Abstract
Aptamers, small oligonucleotides derived from an in vitro evolution process called SELEX, are promising therapeutic and diagnostic agents. Although very effective in vitro, only a few examples are available showing their potential in vivo. We have analyzed the effect of a well characterized pseudoknot RNA aptamer selected for tight binding to human immunodeficiency virus (HIV) type 1 reverse transcriptase on HIV replication. Transient intracellular expression of a chimeric RNA consisting of the human initiator tRNAMet (tRNAMeti)/aptamer sequence in human 293T cells showed inhibition of HIV particle release by >75% when the cells were co-transfected with proviral HIV-1 DNA. Subsequent virus production of human T-lymphoid C8166 cells, infected with viral particles derived from co-transfected 293T cells, was again reduced by >75% as compared with the control. As the observed effects are additive, in this model for virus spread, the total reduction of HIV particle formation by transient intracellular expression of the pseudoknot RNA aptamer amounts to >95%. Low-dose HIV infection of human T cells stably expressing the aptamer did not show any virus replication over a period of 35 days. This is the first example of an RNA aptamer selected against a viral enzyme target to show powerful antiviral activity in HIV-1-permissive human T-lymphoid cell lines.
INTRODUCTION
About a decade ago, a method for in vitro selection of combinatorial oligonucleotide libraries was described (1–3). This technique, generally termed ‘Systematic Evolution of Ligands by EXponential enrichment’ (SELEX), has in recent years led to an explosion in the discovery of potential nucleic acid ligands (aptamers) for target proteins. Important properties of aptamers are tight binding to proteins (KD range from low picomolar to nanomolar) combined with high specificity and virtually no immunogenity. Often, aptamers block the enzymatic activity of their target proteins. Consequently, several concepts of using these molecules as therapeutics and/or diagnostics have been proposed (for a review see 4–6). However, despite the increasing number of aptamers described, there are still few examples of their effectiveness in living cells and in vivo. Obviously, turning an in vitro selected oligonucleotide into a therapeutic agent is no easy task (7). Nevertheless, there are a few promising examples of aptamers selected against extracellular protein targets. One of these aptamers is currently being assessed in clinical trials (8).
However, a far greater number of protein targets are localized within the cell resulting in the need for different strategies of drug delivery (for example 9–11, for a review see 12). One clinically relevant example of interesting intracellular targets is viral proteins. A major challenge in the treatment of AIDS is the fast emergence of resistant human immunodeficiency virus (HIV) mutants (for a review see 13). A second problem is the severe side effects encountered by AIDS patients treated with the available anti-HIV drugs. Theoretically, aptamers are promising candidates to overcome the limitations of current HIV therapy. Because of their high specificity towards the intended target there should be fewer side effects. Consequently, the effect of anti-HIV-1 Rev and anti-HIV-1 Tat decoys/aptamers have been investigated in a cellular environment (14,15). Furthermore, several groups could show inhibition of viral replication in cell culture by anti-HIV-1 Rev decoys/aptamers (16–20). Because of the pivotal role of the reverse transcriptase (RT) in the retroviral life cycle (for a review see 21) it represents an even more attractive target for developing such innovative antiviral strategies. Being an integral component of the released viruses, there is a good chance that an aptamer against this viral enzyme would also be packed into the particles, thus leading to non-infectious viruses. Thus, not only cells expressing the aptamer would be protected.
Applying the SELEX procedure a pool of about 20 RNA aptamers selected against HIV-1 RT could be identified (22). The analysis of the isolated aptamers revealed a consensus sequence that resulted in the formation of an RNA pseudoknot (23,24). Recently, we determined the X-ray structure of one of these aptamers complexed with its target (25). The pseudoknot RNA partially overlaps with the binding site for the natural substrate consistent with the aptamer acting as potent competitive inhibitor. Considering the aptamer covers a much larger area of the target protein (25) than the small drugs currently used in antiviral therapy (for a review see 26), the likelihood of viral escape via single point mutations should be drastically reduced (27). Further, thorough biochemical characterization of this protein/RNA interaction in vitro revealed an extraordinarily tight binding with a KD of ∼25 pM for the complex (28). This is at least 100 times lower than for the natural substrate. Additionally, the inhibitory aptamer is highly specific for HIV-1 RT, with the closely related HIV-2 enzyme showing a binding affinity close to four orders of magnitude lower.
To explore the effect of this RNA aptamer on HIV replication, we transiently expressed the molecule in eukaryotic cells as a polymerase III-driven chimeric gene consisting of the human initiator tRNAMet (tRNAMeti) sequence and the 33 nt pseudoknot sequence. Transient co-transfection of 293T cells with the pseudoknot expressing vector and a proviral HIV-1 DNA resulted in a >75% reduction of viral particle release from these cells. Furthermore, such viral particles showed a strongly reduced intrinsic RT activity, indicating package of the aptamer into the virions. Subsequent infection of T-lymphoid cells with viruses produced in the presence of chimeric tRNAMeti/pseudoknot RNA showed a >75% decline in viral replication as compared with the control. Moreover, a T-lymphoid cell line stably transfected with the aptamer construct did not show any virus replication after low-dose HIV-1 infection. This demonstrates that the aptamer functions as a powerful inhibitor of HIV replication in cell tissue culture (proof of principle).
MATERIALS AND METHODS
In vitro RNA preparation
The 33 nt pseudoknot RNA aptamer (sequence: 5′-GGGAGAUUCCGUUUUCAGUCGGGAAAAACUGAA) and the tRNAMeti/pseudoknot RNA chimera (see Fig. 1B) were prepared in a standard 10 ml T7 reaction mixture for in vitro transcription and purified by gel electrophoresis as described previously (25,29). The RNAs were refolded at a concentration of 200–300 µM at 65°C for 5 min followed by slow cooling to room temperature in 20 mM cacodylate buffer pH 6.5, 25 mM NaCl and 5 mM MgCl2. 5′-End-labeling of the RNA with T4 polynucleotide kinase (New England Biolabs) was performed as described previously (30). Dephosphorylation of the in vitro transcribed RNA prior to end-labeling was carried out according to standard procedures (31).
Figure 1.
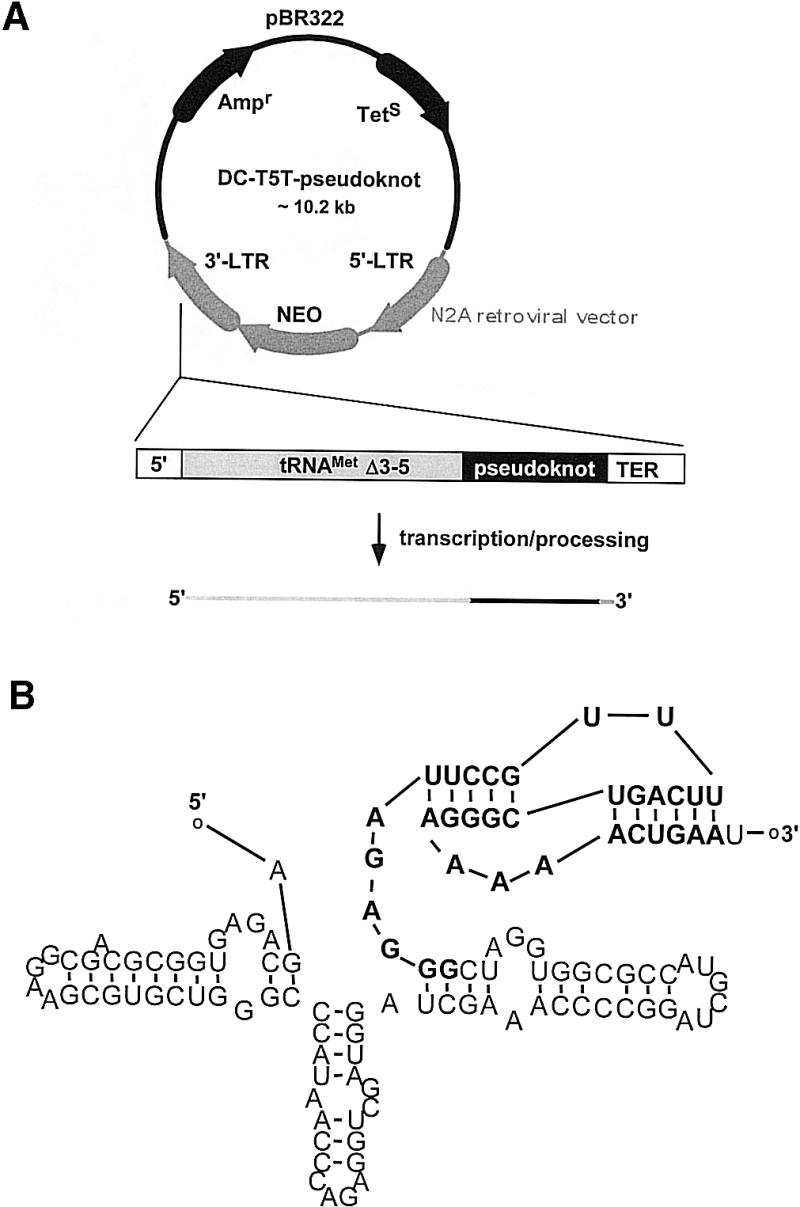
Vector construction. (A) The original retroviral vector DCN2A contains a Δ3′-5 mutant tRNAMeti gene and a polymerase III termination signal (TER) in the U3 region of the 3′ long terminal repeat. An oligonucleotide containing the pseudoknot sequence was cloned downstream of the tRNA gene and upstream of the polymerase III termination signal generating the DCT5T-pseudoknot vector. (B) Secondary structure prediction of the pseudoknot motif within the chimeric tRNA-pseudoknot RNA transcript. The prediction was calculated using the pseudoknot folding software PKNOTS. The pseudoknot part is shown in bold letters. The additional base at the 3′-end of the transcript arises from the termination signal.
RNA secondary structure prediction
The secondary structures of the chimeric RNA transcripts were predicted by using the program PKNOTS (32). Secondary structure models were drawn using the program RnaViz (33).
Protein purification
Recombinant heterodimeric wild type HIV-1 and HIV-2 RT were expressed in Escherichia coli and purified as described before (34,35). Enzyme concentrations were routinely determined using an extinction coefficient at 280 nm of 260 450 M–1 cm–1 (HIV-1 RT) and 238 150 M–1 cm–1 (HIV-2 RT). The purified RTs were free of nuclease contamination.
In vitro binding assay
Protein and 5′-32P-labeled RNA were mixed in standard buffer (50 mM Tris–HCl pH 8.0, 50 mM KCl, 1 mM DTT and 10 mM MgCl2) and incubated at 25°C for 10 min. An aliquot of this mixture was filtered under suction through a prewet (standard buffer) nitro-cellulose filter (Schleicher & Schuell BA85) and rinsed with 4 ml of standard buffer. Radioactivity retained on the filters was measured by scintillation counting.
Polymerase activity determination
RNA-dependent DNA polymerase activity on poly(rA)/oligo(dT)12–18 substrates was measured by a standard assay described previously (36,37) with 2.8 nM of RT for 10 min at 37°C in a buffer containing 50 mM Tris–HCl pH 8.0, 80 mM KCl, 5 mM DTT, 6 mM MgCl2 and 0.05% (v/v) Triton X-100.
Cell lines and cell tissue culture
HS 68 (human newborn foreskin fibroblast) and 293T (human embryonic kidney cells) cell lines were cultured in Dulbecco’s Modified Eagle’s Medium (DMEM; Life Technologies, Karlsruhe, Germany), supplemented with 10% fetal calf serum, 2 mM l-glutamine, 100 IU/ml penicillin and 100 µg/ml streptomycin at 37°C in a humidified atmosphere containing 5% CO2. Jurkat and C8166 cells (both human CD4 positive T-lymphoid cells) were cultured under the same conditions but with RPMI medium instead of DMEM. For transfection and infection experiments the cells were seeded in 48-well culture plates at a density of 105 cells/well.
Northern blot analysis
Nuclear and cytoplasmic RNAs were isolated from HS 68, 293T and Jurkat cell lines using the Trizol reagent (Life Technologies) according to the manufacturers protocol with a few modifications. Briefly, 5 × 106–107 cells were transfected with different double-copy (vector) (DC)T5T constructs using either the LipofectAMINE PLUS™ reagent (Life Technologies) or the calcium phosphate transfection procedure (38). Forty-eight hours post-transfection, cells were incubated in a buffer containing 50 mM Tris–HCl pH 8.0, 140 mM NaCl, 1 mM DTT, 1.5 mM MgCl2 and 0.5 % (v/v) NP-40 for 5 min on ice. Cytoplasmic RNA was separated from nuclear RNA by centrifugation (2 min at 300 g and 4°C). Supernatant containing the cytoplasmic fraction and pellets containing the nuclear fraction were then incubated with the Trizol reagent and RNA purification was performed as described by the manufacturer. The amount of RNA in each sample was determined by OD260 measurements. RNA samples were subjected to electrophoresis in formaldehyde agarose gels (1.2%), transferred to a nylon membrane (Hybond N+, Amersham Pharmacia Biotech) and hybridized with either a 32P-labeled pseudoknot, tRNALys3 or U6 snRNA probe. After extensive washing of the membrane, data were quantified by phosphorimaging (Fuji FLA 3000 radioluminograph scanner).
Transfection and infection experiments
Human 293T kidney cells were transiently co-transfected with proviral HIV-1 DNA [pNL4-3 (39), 150 ng/well], and different DCT5T constructs (250 ng/well): the parental vector (DCT5T), an antisense RNA vector (DCT5T-αrev) and the tRNAMeti-pseudoknot vector (DCT5T-pseudoknot). Transfec tion was performed according to the calcium phosphate procedure (38). Transfection efficiency was determined using the pEGFP-C1 vector (Clontech Laboratories, Inc., Palo Alto, CA). Supernatants were collected 36 h post-transfection and virus production was measured by a commercial p24 ELISA (Innotest™, Innogenetics, Heiden, Germany). In a subsequent step, human CD4-positive T-lymphoid cells (C8166) were infected with these supernatants. Virus concentration, determined via p24 ELISA, was held constant in all infec tion experiments. Forty-eight hours post-infection, virus production was measured by ELISA.
For the generation of stably transfected clones, Jurkat cells were electroporated in the presence of different DCT5T constructs and grown in the presence of G418 for selection as described previously (40). Stable clones were infected with HIV-1 particle containing cell-free supernatants of transiently transfected 293T cells as described above. At given time points virus production was determined via p24 ELISA.
RESULTS
Vector construction
The vector DCT5T originates from a Moloney murine leukemia virus-based, polylinker-modified vector designated N2A (41). This vector contains a Δ3′-5 mutant tRNAMeti gene (42) and a polymerase III termination signal in the U3 region of the 3′ long terminal repeat (43). The deletion within the tRNA gene (18 bp at the 3′-end of the gene and 11 bp of the mature tRNA product) abolishes processing of the primary tRNA transcript. Insertion of a foreign sequence between the Δ3′-5 tRNA gene and a transcription signal generates a chimeric transcript in which the foreign sequence is fused to the 3′-end of the tRNA (42). An oligonucleotide containing the pseudoknot sequence was cloned into the unique BamHI/MluI restriction sites downstream of the tRNA gene and upstream of the polymerase III termination signal, generating the retroviral vector DCT5T-pseudoknot (Fig. 1A). Figure 1B shows a secondary structure prediction of the 117 nt chimeric RNA transcript using the program PKNOTS (32). According to this prediction the pseudoknot motif folds as expected. As described in the next section, these computer-based results were confirmed by biochemical analysis.
Functional evidence for proper folding of chimeric RNA in vitro
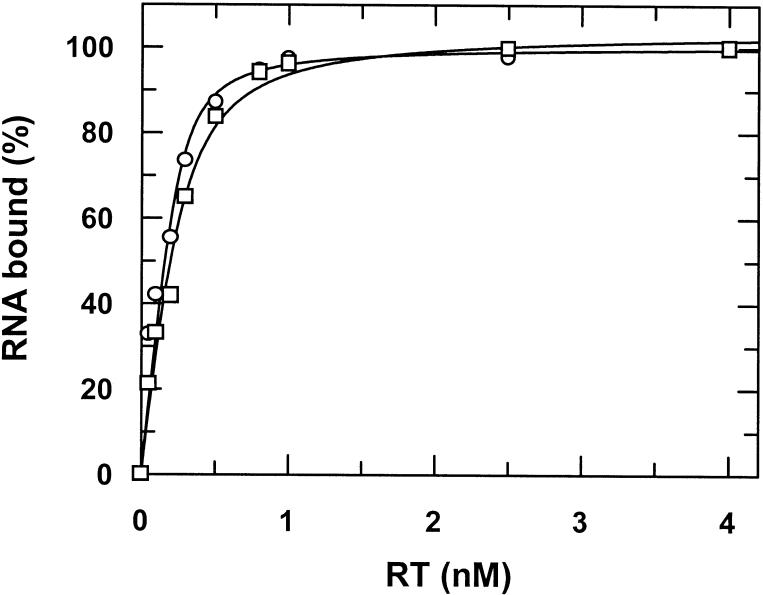
As it cannot be predicted for certain whether or not the pseudoknot motif folds properly in the sequence context of the tRNA based on computer models, we performed in vitro binding studies with this RNA chimera. For this purpose, the corresponding tRNAMeti/pseudoknot transcription unit (see Fig. 1A) was cloned into the T7 RNA transcription vector pBluescript SK (Stratagene, La Jolla, CA) via the KpnI/XbaI restriction sites. Transcription and PAGE purification of the chimeric RNA (see Fig. 1B) was performed as described in Materials and Methods. Because of technical reasons the in vitro transcribed RNA is 137 nt long as compared with the in vivo transcribed RNA with 117 nt. The additional bases are located at the 5′-end of the transcript and originate from the transcription vector pBluescript SK. Figure 2 shows a typical filter binding experiment using radiolabeled RNA. As compared with the aptamer alone the tRNAMeti/aptamer construct shows a slight reduction in binding affinity for HIV-1 RT dropping from 37 to 86 pM. Similar results were obtained applying fluorescence measurements as described recently by Kensch et al. (28) to determine the binding affinity (data not shown). Binding affinities of a chimera consisting of tRNAMeti and the complementary pseudoknot sequence could not be determined accurately. However, the binding is at least three orders of magnitude weaker than for a construct containing the pseudoknot sequence (data not shown). These results clearly indicate proper folding of the pseudoknot motif within the RNA chimera yielding a highly specific interaction with the target protein.
Figure 2.
Binding studies. Filter binding assay of radiolabeled pseudoknot RNA and tRNA-pseudoknot RNA with RT. 5′-32P-labeled RNAs (200 pM) were titrated with increasing amounts of RT. The curves show best fits to a quadratic equation yielding a KD of 37 (±14) and 86 (±17) pM for the pseudoknot (circles) and the tRNA-pseudoknot (squares), respectively.
Effect of the tRNA/pseudoknot chimera on RT polymerase activity
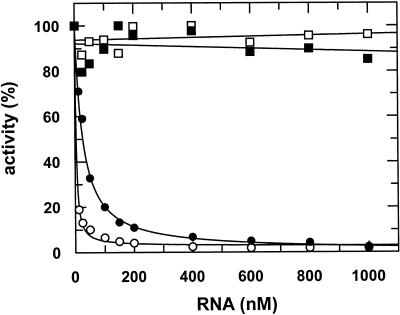
To test the inhibition of the enzymatic activity of the target protein by the chimeric RNA we performed a standard RT assay using poly(rA)/oligo(dT) as substrate. The reactions were started with preincubated RT/RNA complexes. Figure 3 shows strong inhibition of the HIV-1 RT polymerase activity by the pseudoknot RNA alone as compared with the closely related enzyme of HIV-2 (28). The chimeric transcript is somewhat less efficient in blocking the polymerase activity of HIV-1 RT and as found for the aptamer alone shows no effect on HIV-2 RT. Inhibition constants for the pseudoknot and the tRNA-pseudoknot were determined by fitting the experimental data to a hyperbolic equation yielding Ki values (defined as RNA concentration that inhibited polymerase activity by 50%) for HIV-1 RT of 2.8 (±0.24) and 25.5 (±0.03) nM, respectively. No inhibition of polymerase activity was observed with a chimera consisting of tRNAMeti alone or the complementary pseudoknot sequence at concentrations up to 1 µM (data not shown).
Figure 3.
Inhibition of RT activity in vitro. RNA-dependent DNA polymerase activity on poly(rA)/oligo(dT)12–18 substrates was measured for 10 min at 37°C. 2.88 nM HIV-1 (circles) or HIV-2 (squares). RT was preincubated with increasing amounts of pseudoknot (open symbols) or tRNA-pseudoknot RNA (filled symbols).
Intracellular distribution of endogenously expressed aptamer
The polymerase III-driven chimeric tRNA gene for in vivo transcription of the aptamer was primarily chosen for two reasons: transcription efficacy and cytoplasmic distribution of the transcript. The latter being a perquisite for effective inhibition of the target protein that is localized in the cytoplasm. In order to investigate the intracellular localization of the aptamer, we performed northern blot analysis. As shown in Figure 4 the chimeric RNA is essentially found in the cytoplasmic fraction when transiently expressed in different human cell lines. In order to exclude leakage form one compartment to the other during sample preparation, probes against tRNALys3 and U6 snRNA were chosen as cytoplasmic and nuclear controls, respectively.
Figure 4.
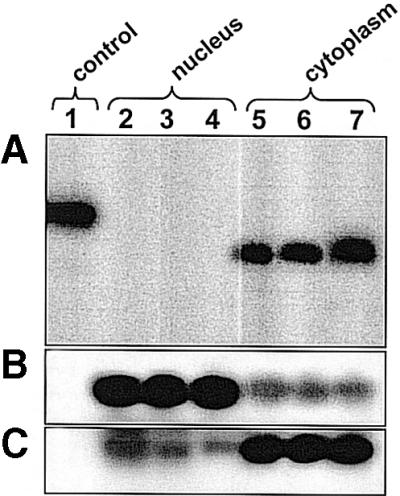
Subcellular distribution of the tRNA-pseudoknot chimera. Total nuclear and cytoplasmic RNAs were prepared from 107 cells (HS 68, 293T and Jurkat) transfected with the DCT5T-pseudoknot vector. Each lane was loaded with the same amount of RNA (20 µg) as determined by OD260 measurements. (A) Pseudoknot RNA, (B) U6snRNA and (C) tRNALys3. Lane 1, control (137 nt RNA chimera prepared by in vitro transcription). Lanes 2–4, nuclear RNA. Lanes 5–7, cytoplasmic RNA from transfected HS 68 (2,5), 293T (3,6) and Jurkat (4,7) cells.
Transient suppression of the production of infective HIV-1 particles by pol III-driven pseudoknot aptamers
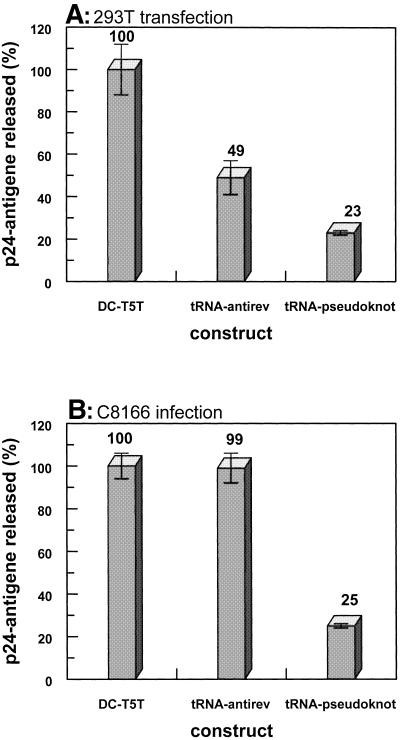
Human 293T kidney cells were transiently co-transfected with infectious proviral HIV DNA and different DCT5T constructs: the parental vector (DCT5T), an antisense RNA vector (DCT5T-αrev) and the tRNAMeti-pseudoknot vector (DCT5T-pseudoknot). The transfection efficacy was monitored by expression of a plasmid-encoded green fluorescence protein and was found to be ∼80% (data not shown). The amount of virus produced and released by these cells was measured 36 h post-transfection via a p24 antigen ELISA (Fig. 5A). Each experiment was performed in triplicate and repeated twice. The parental vector (DCT5T) served as a negative control and the amount of virus produced in the presence of this construct was set to 100%. As a positive control for inhibition, the well characterized antisense construct DCT5T-αrev was used (44). This anti-rev RNA has been shown previously to be a potent inhibitor of HIV-1 replication in transient quantitative co-transfection assays as well as in the long-term experiments after retroviral transduction of HIV-1-susceptible cells and challenge with HIV-1 (40).
Figure 5.
Transient transfection and infection experiments. Each experiment was performed in triplicate and repeated twice. The error bars indicate the standard deviation error from the mean. (A) 293T cells were co-transfected with a HIV-1 proviral DNA and different retroviral constructs: the parental vector (DCT5T), an antisense RNA vector (DCT5T-αrev) and the tRNAMeti-pseudoknot vector (DCT5T-pseudoknot). Virus production was measured 36 h post-transfection via ELISA analysis. (B) Infection of human T-lymphoid cell line (C8166) with viruses produced by the 293T cell line (see Fig. 7A). In each experiment the same amount of virus was applied. HIV-1 replication was measured 48 h post-infection by ELISA.
Thus, this DCT5T-αrev construct was used as internal positive control to calibrate the system. Consistent with earlier measurements, the anti-rev RNA showed an inhibition of virus production under the specific experimental conditions used here by ∼50%. Upon co-transfection with the DCT5T-pseudoknot vector virus production was reduced by ∼77%. Analyzing the RT activity of viruses released by the transient transfected 293T cells, we found ∼70% reduction after normalizing for p24 in case of the aptamer co-expressed (data not shown).
Reduced infectivity of HIV-1 particles generated in the presence of the pseudoknot aptamer
To measure the influence of the RT-binding aptamer on the specific infectivity of HIV-1 particles, human T-lymphoid cells C8166 were infected with constant amounts of HIV produced by co-transfection of 293T cells with proviral HIV DNA and different constructs of the DCT5T vector (see Fig. 5A and previous section). The new cycle of virus replication in C8166 cells was monitored 48 h post-infection via a p24 antigen ELISA (Fig. 5B). When standardized to the DCT5T control, no inhibition of viral replication was observed in the case of the DCT5T-αrev construct. However, viruses assembled in the presence of the tRNAMeti/pseudoknot RNA showed a substantial reduction (∼75%) of their ability to infect and/or replicate in newly infected T-lymphoid cells.
Complete inhibition of viral replication in stably transfected Jurkat cells after low-dose HIV infection
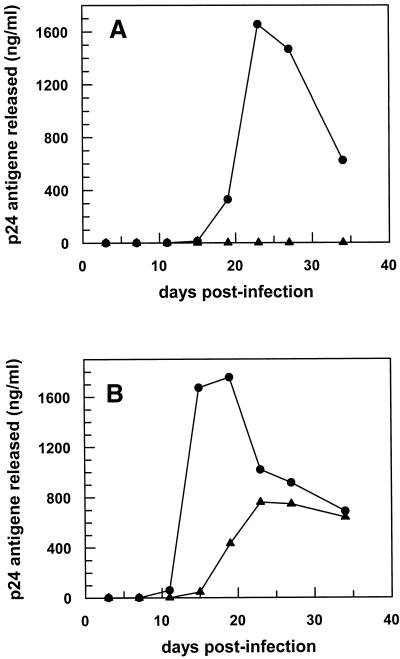
In order to overcome the intrinsic problems of transient transfection assays, we generated stably transfected T-lymphoid cells. Figure 6 shows HIV-1 infection of Jurkat cells either stably transfected with the parental vector DCT5T or the DCT5T-pseudoknot vector. Low-dose HIV infection (190 pg p24/105 cells) of Jurkat cells stably expressing the aptamer did not show any virus replication over a period of 35 days (Fig. 6A). Increasing the initial virus titer by 10-fold, HIV-1 particle production can be observed in the presence of the tRNAMeti/pseudoknot RNA chimera. However, there is a significant reduction of virus production and a delay of virus release of ∼1 week as compared with the control.
Figure 6.
Infection of stably transfected Jurkat cells. Jurkat cell clones either stably transfected with the parental vector DCT5T (circles) or the DCT5T-pseudoknot vector (triangles) were infected with different amounts HIV-1 particles. (A) 190 pg p24 per 105 cells and (B) 1700 pg p24 per 105 cells. At the time points given, viral replication was measured by p24 ELISA.
DISCUSSION
In order to investigate the effect of an inhibitory RNA aptamer, selected for tight binding to HIV-1 RT, on viral replication in vivo, we constructed a retroviral vector for intracellular expression of this aptamer. The pseudoknot RNA sequence was cloned into the DCT5T vector downstream of a polymerase III-driven Δ3′-5 mutant tRNAMeti gene and upstream of a polymerase termination site generating a tRNAMeti/pseudoknot RNA chimera.
Northern blot analysis of 293T cells transiently transfected with this construct revealed the transcript to be predominantly localized in the cytoplasm. This is in agreement with findings reported by others using such a vector for intracellular expression of RNA molecules (45,46). However, it should also be mentioned here that other studies reported opposite results (17,47). The reason for this obvious discrepancy remains unclear. It could be speculated that the overall folding of the transcript influences the interaction with export carriers and thus determines whether or not the tRNA chimera is actually transported into the cytoplasm.
First, we confirmed that the aptamer within the chimeric RNA still specifically binds the target protein. This was investigated by binding studies with in vitro transcribed RNA showing similar affinities for HIV-1 RT compared with the aptamer alone indicating that the pseudoknot motif is properly folded in the sequence context of the tRNAMeti and the extra RNA part at the 5′-end is not interfering with binding to its target. The somewhat lower binding affinity observed for the chimera could be in part explained by the additional base at the 3′-end of the pseudoknot sequence generated in vivo due to the polymerase termination signal. According to the X-ray structure there should not be sufficient space for an additional nucleotide at that position. The binding studies were further supported by inhibition experiments. Here we tested the effects of the chimera on the polymerase activity of RT. As already shown (28) for the pseudoknot alone, the chimera is highly specific for the HIV-1 enzyme compared with HIV-2. This clearly indicates that the strong inhibition of the polymerase activity is due to specific interactions with the pseudoknot part of the chimera. The reduced Ki value in the case of the tRNAMeti/pseudoknot RNA by a factor of about 9 as compared with the pseudoknot alone is most likely caused by the difference in binding affinities as minor changes here have a more pronounced effect in this kind of assay. Here the poly(rA)/oligo(dT)12–18 substrate is in large excess over inhibitor RNA. Therefore, inhibition is due to enzyme trapped by the aptamer as the reaction was started by adding RT/aptamer complexes. Differences of a factor of 2 in the koff values for the enzyme/inhibitor complex as indicated by the KD values could easily cause a difference of 5–10 for the Ki values in the inhibition assay.
To analyze effects of the aptamer in HIV-infected cells we first investigated the production/release of HIV in cells co-transfected with the vector coding for the aptamer chimera and infectious proviral HIV-1 DNA. In this situation no reverse transcription takes place. However, the production of viral particles was reduced by >75% and we cannot be certain at which stage of the replication cycle this aptamer interferes. It is conceivable that the RNA disturbs the viral packaging by binding to the RT within the premature Gag-Pol-polyprotein or the aptamer might even affect the maturation of the gag-pol precursor. There are examples where mutations within the RT gene caused similar effects (48,49). This was attributed to problems during packaging events. A second scenario could be that the binding of the aptamer to the Gag-Pol-polyprotein interferes with the selective packaging of the primer tRNALys3 into viral particles (50).
In a second set of experiments, we infected human T-lymphoid cells with viral particles generated in the presence of the aptamer. In this case reverse transcription of the viral RNA genome has to take place before new viral particles can be released. The replication capability of these viruses was reduced by ∼75%. This is most likely due to an aptamer bound to RT and packed into the virus causing inhibition of reverse transcription during the next replication cycle. This argument is strongly supported by the finding that HIV particles produced by 293T cells in the presence of the aptamer show dramatically reduced polymerase activity. If normalized for p24, those particles show ∼70% lower RT activity as compared with particles produced in the presence of the parental vector (data not shown). This is in perfect agreement with the 75% inhibition observed for the infection experiment. However, currently we cannot entirely rule out that these viruses are also impaired in infecting T-lymphoid cells.
The described effects are additive causing an inhibition of the viral replication rate by >95% in total. It should be noted here that the transfections were transient. This could be important in respect to the maximal inhibitory potential of the aptamer since the transfection efficiency was determined to be in the range of ∼80% in the chosen set up. Therefore, it is conceivable that in the first round where cells were co-transfected with two plasmids they might have taken up only one. If this happens to be the proviral DNA no inhibition would be observed. Consequently, we believe the detected rate of inhibition in this kind of experimental set up to be at the lower limit.
In order to study the effect of the aptamer in a cell culture system closer to the in vivo situation, we performed infection experiments with stably transfected T-lymphoid cells. As already expected from the transient transfections, here we observed complete inhibition of viral replication in the presence of the aptamer after low-dose HIV-1 infection. As anticipated, increasing the tissue culture infectious dose (TCID)50 above a certain threshold level, the amount of aptamer produced by the cells is not sufficient anymore to fully block viral replication. Still, the number of particles released is significantly lower and delayed by ∼1 week as compared with control. Further fine-tuning of the experimental system should enable us to gain insight into the delicate balance between full inhibition versus viral breakthrough.
So far we could not observe any toxic effects caused by the RNA. Using a carrier peptide system to transport in vitro transcribed pseudoknot RNA into human fibroblasts no toxic effects were observed at concentration up to 100 µM (data not shown; T. Restle, manuscript in preparation).
Aptamers are fascinating new reagents to be used as diagnostic tools as well as therapeutics to battle a variety of diseases. However, the main obstacle remains their delivery. This is especially problematic when the target is localized inside the cell. Here we show for the first time that an RNA aptamer selected for tight binding to a viral enzyme is capable of blocking HIV replication. Consequently, we conclude that the application of this aptamer by gene transfer techniques represents a very promising and innovative approach to treat infections with HIV-1. Comparing our results to results obtained by others using anti-HIV-1 Rev aptamers (16–20) is difficult due to the differences in the methodologies applied. It remains to be seen which approach turns out to be more effective. In terms of mechanistic aspects, targeting a viral enzyme could have the advantage of the aptamer being packed into the viral particles and thus rendering them inactive, i.e. decreasing the apparent infectivity.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Roger Goody for continuous support and Paul Rothwell for critical reading of the manuscript. L.C. was supported by a stipend from the Alexander von Humboldt foundation.
REFERENCES
- 1.Robertson D.L. and Joyce,G.F. (1990) Selection in vitro of an RNA enzyme that specifically cleaves single-stranded DNA. Nature, 344, 467–468. [DOI] [PubMed] [Google Scholar]
- 2.Tuerk C. and Gold,L. (1990) Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science, 249, 505–510. [DOI] [PubMed] [Google Scholar]
- 3.Ellington A.D. and Szostak,J.W. (1990) In vitro selection of RNA molecules that bind specific ligands. Nature, 346, 818–822. [DOI] [PubMed] [Google Scholar]
- 4.Brody E.N. and Gold,L. (2000) Aptamers as therapeutic and diagnostic agents. J. Biotechnol., 74, 5–13. [DOI] [PubMed] [Google Scholar]
- 5.Famulok M., Mayer,G. and Blind,M. (2000) Nucleic acid aptamers-from selection in vitro to applications in vivo. Acc. Chem. Res., 33, 591–599. [DOI] [PubMed] [Google Scholar]
- 6.James W. (2001) Nucleic acid and polypeptide aptamers: a powerful approach to ligand discovery. Curr. Opin. Pharmacol., 1, 540–546. [DOI] [PubMed] [Google Scholar]
- 7.White R.R., Sullenger,B.A. and Rusconi,C.P. (2000) Developing aptamers into therapeutics. J. Clin. Invest., 106, 929–934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ruckman J., Green,L.S., Beeson,J., Waugh,S., Gillette,W.L., Henninger,D.D., Claesson-Welsh,L. and Janjic,N. (1998) 2′-Fluoropyrimidine RNA-based aptamers to the 165-amino acid form of vascular endothelial growth factor (VEGF165). Inhibition of receptor binding and VEGF-induced vascular permeability through interactions requiring the exon 7-encoded domain. J. Biol. Chem., 273, 20556–20567. [DOI] [PubMed] [Google Scholar]
- 9.Blind M., Kolanus,W. and Famulok,M. (1999) Cytoplasmic RNA modulators of an inside-out signal-transduction cascade. Proc. Natl Acad. Sci. USA, 96, 3606–3610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shi H., Hoffman,B.E. and Lis,J.T. (1999) RNA aptamers as effective protein antagonists in a multicellular organism. Proc. Natl Acad. Sci. USA, 96, 10033–10038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mayer G., Blind,M., Nagel,W., Bohm,T., Knorr,T., Jackson,C.L., Kolanus,W. and Famulok,M. (2001) Controlling small guanine-nucleotide-exchange factor function through cytoplasmic RNA intramers. Proc. Natl Acad. Sci. USA, 98, 4961–4965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Famulok M., Blind,M. and Mayer,G. (2001) Intramers as promising new tools in functional proteomics. Chem. Biol., 8, 931–939. [DOI] [PubMed] [Google Scholar]
- 13.Richman D.D. (2001) HIV chemotherapy. Nature, 410, 995–1001. [DOI] [PubMed] [Google Scholar]
- 14.Symensma T.L., Giver,L., Zapp,M., Takle,G.B. and Ellington,A.D. (1996) RNA aptamers selected to bind human immunodeficiency virus type 1 Rev in vitro are Rev responsive in vivo. J. Virol., 70, 179–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yamamoto R., Katahira,M., Nishikawa,S., Baba,T., Taira,K. and Kumar,P.K. (2000) A novel RNA motif that binds efficiently and specifically to the Ttat protein of HIV and inhibits the trans-activation by Tat of transcription in vitro and in vivo. Genes Cells, 5, 371–388. [DOI] [PubMed] [Google Scholar]
- 16.Bahner I., Kearns,K., Hao,Q.L., Smogorzewska,E.M. and Kohn,D.B. (1996) Transduction of human CD34+ hematopoietic progenitor cells by a retroviral vector expressing an RRE decoy inhibits human immunodeficiency virus type 1 replication in myelomonocytic cells produced in long-term culture. J. Virol., 70, 4352–4360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Good P.D., Krikos,A.J., Li,S.X., Bertrand,E., Lee,N.S., Giver,L., Ellington,A., Zaia,J.A., Rossi,J.J. and Engelke,D.R. (1997) Expression of small, therapeutic RNAs in human cell nuclei. Gene Ther., 4, 45–54. [DOI] [PubMed] [Google Scholar]
- 18.Konopka K., Duzgunes,N., Rossi,J. and Lee,N.S. (1998) Receptor ligand-facilitated cationic liposome delivery of anti-HIV-1 Rev-binding aptamer and ribozyme DNAs. J. Drug Target, 5, 247–259. [DOI] [PubMed] [Google Scholar]
- 19.Düzgünes N., Pretzer,E., Simoes,S., Slepushkin,V., Konopka,K., Flasher,D. and de Lima,M.C. (1999) Liposome-mediated delivery of antiviral agents to human immunodeficiency virus-infected cells. Mol. Membr. Biol., 16, 111–118. [DOI] [PubMed] [Google Scholar]
- 20.Konopka K., Lee,N.S., Rossi,J. and Duzgunes,N. (2000) Rev-binding aptamer and CMV promoter act as decoys to inhibit HIV replication. Gene, 255, 235–244. [DOI] [PubMed] [Google Scholar]
- 21.Telesnitsky A. and Goff,S.P. (1997) In Coffin,J.M., Hughes,S.H. and Varmus,H.E. (eds), Retroviruses. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 121–160.
- 22.Tuerk C., Macdougal,S. and Gold,L. (1992) RNA pseudoknots that inhibit human immunodeficiency virus type-1 reverse transcriptase. Proc. Natl Acad. Sci. USA, 89, 6988–6992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Green L., Waugh,S., Binkley,J.P., Hostomska,Z., Hostomsky,Z. and Tuerk,C. (1995) Comprehensive chemical modification interference and nucleotide substitution analysis of an RNA pseudoknot inhibitor to HIV-1 reverse transcriptase. J. Mol. Biol., 247, 60–68. [DOI] [PubMed] [Google Scholar]
- 24.Burke D.H., Scates,L., Andrews,K. and Gold,L. (1996) Bent pseudoknots and novel RNA inhibitors of type 1 human immunodeficiency virus (HIV-1) reverse transcriptase. J. Mol. Biol., 264, 650–666. [DOI] [PubMed] [Google Scholar]
- 25.Jäger J., Restle,T. and Steitz,T.A. (1998) The structure of HIV-1 reverse transcriptase complexed with an RNA pseudoknot inhibitor. EMBO J., 17, 4535–4542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.De Clercq E. (2002) Strategies in the design of antiviral drugs. Nature Rev. Drug Discov., 1, 13–25. [DOI] [PubMed] [Google Scholar]
- 27.Fisher T.S., Joshi,P. and Prasad,V.R. (2002) Mutations that confer resistance to template-analog inhibitors of human immunodeficiency virus (HIV) type 1 reverse transcriptase lead to severe defects in HIV replication. J. Virol., 76, 4068–4072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kensch O., Connolly,B.A., Steinhoff,H.J., McGregor,A., Goody,R.S. and Restle,T. (2000) HIV-1 reverse transcriptase-pseudoknot RNA aptamer interaction has a binding affinity in the low picomolar range coupled with high specificity. J. Biol. Chem., 275, 18271–18278. [DOI] [PubMed] [Google Scholar]
- 29.Thrall S.H., Krebs,R., Wöhrl,B.W., Cellai,L., Goody,R.S. and Restle,T. (1998) Pre-steady-state kinetic characterization of RNA-primed initiation of transcription by HIV-1 reverse transcriptase and analysis of the transition to a processive DNA-primed polymerization mode. Biochemistry, 37, 13349–13358. [DOI] [PubMed] [Google Scholar]
- 30.Krebs R., Immendorfer,U., Thrall,S.H., Wöhrl,B.M. and Goody,R.S. (1997) Single-step kinetics of HIV-1 reverse transcriptase mutants responsible for virus resistance to nucleoside inhibitors zidovudine and 3-TC. Biochemistry, 36, 10292–10300. [DOI] [PubMed] [Google Scholar]
- 31.Sambrook J., Fritsch,E.F. and Maniatis,T. (1994) Molecular Cloning—A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York.
- 32.Rivas E. and Eddy,S.R. (1999) A dynamic programming algorithm for RNA structure prediction including pseudoknots. J. Mol. Biol., 285, 2053–2068. [DOI] [PubMed] [Google Scholar]
- 33.De Rijk P. and De Wachter,R. (1997) RnaViz, a program for the visualisation of RNA secondary structure. Nucleic Acids Res., 25, 4679–4684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Müller B., Restle,T., Weiss,S., Gautel,M., Sczakiel,G. and Goody,R.S. (1989) Co-expression of the subunits of the heterodimer of HIV-1 reverse transcriptase in Escherichia coli. J. Biol. Chem., 264, 13975–13978. [PubMed] [Google Scholar]
- 35.Müller B., Restle,T., Kühnel,H. and Goody,R.S. (1991) Expression of the heterodimeric form of human immunodeficiency virus type 2 reverse transcriptase in Escherichia coli and characterization of the enzyme. J. Biol. Chem., 266, 14709–14713. [PubMed] [Google Scholar]
- 36.Restle T., Müller,B. and Goody,R.S. (1990) Dimerization of human immunodeficiency virus type 1 reverse transcriptase. A target for chemotherapeutic intervention. J. Biol. Chem., 265, 8986–8988. [PubMed] [Google Scholar]
- 37.Jacques P.S., Wöhrl,B.M., Ottmann,M., Darlix,J.L. and Le Grice,S.F. (1994) Mutating the ‘primer grip’ of p66 HIV-1 reverse transcriptase implicates tryptophan-229 in template-primer utilization. J. Biol. Chem., 269, 26472–26478. [PubMed] [Google Scholar]
- 38.Wigler M., Pellicer,A., Silverstein,S., Axel,R., Urlaub,G. and Chasin,L. (1979) DNA-mediated transfer of the adenine phosphoribosyltransferase locus into mammalian cells. Proc. Natl Acad. Sci. USA, 76, 1373–1376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Adachi A., Gendelman,H.E., Koenig,S., Folks,T., Willey,R., Rabson,A. and Martin,M.A. (1986) Production of acquired immunodeficiency syndrome-associated retrovirus in human and nonhuman cells transfected with an infectious molecular clone. J. Virol., 59, 284–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sczakiel G. and Pawlita,M. (1991) Inhibition of human immunodeficiency virus type 1 replication in human T cells stably expressing antisense RNA. J. Virol., 65, 468–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hantzopoulos P.A., Sullenger,B.A., Ungers,G. and Gilboa,E. (1989) Improved gene expression upon transfer of the adenosine deaminase minigene outside the transcriptional unit of a retroviral vector. Proc. Natl Acad. Sci. USA, 86, 3519–3523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Adeniyi-Jones S., Romeo,P.H. and Zasloff,M. (1984) Generation of long read-through transcripts in vivo and in vitro by deletion of 3′ termination and processing sequences in the human tRNAimet gene. Nucleic Acids Res., 12, 1101–1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sullenger B.A., Gallardo,H.F., Ungers,G.E. and Gilboa,E. (1990) Overexpression of TAR sequences renders cells resistant to human immunodeficiency virus replication. Cell, 63, 601–608. [DOI] [PubMed] [Google Scholar]
- 44.Junker U., Rittner,K., Homann,M., Bevec,D., Bohnlein,E. and Sczakiel,G. (1994) Reduction in replication of the human immunodeficiency virus type 1 in human T cell lines by polymerase III-driven transcription of chimeric tRNA-antisense RNA genes. Antisense Res. Dev., 4, 165–172. [DOI] [PubMed] [Google Scholar]
- 45.Ilves H., Barske,C., Junker,U., Bohnlein,E. and Veres,G. (1996) Retroviral vectors designed for targeted expression of RNA polymerase III-driven transcripts: a comparative study. Gene, 171, 203–208. [DOI] [PubMed] [Google Scholar]
- 46.Kuwabara T., Warashina,M., Koseki,S., Sano,M., Ohkawa,J., Nakayama,K. and Taira,K. (2001) Significantly higher activity of a cytoplasmic hammerhead ribozyme than a corresponding nuclear counterpart: engineered tRNAs with an extended 3′ end can be exported efficiently and specifically to the cytoplasm in mammalian cells. Nucleic Acids Res., 29, 2780–2788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bertrand E., Castanotto,D., Zhou,C., Carbonnelle,C., Lee,N.S., Good,P., Chatterjee,S., Grange,T., Pictet,R., Kohn,D. et al. (1997) The expression cassette determines the functional activity of ribozymes in mammalian cells by controlling their intracellular localization. RNA, 3, 75–88. [PMC free article] [PubMed] [Google Scholar]
- 48.Yu Q., Ottmann,M., Pechoux,C., Le Grice,S. and Darlix,J.L. (1998) Mutations in the primer grip of human immunodeficiency virus type 1 reverse transcriptase impair proviral DNA synthesis and virion maturation. J. Virol., 72, 7676–7680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mak J., Jiang,M., Wainberg,M.A., Hammarskjold,M.L., Rekosh,D. and Kleiman,L. (1994) Role of Pr160gag-pol in mediating the selective incorporation of tRNA(Lys) into human immunodeficiency virus type 1 particles. J. Virol., 68, 2065–2072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Khorchid A., Javanbakht,H., Wise,S., Halwani,R., Parniak,M.A., Wainberg,M.A. and Kleiman,L. (2000) Sequences within Pr160gag-pol affecting the selective packaging of primer tRNA(Lys3) into HIV-1. J. Mol. Biol., 299, 17–26. [DOI] [PubMed] [Google Scholar]