Abstract
The telomerase ribonucleoprotein reverse transcriptase uses its RNA subunit as a template to synthesize telomeric repeats and maintain telomere tracts on chromosome ends. In the ciliate Euplotes crassus, the core telomerase ribonucleoprotein particle undergoes a developmentally programmed assembly into three higher order complexes after mating. Here, we provide evidence using oligonucleotide-directed affinity purification that all of the E.crassus telomerase complexes contain at least two enzyme active sites. Furthermore, we show using co-immunoprecipitation experiments that EcTERT, the telomerase catalytic subunit, undergoes multimerization in vitro. Two independent interaction domains were identified in EcTERT, one at the N-terminus that spans amino acids 186–354 and one at the C-terminus that spans amino acids 755–857. Unexpectedly, we found that TERT can form head-to-head, tail-to-tail and head-to-tail oligomers in vitro, implying that E.crassus telomerase has the potential to assume different conformations in vivo. Together, these data indicate that oligomerization is a conserved feature of telomerase and that the minimal functional unit of the enzyme is a dimer.
INTRODUCTION
Telomeres are DNA–protein structures that cap the ends of eukaryotic chromosomes, preventing chromosomal degradation and fusion, and allowing for continued cell proliferation (1,2). In most eukaryotes, the telomerase ribonucleoprotein is responsible for both maintenance and de novo synthesis of telomeric DNA (3). Recent studies strongly tie telomere maintenance to the development of human cancer (4), and hence understanding the mechanism of telomerase action and the interactions of its constituents are of considerable interest.
Telomerase is a reverse transcriptase (RT) that uses a short templating sequence contained within its RNA subunit to direct synthesis of telomeric repeats by the catalytic protein subunit TERT (telomerase reverse transcriptase) (3,5). TERT was first purified from the hypotrichous ciliate Euplotes aediculatus (6), and subsequently has been characterized in a variety of other ciliates, yeast, mammals and plants (7–11). All of the known TERT proteins contain seven conserved RT motifs, two telomerase signature motifs (T and T2) and, among ciliates, two ciliated protozoa motifs (CP and CP1) (12,13). Although TERT and the telomerase RNA subunit are sufficient to reconstitute telomerase activity in vitro (10,14,15) telomerase exists as a holoenzyme in vivo (3). A variety of telomerase-associated proteins appear to aid in the assembly or stabilization of the holoenzyme, recruit telomerase to telomeres, modulate enzyme processivity and influence the stability of substrate–template interactions (16 and reference therein).
Emerging data imply that telomerase forms a multimeric complex in vivo. Using an oligonucleotide complementary to the Saccharomyces cerevisiae RNA templating domain, Prescott and Blackburn purified telomerase complexes containing the products of two different enzyme active sites (17). In addition, human telomerase purified by gel filtration chromatography contains two telomerase RNA subunits (18). Telomerase multimerization is mediated, at least in part, by TERT interactions. Human TERT forms oligomers in vitro and in vivo (16,19–21) and two separate catalytically inactive TERT proteins can complement each other in trans to reconstitute enzymatically active particles in vitro (16).
The hypotrichous ciliate Euplotes crassus is a useful biochemical model for telomerase. With approximately 10 million linear DNA molecules in its macronucleus, Euplotes is one of nature’s most abundant sources of telomerase and telomeres. Like other ciliates, E.crassus contains a diploid transcriptionally silent germline micronucleus and a highly polyploid transcriptionally active macronucleus (22). In the sexual phase of the life cycle that occurs upon mating, a new macronucleus is formed from a copy of the micronucleus. During this process, chromosomes undergo extensive fragmentation and telomeres are added de novo by telomerase to stabilize the millions of gene-sized linear DNA molecules that will constitute the mature macronuclear genome. During subsequent vegetative growth, telomerase maintains telomeres on all the ends.
In vegetatively growing cells, telomerase exists as a core particle of 280–400 kDa (23,24), harboring a 123 kDa TERT protein, an ∼65 kDa telomerase RNA and possibly a 43 kDa La homolog, known to associate with the E.aediculatus telomerase RNP (6,25). During macronuclear development, the core particle assembles into three higher order complexes of 550–600, 1600 and ≥5000 kDa, which display structural and biochemical properties distinct from those of the smaller telomerase complex from vegetatively growing cells (24). In contrast to the vegetative enzyme, telomerase in the two largest complexes from mating cells can initiate synthesis on non-telomeric DNA primers. In addition, the product elongation profile is different in mating and vegetatively growing cells: telomerase from mating cells pauses upon incorporation of the third G in the GGGGTTTT repeat, while telomerase from vegetatively growing cells pauses after the addition of the third G and the fourth T (GGGGTTTT) (23). The molecular basis for the differences in telomerase enzymology is unknown.
The sizes of telomerase complexes (280–400, 550–600, 1600 and ≥5000 kDa) raise the possibility that the enzyme undergoes a programmed multimerization during macronuclear development. Here we examine the oligomerization state of Euplotes telomerase. Our data indicate that all the telomerase complexes from Euplotes contain at least two active sites. In addition, we provide evidence that EcTERT can multimerize in vitro to form head-to-head, tail-to-tail and head-to-tail oligomers.
MATERIALS AND METHODS
Cloning and site-directed mutagenesis
The coding region of EcTERT-1 (residues 100–3195, relative to the 5′ telomere) (GenBank accession no. AF528527) was cloned in-frame into pCITE-4b(+) (Novagen) between the NdeI and HincII sites for the expression of a non-tagged version. As will be described in detail elsewhere (Z.Karamesheva, L.Wang and D.E.Shippen, in preparation), site-directed mutagenesis was used to change in-frame TGA and TAA codons to create a single contiguous open reading frame (ORF) for expression of full-length EcTERT. To create T7-tagged versions of EcTERT, inserts were cloned in-frame into pET-28a(+) (Novagen) between BamHI and NotI sites. To generate truncated EcTERT proteins, site-directed mutagenesis was carried out using the QuickChange Site-Directed Mutagenesis Kit (Stratagene) under the following conditions. Aliquots of 50 ng of double-stranded plasmid template and 20 pmol of each forward and reverse mutagenesis primer were mixed with 5 µl of 10× reaction buffer, 1 µl of 10 mM dNTP mix and 1 µl of Pfu DNA polymerase (2.5 U/µl) in a 50 µl reaction. PCR conditions were 30 s at 95°C, followed by 16 cycles of 30 s at 95°C, 1 min at 55°C and 20 min at 68°C. Parental templating plasmid was digested with 1 µl (10 U/µl) of DpnI at 37°C for 1 h. An aliquot of 25 µl of the reaction was then transformed into Epicurian Coli XL1-Blue super-competent cells. Colonies were screened for the desired mutations by ABI automated sequencing.
Expression of EcTERT in rabbit reticulocyte lysate
A TNT Coupled Reticulocyte Lysate System (RRL) (Promega) was used to express recombinant EcTERT proteins. In a 50 µl reaction, 0.5 µg of plasmid (see above) carrying EcTERT or a mutant derivative was incubated with 25 µl of TNT rabbit reticulocyte lysate (RRL), 2 µl of TNT reaction buffer, 1 µl of TNT T7 RNA polymerase, 1 µl of 1 mM amino acid mixture minus methionine, 2 µl of [35S]methionine (>1000 Ci/mmol at 10 mCi/ml) (Amersham Pharmacia Biotech) and 1 µl Rnasin ribonuclease inhibitor (40 U/µl) at 30°C for 1.5 h. The reaction was mixed with 240 µl of BB buffer [20 mM Tris–acetate, pH 7.5, 10% glycerol, 1 mM EDTA, 5 mM MgCl2, 0.2 M NaCl, 0.1 M potassium glutamate, 1% NP-40, 0.5 mM sodium deoxycholate, 1 mM DTT, 0.5 mg/ml lysozyme, 0.5 mg/ml BSA (acetylated, RNase free), 0.05 mg/ml glycogen and 0.2 mg/ml yeast total RNA] and incubated on ice for 15 min. The samples were spun at 16 000 g for 15 min at 4°C and supernatants were collected for immunoprecipitation assays.
Immunoprecipitation
To prepare T7-tag antibody Protein A–Sepharose, 10 µl of Protein A–Sepharose (Amersham Pharmacia Biotech) was rinsed twice with 500 µl of W100 buffer (20 mM Tris–acetate, pH 7.5, 10% glycerol, 1 mM EDTA, 5 mM MgCl2, 0.2 M NaCl, 0.1 M potassium glutamate, 1% NP-40, 0.5 mM sodium deoxycholate and 1 mM DTT) and spun at 1000 g for 1 min between washes. An aliquot of 3 µl of T7-tag monoclonal antibody (Novagen) was mixed with 287 µl of W100 buffer and incubated with the pre-equilibrated Protein A–Sepharose at 4°C overnight with agitation. T7-tag antibody Sepharose beads were washed four times with 500 µl of W100 buffer. The beads were then incubated three times with 500 µl of BB buffer (20 mM Tris–acetate, pH 7.5, 10% glycerol, 1 mM EDTA, 5 mM MgCl2, 0.2 M NaCl, 0.1 M potassium glutamate, 1% NP-40, 0.5 mM sodium deoxycholate, 1 mM DTT, 0.5 mg/ml lysozyme, 1 mg/ml BSA, 0.05 mg/ml glycogen and 0.1 mg/ml yeast RNA) for 15 min at 4°C with agitation. The supernatant from the RRL expression was mixed with blocked beads and agitated for 2 h at 4°C. The beads were washed four times in 500 µl of W300 buffer (20 mM Tris–acetate, pH 7.5, 10% glycerol, 1 mM EDTA, 5 mM MgCl2, 0.2 M NaCl, 0.3 M potassium glutamate, 1% NP-40, 0.5 mM sodium deoxycholate and 1 mM DTT) and twice in 500 µl of TMG buffer (30 mM Tris–acetate, 3 mM MgCl2 and 10% glycerol). The beads were mixed with SDS– PAGE loading buffer, denatured and resolved by SDS–PAGE. Depending upon the size of the proteins examined, 8–12% polyacrylamide gels were used. Autoradiography was carried out using a PhosphorImager (Molecular Dynamics).
Macronuclear extract preparation and fractionation of telomerase complexes
Euplotes crassus strains A1 and A2 were used to obtain macronuclei from mating and vegetatively growing cells as previously described (23). Extracts were purified by Superose 6 gel filtration chromatography and telomerase activity was detected as described previously (24). Superose 6 running buffer was modified to 30 mM Tris–HCl, pH 8.0, 3 mM MgCl2, 10% glycerol, 500 mM potassium glutamate and 0.1% NP-40. Fractions from developing macronuclei that contained proteins in the flow-through (≥5000 kDa) or in the 1600 or 550–600 kDa size range, or from vegetative macronuclear extracts that contained proteins in the size range 280–400 kDa were pooled and stored at –80°C.
Streptavidin affinity purification and telomerase assays
Ultralink Immobilized NeutrAvidin Plus (Pierce) (30 µl/ 100 µl nuclear extract) was washed twice in WB buffer (20 mM Tris–acetate, pH 7.5, 10 mM MgCl2, 1 mM EDTA, 300 mM potassium glutamate, 10% glycerol, 1 mM DTT and 1% NP-40) and spun at 200 g for 1 min between washes. The beads were preblocked three times in WBB buffer (WB buffer containing 0.01% NP-40, 1 mg/ml each of BSA, lysozyme, casein and cytochrome c and 0.2 mg/ml each of glycogen and yeast tRNA) for 15 min at 4°C with agitation, washed twice with WB buffer and resuspended in WB buffer containing 0.05 nmol/µl of each oligonucleotide block 1 d(TAGACCTGTTAGTGTACATTTGAATTGAAGC) and block 2 d(TAGACCTGTTAGGTTGGATTTGTGGGCATCA), 0.2 mg/ml of yeast tRNA and 0.2 mg/ml casein.
Telomerase reactions were carried out with 100 µl of purified telomerase complexes or macronuclear extract in a 200 µl reaction mixture with 1 µg of 5′-biotinylated BT36 d[(G4T4)4G4] or BN36 d(CGACTACGCGATCATCACTATCGACTACGCGATCAT) or the unmodified oligonucleotide T24 d[(G4T4)3] and incubated at 30°C for 10 min. Preblocked Ultralink Immobilized NeutrAvidin Plus (30 µl/100 µl of nuclear extract) was then added along with 5 mM MgCl2, 20 mM EGTA, 50 mM Tris–HCl, pH 8.0, 1 mM spermidine, 1 mM DTT, 0.25 µM [α-32P]dGTP (800 Ci/mmol) and 0.1 mM dTTP and reactions were continued for another 20 min at 30°C followed by a 2 h incubation at 4°C with agitation. With agitation, the beads were washed three times in 300 µl of WB buffer containing 0.1 mg/ml casein, 0.4 M potassium glutamate for 5 min at 4°C and once in 300 µl of WB buffer containing 0.6 M potassium glutamate, 0.1 mg/ml casein for 5 min at 30°C. The beads were then washed three times in 300 µl of WB containing 0.1 mg/ml casein, 0.4 M potassium glutamate for 5 min at room temperature. Bound reaction products were eluted by incubation at 65°C for 15 min with 1% SDS followed by phenol extraction. Products were ethanol precipitated at 4°C overnight in the presence of 5 µl glycogen and 0.2 M NaCl. Urea–polyacrylamide (6%) electrophoresis was carried out to resolve telomerase products.
RESULTS
The E.crassus telomerase RNP has at least two active sites
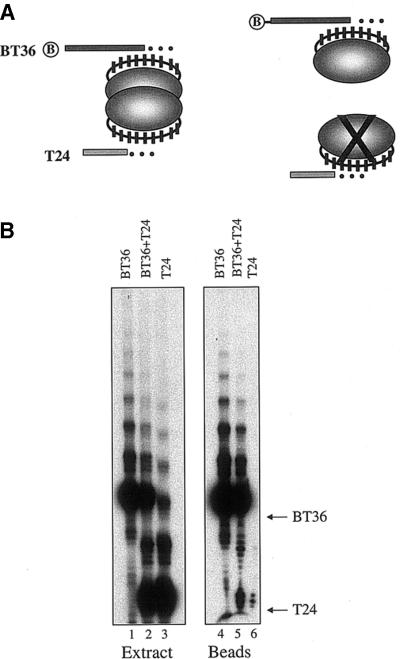
To determine whether the Euplotes telomerase RNP contains more than one active site in vivo, we used the oligonucleotide affinity purification method developed by Prescott and Blackburn (17). Macronuclear extracts from mating cells were incubated with two telomeric oligonucleotides: a biotinylated 36mer (BT36) and a non-biotinylated 24mer (T24). dNTPs were added and a telomerase reaction was initiated. Primer elongation products were purified on streptavidin and, after extensive washing, they were resolved by polyacrylamide gel electrophoresis. As shown in Figure 1A, the products of T24 are expected to co-purify with the products of BT36 if telomerase contains more than one active site. In contrast, if telomerase contains a single active site or is a monomer, only the products of BT36 should be detected. In control reactions, we found that telomerase utilized both oligonucleotides with similar efficiency (Fig. 1B, lanes 1–3). When the extract was incubated with T24 alone and then subjected to purification, only a background level of products was detected (Fig. 1B, lane 6). However, when BT36 was added to the reaction, we observed a reproducible enrichment of products corresponding to extension of T24 (Fig. 1B, lane 5). Furthermore, in contrast to these results with the crude macronuclear extract, affinity purification of isolated telomerase RNP complexes gave a 1:1 ratio of T24 to BT36 products (see below).
Figure 1.
Assay for telomerase oligomerization in vivo. (A) Affinity purification strategy. A 5′ biotinylated telomeric oligonucleotide (BT36) and a non-biotinylated telomeric oligonucleotide (T24) are incubated with telomerase. Following a primer elongation reaction, products are purified on streptavidin. The products of BT36 and T24 should co-purifiy if telomerase harbors more than one active site. (B) Telomerase reactions with macronuclear extract from mating E.crassus in the presence of BT36 alone (lane 1), BT36 and T24 (lane 2) or T24 alone (lane 3). The migration positions of the input BT36 and T24 are indicated. The same set of reactions was subjected to affinity purification and the products associated with the beads were resolved by PAGE (lanes 4–6).
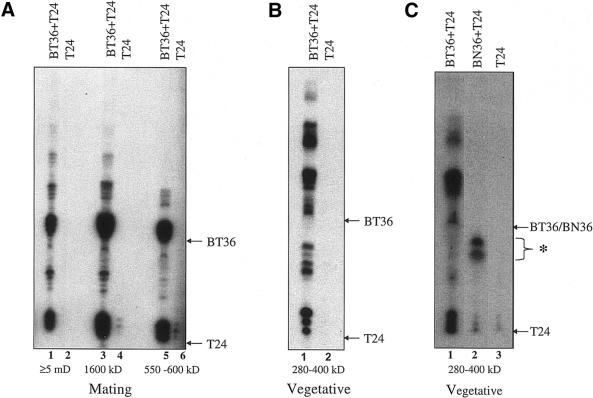
To determine which of the complexes from mating cells contained more than one active site, macronuclear extracts were purified by Superose 6 gel filtration chromatography. Fractions containing enzymatically active telomerase particles of 550–600, 1600 and ≥5000 kDa were collected (data not shown) and then subjected to telomerase assays using BT36 and T24 followed by affinity purification. In each case, T24 products co-purified with BT36 products (Fig. 2A, lanes 1, 3 and 5). Thus, all of the telomerase complexes from mating cells appear to contain at least two active sites.
Figure 2.
Each telomerase RNP complex contains more than one active site. Purified telomerase complexes from mating (A) or vegetative cells (B) were obtained following gel filtration chromatography and subjected to telomerase assays (using primers indicated) followed by affinity purification. (C) Affinity purification of telomerase products generated from isolated vegetative telomerase complexes. Telomerase reactions were carried out with BT36 alone (lane 1), BN36 plus T24 (lane 2) or T24 alone (lane 3). The asterisk denotes the products of a non-specific labeling reaction of BN36. Arrows indicate the migration positions of the input primers.
Two active sites in the telomerase RNP complex from vegetatively growing cells
To determine whether telomerase oligomerization is a developmentally regulated event, we asked whether the RNP particles from vegetatively growing cells contain more than one active site. Macronuclear extracts were subjected to gel filtration chromatography. Fractions in the size range 280– 400 kDa containing active telomerase were assayed with BT36 and T24 and the products were isolated by affinity purification. As with the complexes from mating cells, elongation products of T24 efficiently co-purified with BT36 products (Fig. 2B, lane 1). In the absence of BT36, no elongation products were detected (Fig. 2B, lane 2).
The affinity purification assay requires that both of the oligonucleotides undergo elongation reactions. However, to verify that the co-purification of T24 and BT36 products reflected specific interactions with telomerase instead of non-specific nucleic acid or DNA–protein interactions, telomerase reactions were carried out with purified telomerase RNP particles from vegetatively growing cells using T24 and a biotinylated non-telomeric oligonucleotide (BN36). After affinity purification, only a background level of T24 products could be detected (Fig. 2C, lower bands in lanes 2 and 3) relative to the control reaction that contained BT36 (Fig. 2C, lane 1). The products in lane 2 migrating faster than the input BN36 oligonucleotide (asterisk) reflect a non-specific DNA processing event; similar products were obtained when BN36 was used as a primer in a standard telomerase assay (data not shown). Thus, we conclude that co-purification of telomeric biotinylated and non-biotinylated oligonucleotides reflects specific interactions with telomerase and that the minimal telomerase RNP in E.crassus contains at least two active sites.
Multimerization of EcTERT
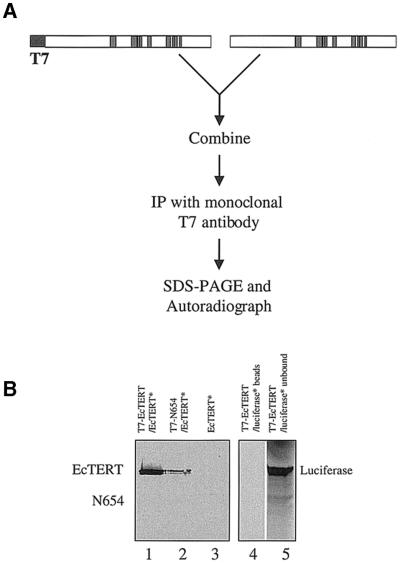
Since recent studies suggest that human TERT can form dimers in vitro and in vivo (16,19), we asked whether oligomerization is a conserved feature of TERT by performing co-immunoprecipitation assays using recombinant EcTERT protein. T7-tagged and non-tagged versions of EcTERT were expressed separately in a transcription and translation-coupled RRL system. The non-tagged TERT was labeled with [35S]methionine. Translation was terminated with cycloheximide, the two EcTERT protein samples were combined and then immunoprecipitated using a T7-tag antibody coupled to Protein A–Sepharose (Fig. 3A). In the presence of the T7-tagged EcTERT, 35S-labeled, non-tagged EcTERT was precipitated (Fig. 3B, lane 1). In control reactions antibody-treated beads did not immunoprecipitate non-tagged EcTERT (Fig. 3B, lane 3). To rule out the possibility that the T7-tagged EcTERT protein was labeled with [35S]methionine after the two translation reactions were combined, a C-terminal truncated version of EcTERT (N654) that is able to interact with the full-length EcTERT (see below) was added to the 35S-labeled EcTERT. Only the full-length EcTERT was detected (Fig. 3B, lane 2), confirming that translation synthesis was halted with cycloheximide treatment. As an additional control, T7-tagged EcTERT was incubated with 35S-labeled, non-tagged luciferase and the reaction was subjected to immunoprecipitation. No interaction was detected; luciferase remained in the unbound fraction (Fig. 3B, lanes 4 and 5). Taken together, the data indicate that EcTERT can undergo specific oligomerization in vitro. As has been reported for human TERT (19), telomerase RNA is not required for EcTERT oligomerization. Inclusion of in vitro transcribed E.crassus telomerase RNA did not alter the efficiency of EcTERT self-association (data not shown).
Figure 3.
EcTERT oligomerizes in vitro. (A) Immunoprecipitation strategy to detect EcTERT multimers. T7-tagged, unlabeled EcTERT and non-tagged, 35S-labeled EcTERT were expressed separately in RRL. The translation reactions were stopped by cycloheximide before the products were combined. Vertical bars denote conserved telomerase and RT motifs. (B) Immunoprecipitation of T7–EcTERT and 35S-labeled EcTERT (lane 1), 35S-labeled EcTERT and T7-tagged N654 (lane 2) and 35S-labeled EcTERT alone (lane 3). Immunoprecipitation of T7-tagged EcTERT and 35S-labeled luciferase (lane 4). An aliquot of 50 µl of the unbound fraction from lane 4 was loaded on lane 5.
The N-terminus is sufficient for EcTERT oligomerization
To define the domain(s) necessary and sufficient for EcTERT self-interaction, we created a series of EcTERT truncation constructs and tested their ability to interact with each other and with full-length EcTERT. The results of these experiments are summarized in Figure 4. While we cannot be certain that all of the constructs generated properly folded proteins, all of the recombinant proteins used in this study were expressed at approximately the same level and were soluble (data not shown).
Figure 4.
Summary of EcTERT oligomerization domains. The interactions of EcTERT proteins with full-length EcTERT and truncation mutants are shown. nd, not determined.
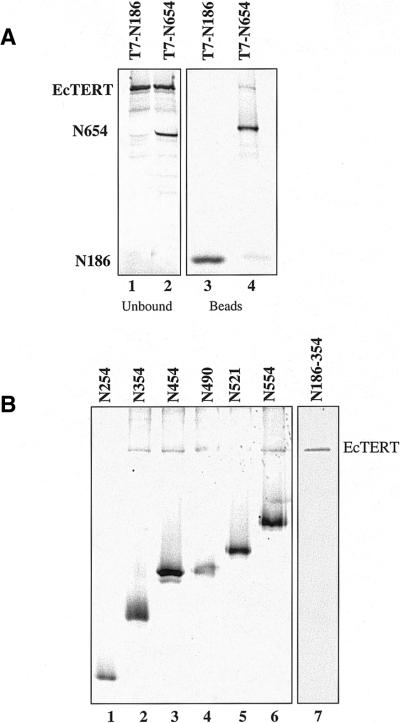
We initially examined two constructs, N186, which has a C-terminal truncation and lacks all of the conserved RT and telomerase-specific motifs, and N654, which contains the CP motif, T motif and RT motifs 1, 2 and A (Fig. 4). Non-tagged, full-length EcTERT was co-expressed with T7-tagged N186 or N654 in the presence of [35S]methionine and the proteins were subjected to immunoprecipitation. Full-length EcTERT did not co-purify with N186 and remained in the unbound fraction (Fig. 5A, lane 1). In contrast, full-length EcTERT was immunoprecipitated in the presence of N654 (Fig. 5A, lane 4). The efficiency of binding was lower than the full-length EcTERT self-interaction (Fig. 3B, lanes 1 and 2; data not shown), suggesting that the C-terminus contributes to EcTERT oligomerization.
Figure 5.
Identification of an N-terminal oligomerization domain in EcTERT. (A) Untagged full-length EcTERT was co-expressed with T7-tagged N186 and N654 in the presence of [35S]methionine. Immuno precipitates (lanes 3 and 4) and unbound fractions (lanes 1 and 2) are shown. (B) T7-tagged C-terminal truncation mutants were co-expressed with full-length EcTERT in the presence of 35S and then subjected to immunoprecipitation (lanes 1–6). In lane 7, T7-tagged, unlabeled N186–354 was incubated with untagged, 35S-labeled full-length EcTERT, subjected to immunoprecipitation and run on a separate gel.
Additional truncation constructs were tested to define the boundaries of the N-terminal interaction domain (Fig. 5B). Full-length EcTERT was immunoprecipitated in the presence of N354, but not N254 (Fig. 5B, lanes 1 and 2). To define the minimal region necessary to bind full-length EcTERT, we created an ORF that encodes the 168 amino acids between residues 186 and 354 (N186–354) (Fig. 4). This protein bound the full-length protein (Fig. 5B, lane 7; note that in this experiment N186–354 was not labeled with [35S]methionine). Hence, the region between residues 186 and 354 defines an interaction domain in the N-terminus of EcTERT.
A domain in the C-terminus of EcTERT contributes to oligomerization
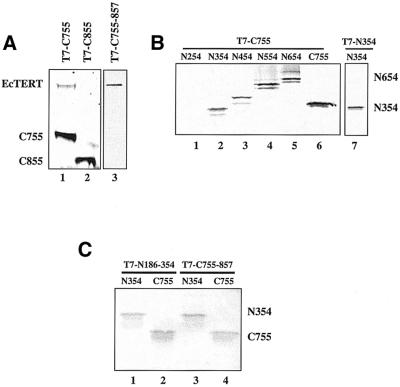
We next investigated the role of the C-terminus (residues 655–1032) in EcTERT oligomerization using a series of N-terminal truncations (Fig. 4). C655 and C755, but not C855, were capable of interacting with full-length EcTERT (Fig. 6A, lanes 1 and 2; data not shown). To further define the C-terminal interaction domain, we generated C755–857 and tested its ability to bind full-length EcTERT. As shown in Figure 6A, lane 3, this region harbors a domain sufficient for interaction with full-length TERT. Together, these findings argue that EcTERT contains two independent interaction domains, one at the C-terminus and one at the N-terminus.
Figure 6.
Identification of a C-terminal oligomerization domain and evidence for multiple EcTERT conformations in vitro. (A) A C-terminal oligomerization domain. T7-tagged truncation mutants were co-expressed with untagged full-length EcTERT in the presence of 35S. In lane 3, T7-C755-857 was unlabeled. (B) Evidence for multiple EcTERT conformations. Unlabeled, T7-tagged C755 was incubated with 35S-labeled truncation mutations prior to immunoprecipitation (lanes 1–6). In lane 7, unlabeled, T7-tagged N354 was tested for homodimerization with untagged, 35S-labeled N354 and run in a separate gel. (C) Interactions of the minimal N- and C-terminal oligomerization domains. Unlabeled, T7-tagged N186–354 and T7-tagged C755–857 were incubated with 35S-labeled N354 (lanes 1 and 3) or C755 (lanes 2 and 4) prior to immunoprecipitation.
EcTERT can oligomerize in a head-to-head, tail-to-tail or head-to-tail orientation in vitro
In the experiments described thus far, we examined the ability of truncated EcTERT proteins to interact with the full-length protein. Since human TERT has been shown to form a head-to-tail dimer (19), we asked whether this is a conserved feature of TERT proteins by assaying the ability of N-terminal and C-terminal constructs to interact with each other. We found that C755 interacts with N354 and N186–354, but not N254 (Fig. 6B, lanes 1 and 2; data not shown). In addition, the minimal C-terminal domain C755–857 binds N354 (data not shown). Together, these observations not only indicate that EcTERT is capable of forming head-to-tail oligomers in vitro, but also demonstrate that the N- and C-terminal interaction domains act independently.
We next asked whether EcTERT can assume other conformations in vitro by assaying for head-to-head and tail-to-tail interactions. Co-immunoprecipitation experiments carried out with 35S-labeled, non-tagged C755 and T7-tagged, unlabeled C755–857 revealed that EcTERT has the potential to form tail-to-tail multimers (Fig. 6C, lane 4). A similar experiment performed with N354 and T7–N186–354 also showed the potential for head-to-head interactions (Fig. 6B, lane 7). From these findings we conclude that EcTERT has the potential to form multiple conformations in vitro.
As discussed above, we found that minimal fragments capable of interacting with full-length EcTERT are N186–354 and C755–857. Although these two fragments are capable of interacting with N354 and C755 (Fig. 6C), they are incapable of forming homo- or heterodimeric complexes with each other (data not shown). We interpret this result to mean that the two minimal interaction domains require additional flanking segments in their binding partners to establish a stable interaction.
DISCUSSION
The E.crassus telomerase RNP is unique among this class of RTs in its ability to undergo developmentally programmed assembly into higher order complexes (24). In this study, we investigated the oligomerization state of E.crassus telomerase particles using a combination of immunoprecipitation and affinity purification approaches. Both lines of experimentation indicate that the minimal telomerase RNP contains more than one active site.
Our affinity strategy, which was adapted from Prescott and Blackburn’s analysis of yeast telomerase (17), was designed to detect the products of two active sites in a single RNP complex. The assay uses two different primers and demands that both serve as substrates for template-directed nucleotide addition. As in the standard telomerase assay, the primers are present in a large excess over the enzyme. Accordingly, elongation products generated in the reaction almost certainly result from the initial binding event (5). We cannot rule out the possibility that a telomerase monomer sequentially extends two primers at a single active site, since all telomerases, including the E.crassus enzyme, are known to bind DNA at two sites: a proteinaceous anchor site in TERT which mediates enzyme processivity and the RNA template (5,26). However, our affinity purification and in vitro protein interaction data are consistent with the interpretation that E.crassus telomerase exists as a multimer. Furthermore, several recently published reports demonstrate dimerization of yeast and human telomerases using alternative experimental approaches (16,18–21). Therefore, we propose that oligomerization is a conserved feature of the telomerase RNP.
Our data suggest that even the smallest E.crassus telomerase RNP complex found in vegetatively growing cells contains two enzyme active sites, but is this complex actually large enough to accommodate a telomerase dimer? Based on biochemical analysis of telomerase from E.aediculatus and S.cerevisiae, the ratio of TERT to telomerase RNA is likely to be 1:1 (6,17). Accordingly, the size of a dimeric E.crassus vegetative RNP should be ∼376 kDa with two TERTs (∼123 kDa) plus two telomerase RNA subunits (∼65 kDa). Telomerase from vegetatively growing E.aediculatus also contains a 43 kDa protein, which appears to be a La homolog (6,25). Although polyclonal antibodies directed against the E.aediculatus p43 protein failed to cross-react with the E.crassus protein (Z.Karamysheva and D.Shippen, unpublished results), the close phylogenetic relationship between the two organisms implies that E.crassus telomerase may also contain one or two p43 subunits. Thus, a dimeric core RNP particle may approach 460 kDa in size.
Size estimates for vegetative E.crassus telomerase range from 280 kDa, based on Superose 6 gel filtration data (24), to 375–400 kDa, based on glycerol gradient sedimentation (23). Furthermore, closer inspection of the elution profile from Superose 6 chromatography reveals that telomerase activity actually spreads across fractions corresponding to molecular weights of ∼200–440 kDa (L.Wang and D.E.Shippen, unpublished data; 23). Thus, while we have no definitive biochemical data demonstrating that the vegetative telomerase complex from E.crassus is not a monomer, these observations taken in the context of the inherent inaccuracies of molecular mass estimates from gel filtration chromatography indicate that the core E.crassus telomerase RNP is large enough to accommodate a dimer.
The constituents of the larger telomerase complexes found in mating cells are under investigation. We have recently shown that all the telomerase RNPs from mating E.crassus cells are physically associated with the DNA polα/primase machinery (27). This physical coupling presumably acts to biochemically couple de novo G-strand synthesis by telomerase with synthesis of the C-strand by the conventional replication machinery. These findings in conjunction with the data presented here suggest that higher order telomerase complexes in developing macronuclei may be comprised of a dimeric core telomerase RNP particle onto which other protein machineries assemble.
Telomerase multimerization appears to be mediated through TERT interactions. Using a co-immunoprecipitation assay, we found two independent regions in the EcTERT N- and C-termini (amino acids 186–354 which span the CP motif and 755–857 which cover the C, D and E RT motifs, respectively) that are sufficient for interaction in vitro. While all of the recombinant proteins used in our study were soluble, not all may retain their native conformation. Evidence supporting this conclusion is that the two minimal regions in the N- and C-termini sufficient for interaction with the full-length EcTERT protein fail to form homo or heterodimers with each other in vitro. Thus, flanking segments in these regions are likely to be important to stabilize binding.
The interaction domains we identified overlap with the two regions in human TERT defined by Arai et al. as sufficient for head-to-tail dimer formation in vivo (19). The hTERT N-terminal interaction domain (amino acids 301–538) lies upstream of the T motif and in a region that harbors the CP motif in ciliates, while the C-terminal interaction domain (amino acids 914–228) covers RT motif E. As with the HIV-1 RT (28), hTERT does not appear to be capable of forming head-to-head or tail-to-tail dimers in vivo (19). Intriguingly, our data suggest that E.crassus telomerase has the potential to assume head-to-tail as well as head-to-head and tail-to-tail configurations. It is possible that one complex is favored in vivo; our in vitro immunoprecipitation assay would not be able to distinguish the difference.
Two models have been proposed for the coordinated action of a telomerase dimer (17,18). One model proposes that the two active sites bind to two separate substrates simultaneously, while the second model proposes that the two catalytic sites act sequentially on a single substrate in a template switching hand-off mechanism (18). These models are not mutually exclusive and in E.crassus, where telomerase is known to undergo dramatic changes in structure and biochemical properties during development (23,24,29), both configurations may exist. In mating cells, for example, the ability of a single RNP particle to associate with two different chromosome ends simultaneously could significantly increase the efficiency of de novo telomere formation. Since thousands of new telomeres must be formed in a short developmental window (22), such a mechanism could rapidly stabilize the maturing macronuclear genome. In vegetatively growing cells, the template switching configuration may exist. Frequent ‘programmed’ template switching events could provide an explanation for the additional ‘pause’ in the product elongation profile associated with telomerase in this stage of the life cycle (23).
Acknowledgments
ACKNOWLEDGEMENTS
We thank Gary Kunkel, Jeff Kapler and Zemfira Karamysheva for helpful comments on the manuscript. This work was supported by National Institutes of Health grant GM49157 to D.E.S.
DDBJ/EMBL/GenBank accession no. AF528527
REFERENCES
- 1.McEachern M.J., Krauskopf,A. and Blackburn,E.H. (2000) Telomeres and their control. Annu. Rev. Genet., 34, 331–358. [DOI] [PubMed] [Google Scholar]
- 2.Shore D. (2001) Telomeric chromatin: replicating and wrapping up chromsoome ends. Curr. Opin. Genet. Dev., 11, 189–198. [DOI] [PubMed] [Google Scholar]
- 3.Nugent C.I. and Lundblad,V. (1998) The telomerase reverse transcriptase: components and regulation. Genes Dev., 12, 1073–1085. [DOI] [PubMed] [Google Scholar]
- 4.Stewart S.A. and Weinberg,R.A. (2000) Telomerase and human tumorigenesis. Semin. Cancer Biol., 10, 399–406. [DOI] [PubMed] [Google Scholar]
- 5.Collins K. (1999) Ciliate telomerase biochemistry. Annu. Rev. Biochem., 68, 187–218. [DOI] [PubMed] [Google Scholar]
- 6.Lingner J. and Cech,T.R. (1996) Purification of telomerase from Euplotes aediculatus: requirement of a primer 3′ overhang. Proc. Natl Acad. Sci. USA, 93, 10712–10717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Meyerson M., Counter,C.M., Eaton,E.N., Ellisen,L.W., Steiner,P., Caddle,S.D., Ziaugra,L., Beijersbergen,R.L., Davidoff,M.J., Liu,Q., Bacchetti,S., Haber,D.A. and Weinberg,R.A. (1997) hEST2, the putative human telomerase catalytic subunit gene, is up-regulated in tumor cells and during immortalization. Cell, 90, 785–795. [DOI] [PubMed] [Google Scholar]
- 8.Nakamura T.M., Morin,G.B., Chapman,K.B., Weinrich,S.L., Andrews,W.H., Lingner,J., Harley,C.B. and Cech,T.R. (1997) Telomerase catalytic subunit homologs from fission yeast and human. Science, 277, 955–959. [DOI] [PubMed] [Google Scholar]
- 9.Bryan T.M., Sperger,J.M., Chapman,K.B. and Cech,T.R. (1998) Telomerase reverse transcriptase genes identified in Tetrahymena thermophila and Oxytricha trifallax. Proc. Natl Acad. Sci. USA, 95, 8479–8484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Collins K. and Gandhi,L. (1998) The reverse transcriptase component of the Tetrahymena telomerase ribonucleoprotein complex. Proc. Natl Acad. Sci. USA, 95, 8485–8490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fitzgerald M.S., Riha,K., Gao,F., Ren,S., McKnight,T.D. and Shippen,D.E. (1999) Disruption of the telomerase catalytic subunit gene from Arabidopsis inactivates telomerase and leads to a slow loss of telomeric DNA. Proc. Natl Acad. Sci. USA, 96, 14813–14818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lingner J., Hughes,T.R., Shevchenko,A., Mann,M., Lundblad,V. and Cech,T.R. (1997) Reverse transcriptase motifs in the catalytic subunit of telomerase. Science, 276, 561–567. [DOI] [PubMed] [Google Scholar]
- 13.Miller M.C., Liu,J.K. and Collins,K. (2000) Template definition by Tetrahymena telomerase reverse transcriptase. EMBO J., 19, 4412–4422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Weinrich S.L., Pruzan,R., Ma,L., Ouellette,M., Tesmer,V.M., Holt,S.E., Bodnar,A.G., Lichtsteiner,S., Kim,N.W. and Trager,J.B. (1997) Reconstitution of human telomerase with the template RNA component hTR and the catalytic protein subunit hTRT. Nature Genet., 17, 498–502. [DOI] [PubMed] [Google Scholar]
- 15.Beattie T.L., Zhou,W., Robinson,M.O. and Harrington,L. (1998) Reconstitution of human telomerase activity in vitro. Curr. Biol., 8, 177–180. [DOI] [PubMed] [Google Scholar]
- 16.Beattie T.L., Zhou,W., Robinson,M.O. and Harrington,L. (2001) Functional multimerization of the human telomerase reverse transcriptase. Mol. Cell. Biol., 21, 6151–6160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Prescott J. and Blackburn,E.H. (1997) Functionally interacting telomerase RNAs in the yeast telomerase complex. Genes Dev., 11, 2790–2800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wenz C., Enenkel,B., Amacker,M., Kelleher,C., Damm,K. and Lingner,J. (2001) Human telomerase contains two cooperating telomerase RNA molecules. EMBO J., 20, 3526–3534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Arai K., Masutomi,K., Khurts,S., Kaneko,S., Kobayashi,K. and Murakami,S. (2002) Two independent regions of human telomerase reverse transcriptase are important for its oligomerization and telomerase activity. J. Biol. Chem., 277, 8538–8544. [DOI] [PubMed] [Google Scholar]
- 20.Armbruster B.N., Banik,S.S., Guo,C., Smith,A.C. and Counter,C.M. (2001) N-terminal domains of the human telomerase catalytic subunit required for enzyme activity in vivo. Mol. Cell. Biol., 21, 7775–7786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Moriarty T.J., Huard,S., Dupuis,S. and Autexier,C. (2002) Functional multimerization of human telomerase requires an RNA interaction domain in the N-terminus of the catalytic subunit. Mol. Cell. Biol., 22, 1253–1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Prescott D.M. (1994) The DNA of ciliated protozoa. Microbiol. Rev., 58, 233–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bednenko J., Melek,M., Greene,E.C. and Shippen,D.E. (1997) Developmentally regulated initiation of DNA synthesis by telomerase: evidence for factor-assisted programmed telomere formation. EMBO J., 16, 2507–2518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Greene E.C. and Shippen,D.E. (1998) Developmentally programmed assembly of higher order telomerase complexes with distinct biochemical and structural properties. Genes Dev., 12, 2921–2931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Aigner S., Lingner,J., Goodrich,K.J., Grosshans,C.A., Shevchenko,A., Mann,M. and Cech,T.R. (2000) Euplotes telomerase contains a La motif protein produced by apparent translational frameshifting. EMBO J., 19, 6230–6239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Melek M., Greene,E.C. and Shippen,D.E. (1996) Processing of non-telomeric 3′ ends by telomerase: default template alignment and endonucleolytic cleavage. Mol. Cell. Biol., 16, 3437–3445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ray S., Karamysheva,Z., Wang,L., Shippen,D.E. and Price,C.M. (2002) Telomerase interacts with DNA replication proteins to physically associate de novo telomere formation and chromosomal replication machinery. Mol. Cell. Biol., 22, 5859–5868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kohlstaedt L.A., Wang,J., Rice,P.A., Friedman,J.M. and Steitz,T.A. (1993) The structure of HIV-1 reverse transcriptase. In Skalka,A.M. and Goff,S.P. (eds), Reverse Transcriptase. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 223–249.
- 29.Bednenko J., Melek,M. and Shippen,D. (1998) Reiterative dG addition by Euplotes crassus telomerase during extension of non-telomeric DNA. Nucleic Acids Res., 26, 3998–4004. [DOI] [PMC free article] [PubMed] [Google Scholar]