and HepG2 libraries, respectively).
A three-step procedure of cDNA size selection resulted in an increased number of clones displaying authentic translation products as it reduced the probability of cloning short DNA fragments, and possibly read through non-natural coding frames. This supposition is also supported by the 5-fold increase of in-frame fusion phage observed in the HepG2 library, where the second step of gel filtration chromatography was omitted, resulting in an increased percentage of clones containing inserts below 300 bp (data not shown).
With the aim of developing a general strategy for screening phage-displayed cDNA libraries with ligates of a different chemical nature, we chose as tag a 13 amino acid long peptide previously identified by peptide library screening as an optimal target of the E.coli BirA enzyme (21). The rationale for this choice was to ensure purification of ORF-encoding clones from the rest of the library through a rapid, efficient and highly specific affinity chromatography method, with low probability of carrying over phage displaying peptide sequences that cross-react with the affinity matrix. The panning procedure using SA as a ligate would be expected to enrich three classes of background clones: one encoding for biotag functional mimotopes arising from out-of-frame cDNA fusions, a second group of natural biotin domains and a third type of phage which are not biotinylated, but can display peptides with significant affinity for SA. However, the likelihood of selecting large numbers of such clones during phage-displayed ORF purification should be very low for a number of reasons.
First, the chosen biotag represents a consensus sequence resulting from the comparative analysis of several sequences identified by iterative cycles of selection and optimization of three peptide libraries, each consisting of billions of peptides. This consensus peptide also corresponds to the minimal synthetic BirA substrate showing biotinylation kinetics comparable to the much longer natural protein substrates (37), making it very difficult to fortuitously generate functional biotag mimotopes during construction of the lambda display cDNA libraries. Secondly, although the presence of biotin-dependent enzymes is ubiquitous in nature, biotinylation is a relatively rare modification, with between one and five biotinylated protein species being found in different organisms (38). Finally, unbiotinylated peptides bind to SA at a site overlapping with the biotin recognition site, and have a lower affinity for ligates than biotin whose dissociation constant is in the low femtomolar range (39–41). This line of reasoning, together with the well established avidin/SA technology, further supported our choice.
In E.coli with endogenous biotin ligase, biotin domain proteins expressed at low or moderate levels are typically biotinylated at ∼10–30% efficiency (42). This can be remedied by co-overexpressing the ligase. Testing the efficiency of in vivo biotinylation of a λD-biotag phage with or without additional BirA overexpressed from a compatible replicon revealed that lambda phage particles are modified even without exogenous enzyme being overexpressed in the infected cell. Furthermore, the selection efficiency of biotinylated phage compared with SA affinity resin was very high, with an enrichment factor of biotinylated versus unmodified phage of >1000-fold. We did not accurately measure the extent of biotinylation per phage particle, nonetheless, no significant difference in the recognition efficiency by SA was observed between phage preparations obtained with or without the BirA expression plasmid, suggesting that in our experimental conditions the amount of endogenous BirA is not a limiting factor for in vivo modification of most or all phage.
Pre-selection of three different libraries with SA increased the frequency of clones containing ORF cDNA inserts by a factor of 10–50 fold, with up to 95% of the output phage displaying tagged D/cDNA fusion proteins, as judged by immunological screening with an antibody raised against the biotag peptide. The specificity of this procedure and the selective in vivo biotinylation of all purified ORF-encoding phage was confirmed by the total overlap of results observed when testing the same clones for their reactivity to SA. In most cases, the relative recognition efficiency of the tested clones by the two probes was very different, meaning that each phage has its own susceptibility to biotinylation which is not necessarily determined by the level of displayed recombinant fusions.
A critical issue in the λD-bio system, especially when dealing with libraries, is the efficiency of phage recovery after SA purification. For this reason we employed magnetic beads with DNA-linked SA as an affinity matrix because the high affinity of biotin for SA would make phage elution by ligand competition almost impossible. In fact, the number of phage rescued by infection after DNase treatment was in the same order of magnitude as the input of biotinylated virus. Furthermore, even if some loss of biotinylated phage occurred during purification, we used an excess of input virus with respect to the library complexity (1000-fold) to ensure that no interesting species would be lost at this stage.
The major goal of our work was to use the λD-bio vector system to improve the efficiency of selecting natural protein ligands from complex cDNA expression repertoires. To this end we used a mAb against the GAP-43 protein to screen an equal number of phage from the human brain library before and after SA purification of ORF-encoding clones. This choice was based on previous experience where affinity selection of a similar repertoire displayed on lambda phage yielded phage displaying GAP-43 peptides, but also clones not containing GAP-43 sequences while still reacting with the antibody, which were therefore considered GAP-43 antigenic mimics (17). Consistently, mAb GAP-43 affinity selection of the λDHB library led to the isolation of a large number of mimotopes, and several clones had to be sequenced to find one GAP-43-encoding phage. In contrast, by pre-selecting ORF-displaying phage, a much higher prevalence of GAP-43 isolates was observed in the mAb GAP-43-enriched population after an equal number of affinity selection cycles.
The overall efficiency of the λDHB-bio library in yielding authentic polypeptides was very high, since 74% of the clones specifically recognized by the antibody after two selection cycles encoded protein fragments from known proteins in the correct frame and orientation. In fact, besides GAP-43-displaying phage, several other isolates encoded overlapping regions of the human homolog of rat myomegalin, a recently identified scaffold protein required for localization to the Golgi of signaling molecules involved in the cAMP-dependent pathway (43). These phage were confirmed to be specifically recognized by the mAb GAP-43 in ELISA and were able to compete for mAb recognition by the GAP-43-displaying phage. Similar behavior was displayed by a few additional selected phage displaying peptide fragments from the neurofilament triplet M protein or another unclassified human ORF. Antibody cross-reactivity and polyspecificity is not an unprecedented finding, and it has been observed since the earliest immunochemical studies, even though the structural basis and biological relevance of this phenomenon are still open questions (44–48).
Sequence analysis of seven GAP-43 isolates from the pool of λDHB-bio-enriched phage, and one isolate from the λDHB-enriched population, revealed that they corresponded to six different clones all of which shared a common sequence previously shown to contain the mAb GAP-43 epitope (17). Similarly, sequencing 20 h-myomegalin-displaying phage showed that they represented 14 different clones, all of which containing a 38 amino acid long fragment, presumably containing the protein determinant responsible for mAb GAP-43 recognition. Finally, the two neurofilament triplet M protein phage also displayed different, but overlapping peptide sequences. These findings underline the quality of the library as they give a measure of the redundancy of clones encompassing the same protein sequence present in the repertoire.
Among phage selected from the ORF-enriched λDHB-bio repertoire, a low percentage of mimotopes was still present (seven out of 34 analyzed). Four clones contained several stop codons which did not display the biotag and were not removed during the SA purification. This was our first attempt to eliminate non-biotinylated phage, yielding 70% of biotag-displaying phage after SA selection. We subsequently optimized the binding and washing steps reaching frequencies of ORF-expressing phage up to 95%.
The remaining three mimotopes displayed the biotag and contained ORFs whose length ranged from 40 to 60 amino acids. This size is around the lower limit of the library inserts, thus a more stringent size selection procedure of the fragments prior to cloning into the lambda vector would probably lead to further reduction of this type of background.
The success of a selection experiment greatly depends on the quality of the library, both in terms of complexity as well as proportion of clones expressing correct proteins. By using the λD-bio vector system a large number of irrelevant clones were eliminated by a simple purification step over SA beads, leading to the rapid and non-ambiguous isolation of the natural ligands of the selector molecule. The λD-bio system represents a general strategy for the identification of natural protein ligands to virtually any type of ligate, as this same procedure was applied to the selection of DNA-binding proteins using large DNA fragments from the regulatory region of mammalian genes as ligates (C.Cicchini, H.Ansuini, L.Amicone, T.Alonzi, A.Nicosia, R.Cortese, M.Tripodi and A.Luzzago, manuscript submitted).
Acknowledgments
ACKNOWLEDGEMENTS
We thank J. Clench for linguistic revision. The work was supported in part by the Associazione Italiana Ricerca sul Cancro (AIRC), by the Ministero dell’Università e Ricerca Scientifica (MURST) and by a CNR target project on Biotechnology.
REFERENCES
- 1.Mendelsohn A.R. and Brent,R. (1999) Protein interaction methods—toward an endgame. Science, 284, 1948–1950. [DOI] [PubMed] [Google Scholar]
- 2.Pelletier J. and Sidhu,S. (2001) Mapping protein–protein interactions with combinatorial biology methods. Curr. Opin. Biotechnol., 12, 340–347. [DOI] [PubMed] [Google Scholar]
- 3.Amstutz P., Forrer,P., Zahnd,C. and Plückthun,A. (2001) In vitro display technologies: novel developments and applications. Curr. Opin. Biotechnol., 12, 400–405. [DOI] [PubMed] [Google Scholar]
- 4.Cortese R., Monaci,P., Nicosia,A., Luzzago,A., Santini,C., Bartoli,F., Cortese,I., Fortugno,P., Galfre,G., Nicosia,A. and Felici,F. (1996) Identification of biologically active peptides using random libraries displayed on phage. Curr. Opin. Biotechnol., 7, 616–621. [DOI] [PubMed] [Google Scholar]
- 5.Dunn I.S. (1996) Phage display of proteins. Curr. Opin. Biotechnol., 7, 547–553. [DOI] [PubMed] [Google Scholar]
- 6.Model P. and Russel,M. (1988) Filamentous bacteriophage. In Calendar,R. (ed.), The Bacteriophages. Plenum Press, University of California, Berkeley, Vol. 2, pp. 375–456.
- 7.Malik P., Terry,T.D., Gowda,L.R., Petukhov,A.L., Symmons,M.F., Welsh,L.C., Marvin,D.A. and Perham,R.N. (1996) Role of capsid structure and membrane protein processing in determining the size and copy number of peptides displayed on the major coat protein of filamentous bacteriophage. J. Mol. Biol., 260, 9–21. [DOI] [PubMed] [Google Scholar]
- 8.Maruyama I.N., Maruyama,H.I. and Brenner,S. (1994) λfoo: a λ phage vector for the expression of foreign proteins. Proc. Natl Acad. Sci. USA, 91, 8273–8277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dunn I.S. (1995) Assembly of functional bacteriophage lambda virions incorporating C-terminal peptide or protein fusions with the major tail protein. J. Mol. Biol., 248, 497–506. [DOI] [PubMed] [Google Scholar]
- 10.Sternberg N. and Hoess,R.H. (1995) Display of peptide and proteins on the surface of bacteriophage λ. Proc. Natl Acad. Sci. USA, 92, 1609–1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mikawa Y.G., Maruyama,I.N. and Brenner,S. (1996) Surface display of proteins on bacteriophage λ heads. J. Mol. Biol., 262, 21–30. [DOI] [PubMed] [Google Scholar]
- 12.Kuwabara I., Maruyama,H., Mikawa,Y.G., Zuberi,R.I., Liu,F.T. and Maruyama,I.N. (1997) Efficient epitope mapping by bacteriophage λ surface display. Nat. Biotechnol., 15, 74–78. [DOI] [PubMed] [Google Scholar]
- 13.Sche P.P., McKenzie,K.M., White,J.D. and Austin,D.J. (1999) Display cloning: functional identification of natural product receptors using cDNA-phage display Chem. Biol., 6, 707–716. [DOI] [PubMed] [Google Scholar]
- 14.Zozulya S., Lioubin,M., Hill,R.J., Abram,C. and Gishizky,M.L. (1999) Mapping signal transduction pathways by phage display. Nat. Biotechnol., 17, 1193–1198. [DOI] [PubMed] [Google Scholar]
- 15.Zucconi A., Dente,L., Santonico,E., Castagnoli,L. and Cesareni,G. (2001) Selection of ligands by panning of domain libraries displayed on phage lambda reveals new potential partners of Synaptojanin 1. J. Mol. Biol., 307, 1329–1339. [DOI] [PubMed] [Google Scholar]
- 16.Santini C., Brennan,D., Mennuni,C., Hoess,H.R., Nicosia,A., Cortese,R. and Luzzago,A. (1998) Efficient display of an HCV cDNA expression library as C-terminal fusion to the capsid protein D of bacteriophage lambda. J. Mol. Biol., 282, 125–135. [DOI] [PubMed] [Google Scholar]
- 17.Santi E., Capone,S., Mennuni,C., Lahm,A., Tramontano,A., Luzzago,A. and Nicosia,A. (2000) Bacteriophage lambda display of complex cDNA libraries: a new approach to functional genomics. J. Mol. Biol., 296, 497–508. [DOI] [PubMed] [Google Scholar]
- 18.Amicone L., Spagnoli,F.M., Spath,G., Giordano,S., Tommasini,C., Bernardini,S., De Luca,V., Della Rocca,C., Weiss,M.C., Comoglio,P.M. and Tripodi,M. (1997) Transgenic expression in the liver of truncated Met blocks apoptosis and permits immortalization of hepatocytes. EMBO J., 16, 495–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Spagnoli F.M., Cicchini,C., Tripodi,M. and Weiss,M.C. (2000) Inhibition of MMH (Met murine hepatocyte) cell differentiation by TGF(beta) is abrogated by pre-treatment with the heritable differentiation effector FGF1. J. Cell Sci., 113, 3639–3647. [DOI] [PubMed] [Google Scholar]
- 20.Pessi A., Bianchi,E., Chiappinelli,L., Bonelli,F., Tougne,C., Lambert,P.H. and Del Giudice,G. (1991) Multiple antigen peptides (MAPs) as candidate vaccines against malaria. Parassitologia, 33, 79–84. [PubMed] [Google Scholar]
- 21.Schatz P.J. (1993) Use of peptide libraries to map the substrate specificity of a peptide-modifying enzyme: a 13 residues consensus peptide specifies biotinylation in Escherichia coli. Biotechnology, 11, 1138–1143. [DOI] [PubMed] [Google Scholar]
- 22.Davis B.D. (1990) Protein synthesis and localization. In Davis,B.D., Dulbecco,R.N., Eisen,H.N. and Ginsberg,H.S. (eds), Microbiology. J.B. Lippincott Company, Philadelphia, Chapter 6, pp. 105–121.
- 23.Kwong-Kwok W., Stillwell,L.C., Dockery,C.A. and Saffer,J.D. (1996) Use of tagged random hexamer amplification (TRHA) to clone and sequence minute quantities of DNA-application to a 180 kb plasmid isolated from Sphingomonas F199. Nucleic Acids Res., 24, 3778–3783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jacobson R.D., Virag,I. and Skene,J.H. (1986) A protein associated with axon growth, GAP-43, is widely distributed and developmentally regulated in rat CNS. J. Neurosci., 6, 1843–1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.De la Monte S.M., Federoff,H.J., Ng,S.C., Grabczyk,E. and Fischman,M.C. (1989) GAP-43 gene expression during development: persistence in a distintive set of neurons in the mature central nervous system. Brain Res. Dev. Brain Res., 46, 161–168. [DOI] [PubMed] [Google Scholar]
- 26.Meiri K.F., Bickerstaff,L.E. and Schwob,J.E. (1991) Monoclonal antibodies show that kinase C phosphorylation of GAP-43 during axonogenesis is both spatially and temporally restricted in vivo. J. Cell Biol., 112, 991–1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Seki N., Ohira,M., Nagase,T., Ishikawa,K., Miyajima,N., Nakajima,D., Nomura,N. and Ohara,O. (1997) Characterization of cDNA clones in size-fractionated cDNA libraries from human brain. DNA Res., 4, 345–349. [DOI] [PubMed] [Google Scholar]
- 28.Soejima H., Kawamoto,S., Akai,J., Miyoshi,O., Arai,Y., Morohka,T., Matsuo,S., Niikawa,N., Kimura,A., Okubo,K. and Mukai,T. (2001) Isolation of novel heart-specific genes using the BodyMap database. Genomics, 15, 115–120. [DOI] [PubMed] [Google Scholar]
- 29.Myers M.W., Lazzarini,R.A., Lee,V.M., Schlaepfer,W.W. and Nelson,D.L. (1987) The human mid-size neurofilament subunit: a repeated protein sequence and the relationship of its gene to the intermediate filament gene family. EMBO J., 6, 1617–1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Seehaus T., Breitling,F., Dubel,S., Klewinghaus,I. and Little,M. (1992) A vector for the removal of deletion mutants from antibody libraries. Gene, 114, 235–237. [DOI] [PubMed] [Google Scholar]
- 31.Davis C.A. and Benzer,S. (1997) Generation of cDNA expression libraries enriched for in-frame sequences. Proc. Natl Acad. Sci. USA, 94, 2128–2132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hawke N.A., Strong,S.J., Haire,R.N. and Litman,G.W. (1997) Vector for positive selection of in-frame genetic sequences. Biotechniques, 23, 619–621. [DOI] [PubMed] [Google Scholar]
- 33.Jacobsson K. and Frykberg,L. (1998) Gene VIII-based, phage-display vectors for selection against complex mixtures of ligands. Biotechniques, 24, 294–301. [DOI] [PubMed] [Google Scholar]
- 34.Holz C., Lueking,A., Bovekamp,L., Gutjahr,C., Bolotina,N., Lehrach,H. and Cahill,D.J. (2001) A human cDNA expression library in yeast enriched for open reading frames. Genome Res., 11, 1730–1735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Niwa M., Maruyama,H., Fujimoto,T., Dohi,K. and Maruyama,I.N. (2000) Affinity selection of cDNA libraries by lambda phage surface display. Gene, 256, 229–236. [DOI] [PubMed] [Google Scholar]
- 36.Makalowski W. and Boguski,M.S. (1998) Evolutionary parameters of the transcribed mammalian genome: an analysis of 2820 orthologous rodent and human sequences. Proc. Natl Acad. Sci. USA, 95, 9407–9412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Beckett D., Kovaleva,E. and Schatz,P.J. (1999) A minimal peptide substrate in biotin holoenzyme synthetase-catalyzed biotinylation. Protein Sci., 8, 921–929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cronan J.E. Jr (1990) Biotination of proteins in vivo. A post-translational modification to label, purify, and study proteins. J. Biol. Chem., 265, 10327–10333. [PubMed] [Google Scholar]
- 39.Devlin J.J., Panganiban,L.C. and Devlin,P.E. (1990) Random peptide libraries: a source of specific protein binding molecules. Science, 249, 404–406. [DOI] [PubMed] [Google Scholar]
- 40.Lam K.S., Salmon,S.E., Hersh,E.M., Hruby,V.J., Kazmierski,W.M. and Knapp,R.J. (1991) A new type of synthetic peptide library for identifying ligand-binding activity. Nature, 354, 82–84. [DOI] [PubMed] [Google Scholar]
- 41.Green N.M. (1990) Avidin and streptavidin. Methods Enzymol., 184, 51–67. [DOI] [PubMed] [Google Scholar]
- 42.Cull M.G. and Schatz,P.J. (2000) Biotinylation of proteins in vivo and in vitro using small peptide tags. Methods Enzymol., 326, 430–440. [DOI] [PubMed] [Google Scholar]
- 43.Verde I., Pahlke,G., Salanova,M., Zhang,G., Wang,S., Coletti,D., Onuffer,J., Jin,S.L. and Conti,M. (2001) Myomegalin is a novel protein of the golgi/centrosome that interacts with a cyclic nucleotide phosphodiesterase. J. Biol. Chem., 276, 11189–11198. [DOI] [PubMed] [Google Scholar]
- 44.Van Regenmortel M.H.V. (1994) The recognition of proteins and peptides by antibodies. In van Oss,C.J. and van Regenmortel,M.H.V. (eds), Immunochemistry. Marcel Dekker, New York, pp. 277–300.
- 45.Webster D.M., Henry,A.H. and Rees,A.R. (1994) Antibody–antigen interactions. Curr. Opin. Struct. Biol., 4, 123–129. [DOI] [PubMed] [Google Scholar]
- 46.Wilson I.A. and Stanfield,R.L. (1994) Antibody–antigen interactions. Curr. Opin. Struct. Biol., 4, 857–867. [DOI] [PubMed] [Google Scholar]
- 47.Mariuzza R.A. and Poljak,R.J. (1993) The basics of binding: mechanisms of antigen recognition and mimicry by antibodies. Curr. Opin. Immunol., 5, 50–55. [DOI] [PubMed] [Google Scholar]
- 48.Kramer A., Keitel,T., Winkler,K., Stocklein,W., Hohne,W. and Schneider-Mergener,J. (1997) Molecular basis for the binding promiscuity of an anti-p24 (HIV-1) monoclonal antibody. Cell, 91, 799–809. [DOI] [PubMed] [Google Scholar]